In this section, we first introduce the variational autoencoder and genetic algorithm and then describe the proposed VAEGA model.
3.1. Variational Autoencoder
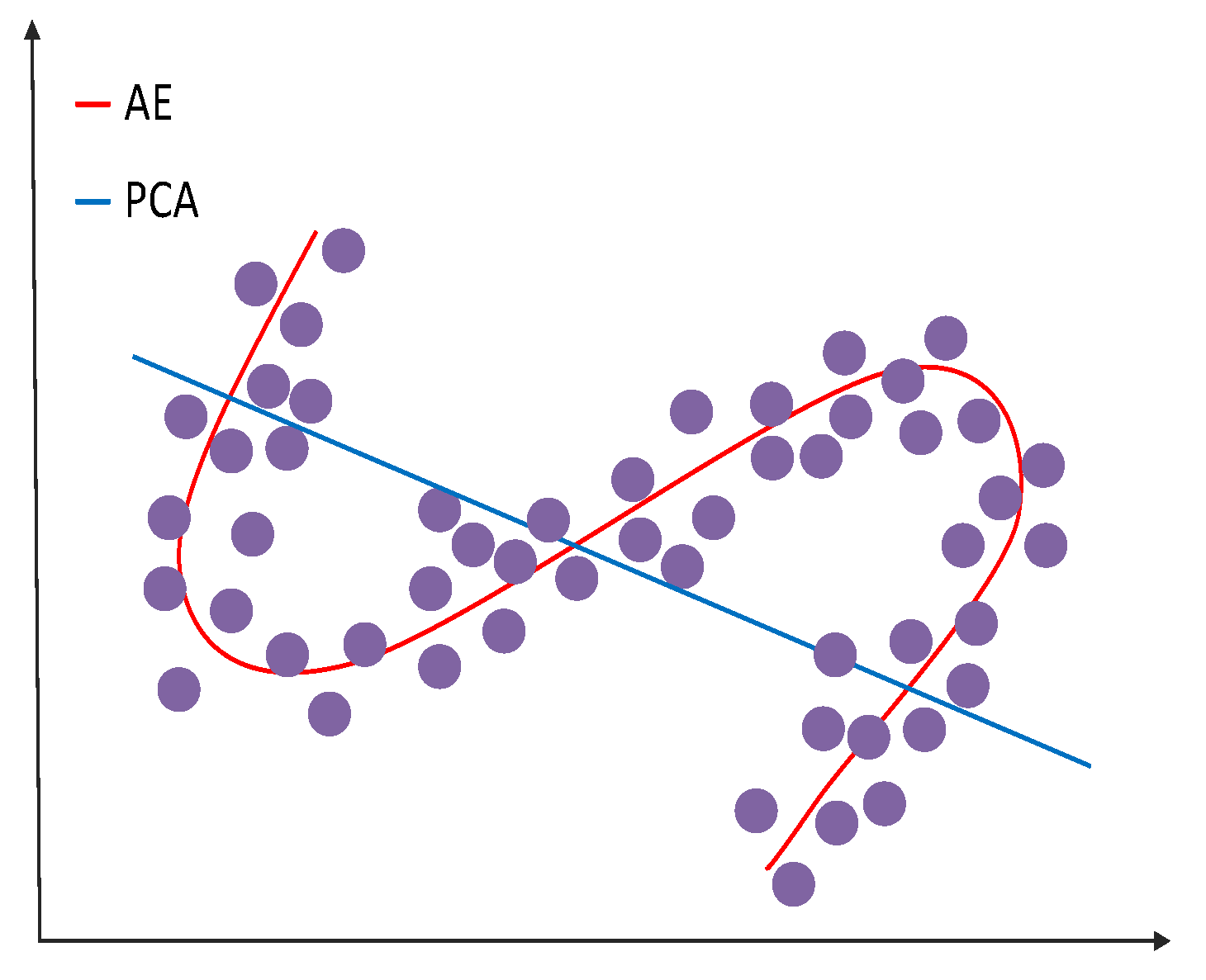
In neural networks, VAEs are feedforward acyclic neural networks. They are an unsupervised machine learning method. They can effectively extract very good data features and reduce the dimension of high-dimensional data. As opposed to AE, VAE’s latent space consists of a probability distribution of approximate data, while AE’s consists of a specific encoding of the input data.
Figure 2 shows a sample of a prior distribution in Euclidean space of VAE latent vectors.
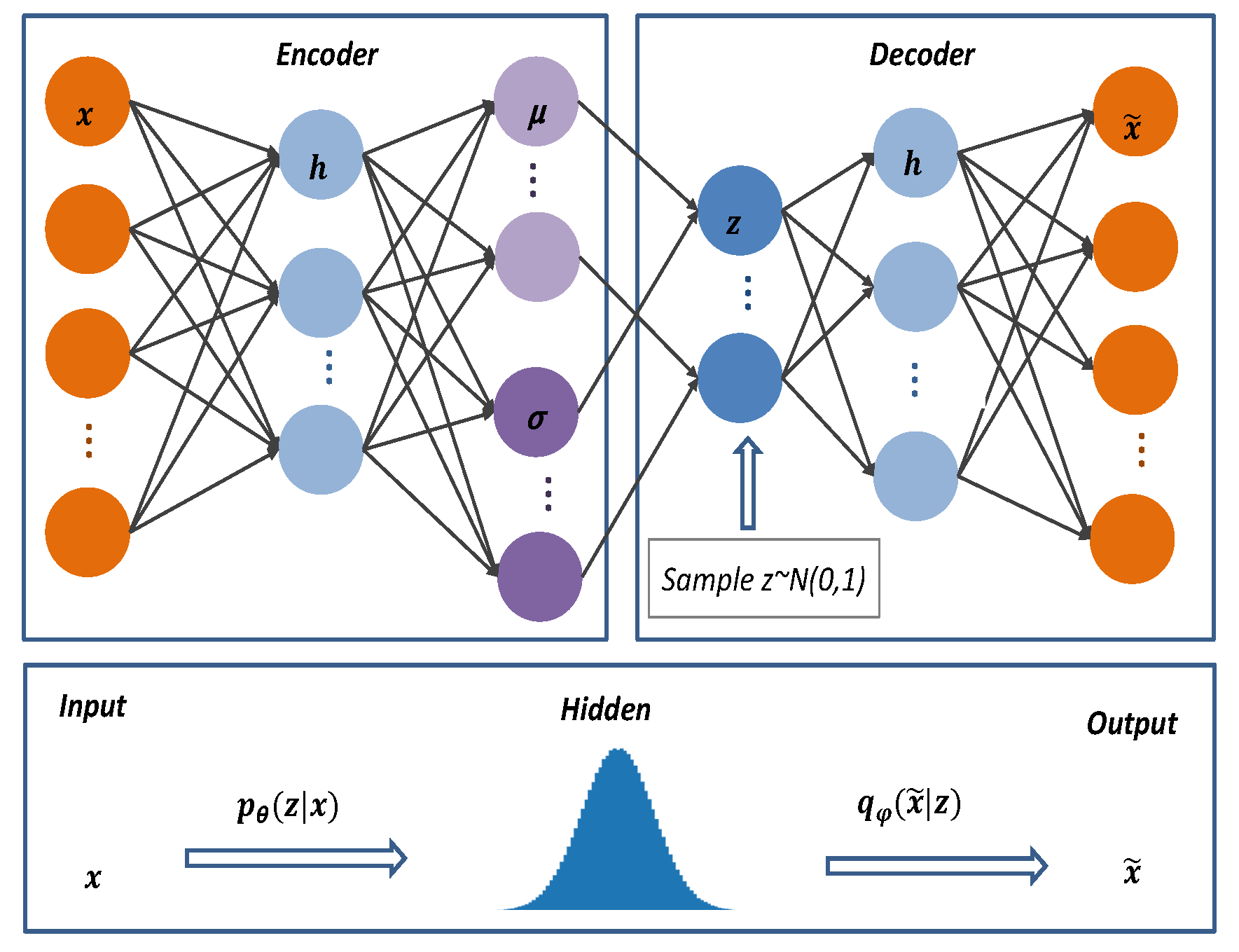
VAE’s structure resembles classic autoencoders with encoders, decoders, and latent spaces. The network structure of VAE is shown in
Figure 3.
From
Figure 3,
is the inferred network model known as the encoder. The original input data
is mapped from the current space
to the hidden space
to obtain the variational probability distribution of the hidden layer
z. As input
x and hidden space
Z are connected, the inferred model learns the joint probability distribution between them.
is the generative network model, that is, the decoder. It performs sampling from the probability distribution
of the hidden layer
Z, and after sampling the hidden vector,
z is decoded to produce an approximation of the target distribution
.
We applied the Bayesian formula as in Equation (
1) to compute the probability distribution
of the hidden variable
Z given the data
X.
We applied variational inference to approximate an unresolvable conditional probability distribution
with a solvable distribution
since the input data distribution
is not computed as the dimensionality increases. We fitted the probability distributions
and
, applying KL divergence to determine the disparity between them and to minimize the difference as best as possible. As shown in Equation (
2),
The model parameters are trained by minimizing the difference between the two probability distributions. A smaller difference shows better parameters are obtained from training the VAE. Since
X is certain, the first term in the equation
is a fixed value, so the training objective becomes to minimize the last two items
of the equation, which is denoted as
L. Then
−L represents the lower bound of evidence
. Here, minimizing
L gives the maximized lower bound of evidence (ELBO), as shown in Equation (
3):
The first term of the equation is:
, which indicates the continuous random samples of the probability distribution of the hidden space
z. The second term reveals that the latent vector
z is continuously sampled to ascertain that the probability of reconstructing the sample
x is maximized, so that the post-validation distribution
and the prior distribution of
are as close as possible. In order to construct the loss function of the model from this, we first consider the similarity between the input and the output and then make use of reconstruction loss to measure the difference between them. Secondly, due to the peculiarity of its coding layer, the variational autoencoder uses the latent loss to measure the “fitness” between the true probability distribution and the standard normal distribution. Consequently, the loss function of VAE ultimately consists of two items, as shown in Equation (
4):
3.3. The Proposed VAEGA Model
This section presents our proposed VAEGA model in detail. The model is applied to reduce the dimensionality challenges encountered by outlier detection in high-dimensional data. While focusing on improving the effectiveness of the outlier detection model from the perspective of data subspace, the heuristic optimization algorithm is used to obtain the abnormal subspace of outliers to solve the problem that high-dimensional outlier detection models generally lack interpretability.
The VAEGA model is defined as a model that combines a variational autoencoder and a genetic algorithm for the purpose of detecting outliers in high-dimensional data and searching for abnormal subspaces.
Figure 5 shows the framework of the model.
The VAEGA model is divided into two modules. The first part is the VAE model, which executes the process of quickly filtering out the outliers. In the training phase, the VAE module uses a neural network to build a variational encoder
and decoder
. The unlabeled data are used as input for iterative optimization to learn the best encoding and decoding scheme and obtain the mean
, and standard deviation
, of the input data
x. This step effectively represents the distribution of high-dimensional input data and ameliorates the representation of the high-dimensional data. For real-valued sample data, assuming that the distribution function is a multi-dimensional Gaussian distribution, its indicators are shown in Equations (
5)–(
7) as follows:
To optimize the weight parameters, we fit the data’s true probability distribution with a standard normal distribution. Through the use of stochastic gradient descent and backpropagation, the loss function of the model converged to a stable minimum value. The loss function of the VAE neural network is shown in Equation (
8), and Equation (
9) shows the
KL divergence used to measure the two probability distributions.
where a random variable
X is normally distributed with a mean
and standard deviation
,
,
is the trace of the matrix, and
is the input data dimension. In Equation (
8), the first item depicts the space between the reconstructed data and the input data. As a “reconstruction item” it tends to produce high performance in the encoding and decoding schemes. Regarding the second item, the KL divergence can be viewed as a penalty that regulates the hidden space.
It is important that we sample the latent vector
z from
, which obeys the Gaussian distribution
. The mean
and variance
of
z can be computed by the model, and the neural network relies on this sampling process to reverse optimize the variational autoencoder model. In this case, backpropagation for the model is impossible because sampling is not derivable. We then take into account that the results of sampling are derivable, which can be overcome by employing the reparameterization trick. The random variable
z is sampled from the standard normal distribution
N(0, 1) instead of the original distribution, and then the random variable
undergoes the following transformation, as shown in Equation (
10).
From to z involves linear operations, which are derivable. The distribution of is deterministic and does not need to be learned. The sampled value of participates in the gradient descent, while the operation of sampling does not.
Using the trained VAE network model, we input the test data. The feature information of the normal data can be represented by latent vectors in latent space and reconstructed with minimal loss. However, outliers are difficult to represent in the latent space, resulting in huge errors between the reconstructed data and the original input data. Therefore, we can combine the reconstruction probability error and latent space information as a measure of the outlier degree of the data, denoted as
, as shown in Equation (
11).
For comparability, we normalize the anomaly score
of each point to the range [0, 1] and re-denote it as
, as shown in Equation (
12). As the anomaly score approaches 1, the abnormality of the data increases, making it more likely that it is an outlier. On the contrary, it indicates that the characteristics of the data are more normal.
It is then compared to a threshold , where is a metric used to control the model’s sensitivity to outliers. If the abnormal score of the candidate data is less than the threshold , it means that the candidate data has a high degree of similarity with the normal sample and is marked as a normal value. If the abnormal score of the candidate data is greater than or equal to the threshold , it means that the difference between the current data to be tested and the normal sample is large. This is then recorded as an outlier.
The second part of the model is the GA module, which takes as input the high-dimensional abnormal data obtained by the previous module and trains the genetic algorithm to search the abnormal subspace of high-dimensional outliers. The abnormal subspace of the outliers can provide the basis for the analysis of abnormal causes.
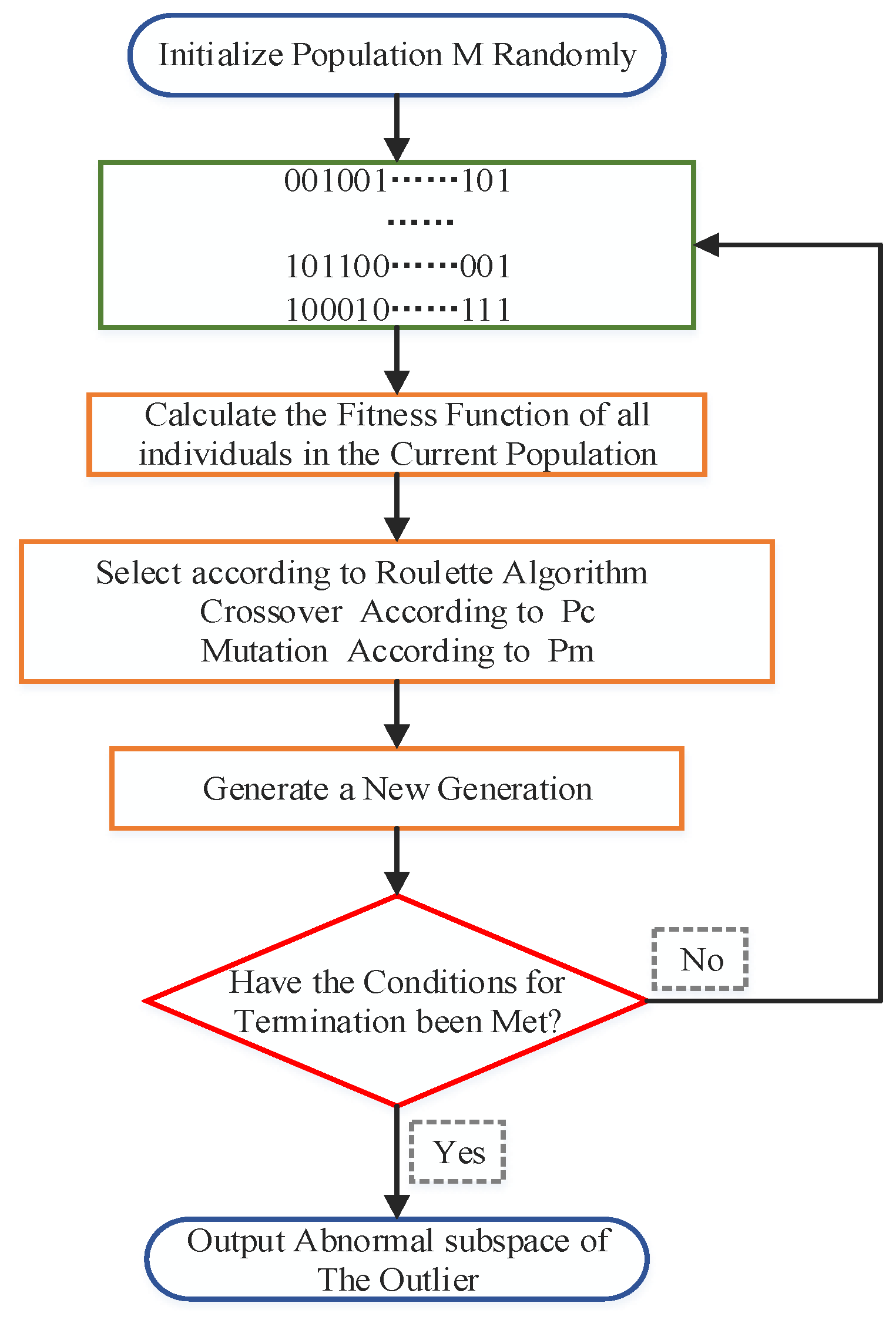
Figure 4 shows the flow of its search algorithm.
By analyzing the subspace of the outliers, we found that there are only two states of the subspace feature components, so the binary encoding rules were chosen to establish the mapping relationship between data subspaces and encoding strings. The problem of solving the abnormal subspace of data is transformed into the problem of searching for the optimal individual through genetic algorithm. Firstly,
individuals containing
genes are randomly generated to form an initial population representing feasible solutions of the abnormal subspace, where
is the population size and
is the dimension of the full data space. Each chromosome consists of {0,1}, where 1 means that the feature component was selected. The initial population generation is shown in
Figure 6.
To determine the fitness function for the target problem, the second part analyzes the target problem. We redefine the metric to be the fitness function of the genetic algorithm since our goal is to obtain the anomalies of the current candidate subspace to determine whether it has been eliminated.
Definition 1. Subspace Outlying Degree, SOD. In this work, (the distance between the point to be measured and the k-th nearest neighbor) is used as an outlier metric, which is called the subspace outlying degree. Since there is likely to be a high numerical outlier distance within a subspace, it is difficult to compare outliers between different subspaces. Therefore, to improve the comparability of the abnormal subspace SOD, the subspace abnormality SOD is defined as the ratio of at a given point p to the average(avg) in the data set denoted as Data in the same subspace s, as shown below: The higher the ratio, the higher the for the point sample p compared to other points, so the higher the outlier value of p, and vice versa. Our definition of SOD derives from the definition of the outlier subspace. Given the input data set denoted as Data, the parameter denoted as n is the dimension of the data set, and k is the number of adjacent data points. If there is no subspace s’, such that SOD(s’, p) > SOD(s, p), then the abnormal subspace of a given data point p is s.
The fitness function SOD is calculated for all individuals in the current population and sorted by size. Determine whether the chromosome corresponding to
in the current generation population satisfies the stopping condition. If so, decode the chromosome to obtain the abnormal subspace of candidate outliers. Otherwise, the VAEGA model will utilize genetic operators to generate a new generation of subspace populations. The specific steps are as follows. We calculate the fitness function SOD of each chromosome and then use the current ratio of each individual’s
value to the sum
as the individual’s selection probability
, as shown in Equation (14).
The larger the value, the higher the abnormal degree of the candidate outlier in the current subspace, and the higher the probability of the individual being selected to be inherited by the next generation.
The selection of individuals is a random process, which means individuals with high SOD values may be lost in the selection process. Hence, we add a mechanism for optimal retention selection, which directs the top N outstanding individuals with SOD values in the previous generation into the new generation population.
Then we use the non-replacement remainder rule to select population N chromosomes to inherit into the next generation population. In the new generation population, our work randomly selects an even number of parent chromosomes without repetition and performs a uniform crossover operation according to the crossover probability
. The resulting child chromosomes will be put into a new generation of populations. The binary uniform crossover operation is shown in
Figure 7.
It is assumed that the random probability of genes at positions 2, 3, 5, 8, and 9 is greater than the crossover probability
. Then, the genes at positions 2, 3, 5, 8, and 9 are exchanged to form two new child chromosomes in
Figure 8.
To generate distinct evolutionary directions for populations, some children chromosomes are picked at random for single point mutations based on mutation probability . The new generated population continues to repeat the above process until the algorithm converges or the stopping condition is satisfied.
By doing so, an optimal abnormal subspace solution that meets the search objective is obtained.
The most challenging step in using the genetic algorithm to search for anomalous subspaces is to calculate the fitness of each individual subspace. When we calculate the fitness SOD of candidate values in the current subspace, we need to scan the entire dataset, but the scale of high-dimensional datasets is usually large. For severely imbalanced datasets, we use a random multiple sampling technique, replacing the entire dataset with a randomly sampled dataset. By using random multiple sampling, the subjective bias of the samples obtained by single sampling can be reduced to a certain extent, so that the sample subset can represent the entire data set. By applying the random sampling method, it is possible to more efficiently calculate the abnormality degree of the current abnormal point subspace to measure and evaluate fitness more quickly. Nevertheless, we expect that the quality of the search results may be slightly affected, as will be verified in the experimental section.