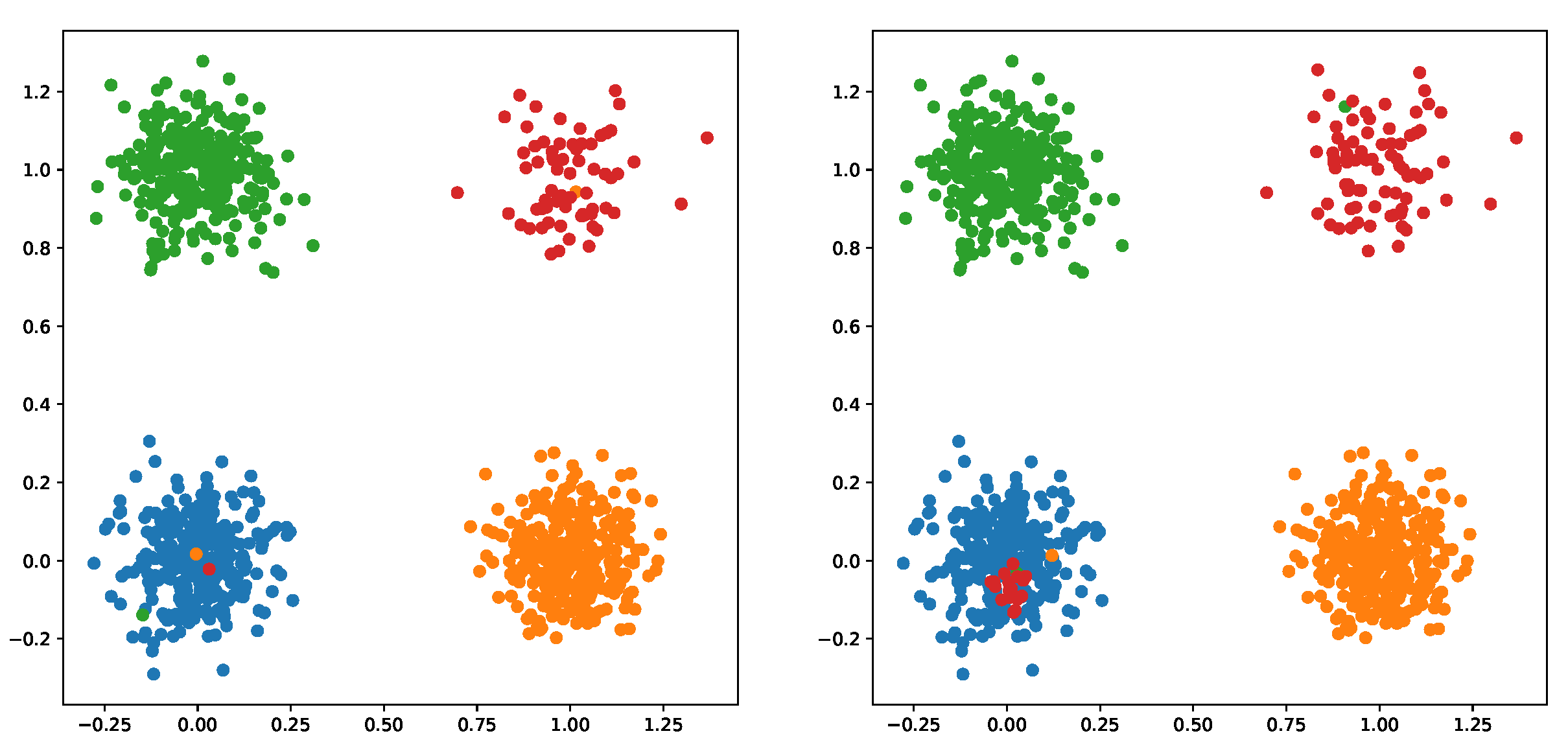
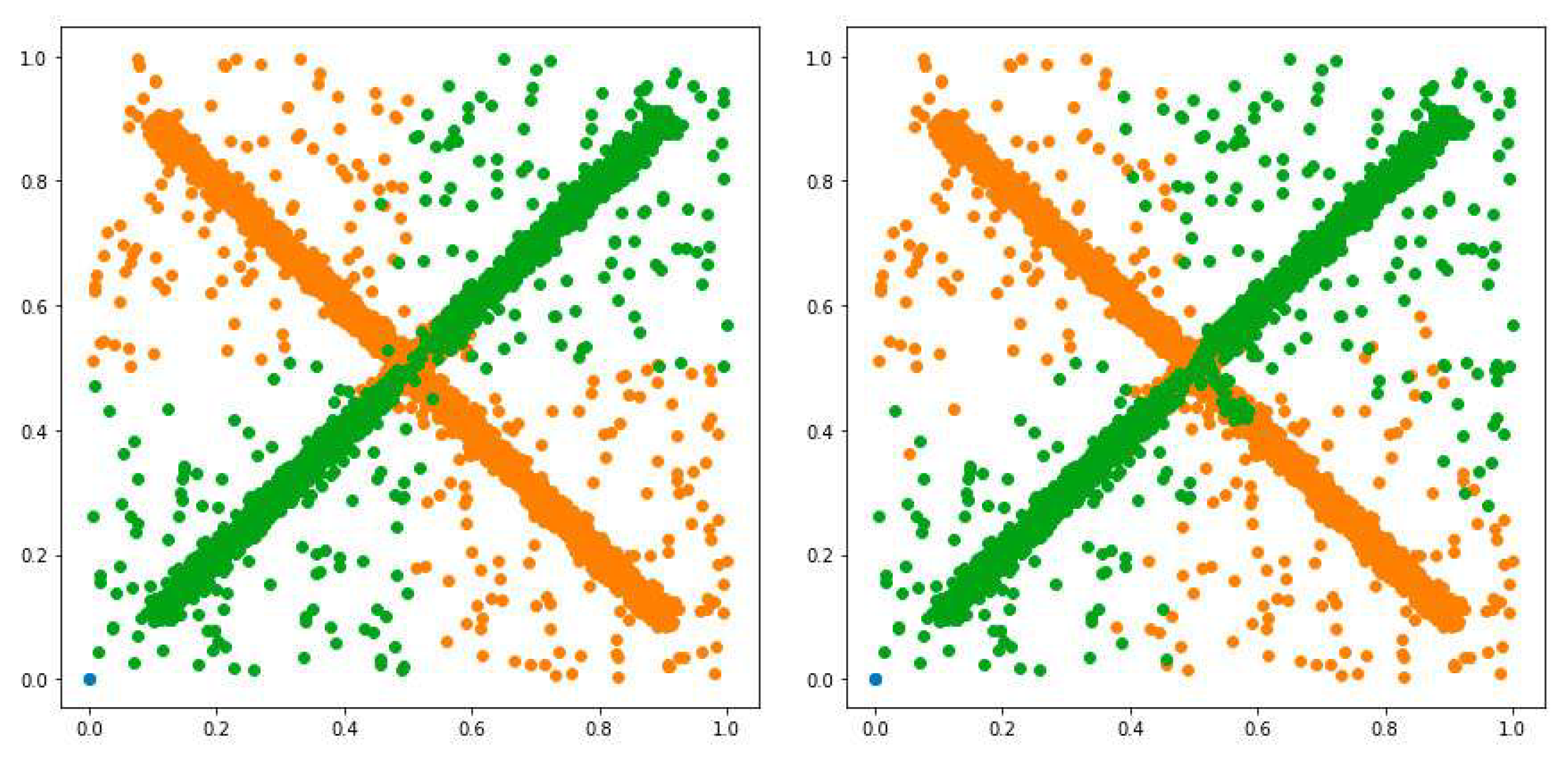
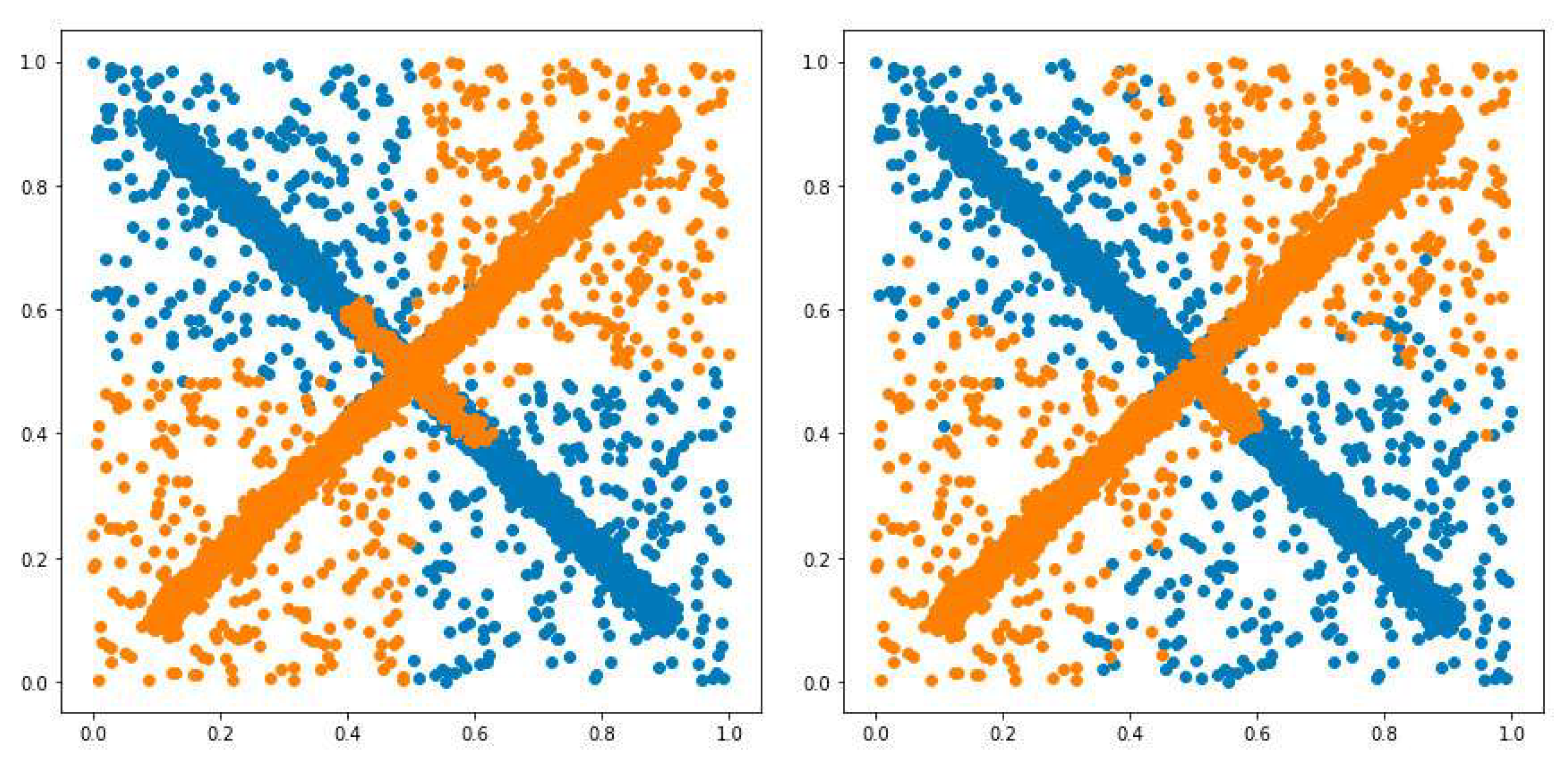
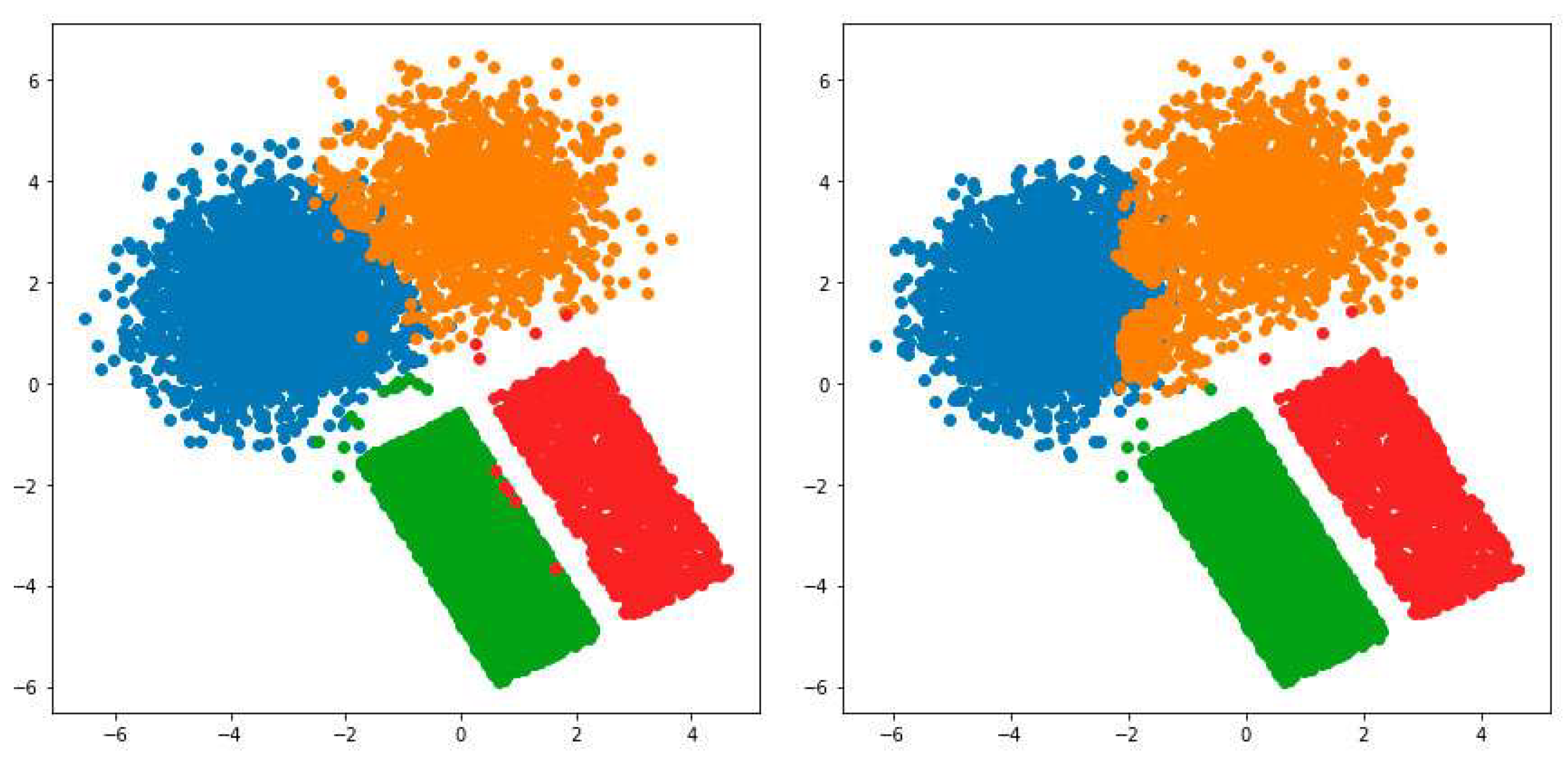
3.1. Possibilistic Fuzzy C-Means Method
Let us consider clustering of a given dataset
(where
is the
i-th data instance and
p is the number of features describing instances) as a partitioning of
X into
subgroups such that each subgroup represents "natural" substructure in
X. In [
8], this partition of
X, denoted as
M, is a set of
values that can be conveniently arrayed as two matrices
and
:
Equation (
1) defines the set of possibilistic fuzzy partition of
X. Here,
U is the fuzzy matrix while
T is the possibilistic one. Both matrices are mathematically identical, having entries between 0 and 1. These matrices are interpreted according to their entries. For the fuzzy matrix,
U, each entry
is taken as the membership of
in the
i-th cluster of
M, and for the probabilistic matrix each entry
is usually the (posterior) probability
that, given
, it came from cluster
i. It should be noted that
is a measure that shows how typical a given entry is for
i-th cluster.
In [
71], the authors explained why both possibilistic and fuzzy matrices should be used for clustering and in [
8] they also listed the disadvantages of both Fuzzy
C-Means (FCM) [
72] and Possibilistic
C-Means (PCM) [
73] methods. Originally, there was a constraint on the typicality values (the sum of the typicality values over all data points in a particular cluster was set to 1). Thus, in [
8] the authors relax this constraint, namely, in their algorithm, the row sum of typicality values is equal to 1, but the column constraint on the membership values is retained. This leads to the following optimization problem:
where
is the set of cluster centres,
,
,
and
are algorithm’s parameters chosen by the end-user,
is the standard Euclidean distance, and
(
) is also a user-defined constant. In (2)
,
,
, have the same meaning as membership values in FCM, and
,
,
, have the same interpretation as typicality values in PCM. The PFCM approach is iterative, and
U,
T and
V are updated over each iteration according to the rules presented in [
8].
3.2. Possibilistic Fuzzy Multi-Class Novelty Detector
The PFuzzND approach [
5] is used for the dynamic novelty detection learning scenario. It is an improved version of the Fuzzy multi-class Novelty Detector for data streams [
74], which is a fuzzy generalization of the offline–online framework presented in [
11] for the MINAS approach. All the mentioned algorithms can be divided into two stages. During the first stage, called offline, the model is built using a labelled training dataset. The second stage, during which new classes may emerge and disappear and the old classes may also drift, is referred to as the online or the prediction stage.
In the offline step of the PFuzzND algorithm, a decision model is learned from a labelled set of examples using the PFCM clustering method. To be more specific, for each known class a given number of clusters, , is designed, and this number is a user-defined parameter that can only increase (differently for each class) during the online stage until it reaches the maximum possible value . It also should be noted that is the same for all known classes during the offline phase, and each class cannot be divided into more than clusters. Furthermore, the maximum number of clusters, , that can be created for each class (normal or novel), is another user-defined parameter, which does not change during operation of the algorithm. Finally, it is assumed that generally only the portion of instances from the dataset are labelled, and, thus, they are used during the offline stage.
Thus, let us denote the number of the classes, known at the offline stage, as , then at the beginning of the online stage the decision model is defined as the set of clusters found for all different known classes. Additionally, an empty external set, called short memory, is created at the end of the offline phase. Examples labelled during the online stage as unknown are stored in the short memory for a time period , after this time limit, these instances are removed from the mentioned set. The latter can occur if it is established that they do not belong to the existing classes, and do not form novel classes either. Thus, these instances are considered as anomalies.
Moreover, there are four additional user-defined parameters for the online phase of the PFuzzND approach:
The minimum number of instances (denoted as ) in the short memory to start the novelty detection procedure.
The initial threshold, , to launch the classification process of the unlabelled instances.
Two adaptation thresholds, and , used during the classification step.
During the online step, first, for each new instance,
, its membership and typicality values related to all clusters, known at the moment, are calculated. Typicality values have more influence here as they are used to determine whether the instance
will be labelled by one of the existing classes or will be marked as unknown. For this purpose, the highest typicality value of the
j-th instance and the corresponding existing class,
(
), are determined. Subsequently, the maximum typicality values are found for all instances considered before the new one arrives (and these instances should belong to the class
), and the mean value of these typicalities is calculated. Let us denote this mean value as
, where
t is a timestamp, which shows how many instances were processed after the offline stage, and, therefore, is equal to 1 at the beginning of the online stage. If the highest typicality of the new instance,
, is greater than the difference between the obtained mean value
and the adaptation threshold
, then the instance
belongs to the class
[
5]. It should be noted that the highest typicality of the first instance processed after the offline phase is compared to the initial threshold
.
If the instance
is labelled, then its typicality value is used to update
, while the PFCM approach is used to update the clusters of the class
. Otherwise, the highest typicality of the new instance
is compared to the difference between
and the adaptation threshold
. If the highest typicality is greater than the mentioned difference, then instance
belongs to the class
, but a new cluster with
as its centroid should be created [
5]. If instances belonging to the class
are already divided into
clusters, then the oldest cluster is removed and the new cluster with
as its centre is generated instead of it.
If neither conditions are met, then the instance
will be marked as unknown and stored in the short memory until the novelty detection step will not be executed. The latter happens if the number of instances marked as unknown reaches
. In this case, firstly, the PFCM approach is applied to all the instances in the short memory and the pre-determined number
of clusters is designed. After that, for each generated cluster its fuzzy silhouette [
75] is calculated and if the obtained value is greater than 0 and the considered cluster is not empty, then this cluster is evaluated as the valid one. All validated clusters of short memory represent new patterns or novelties.
The next step of the novelty detection procedure consists of calculating the similarity between these validated clusters to the ones already existing (clusters belonging to the known classes), which is achieved by using the fuzzy clustering similarity metric introduced in [
9]. Thus, the known cluster, that is the most similar to the examined cluster from the short memory, is determined. If the value of the mentioned metric for these two clusters is greater than
(which is another parameter of the PFuzzND approach), then all instances from the examined cluster are labelled the same way as instances of the considered known cluster. Consequently, clusters of the corresponding class are updated by the PFCM algorithm. In this case, the known class has evolved, and we are referring to the concept drift. Otherwise, a new class is created, therefore, we should increment
by one, and instances from the examined cluster are labelled as belonging to the new class.
If one of the short memory clusters is not validated, then it is discarded and its instances remain in the short memory until the model executes the novelty detection procedure again or decides to remove them all, which can happen if these instances are in the short memory for iterations.
3.3. Evolutionary Algorithms
Differential Evolution or DE is a well-known population-based algorithm introduced in [
76] for real-valued optimization problems. DE maintains a population of solutions (also called individuals or target vectors) during the search process and the key idea of this algorithm comprises the usage of difference vectors calculated between the individuals in the current populations. These difference vectors are applied to the members of the population to generate mutant solutions. DE contains only three parameters, namely the population size
, scaling factor for mutation
F and crossover probability
.
Generally, the initialization step is performed by randomly generating
points
,
,
, in the search space with uniform distribution within
. Here,
P is the dimensionality of the search space. Mutation, crossover and selection operators are then iteratively applied to the generated population. Originally, DE used the rand/1 mutation strategy [
76]; however, most recent DE approaches, including L-SHADE [
7], often use the current-to-pbest/1 strategy, which was initially proposed for the JADE algorithm [
77]. The current-to-pbest strategy works as follows:
where
is an index of one of the best individuals (the quality of individuals is estimated by their fitness values),
k and
l are randomly chosen indexes from the population, and scaling factor
F is usually in the range
. Indexes
,
k and
l are generated in such a way that they are mutually different and are not equal to
i.
The crossover operation is performed after mutation to generate a trial vector by combining the information contained in the target and mutant vectors. Specifically, during the crossover, the trial vector
,
, receives randomly chosen components from the mutant vector
with probability
as follows:
where
is a randomly chosen index from
, which is required to ensure that the trial vector is different from the target vector to avoid unnecessary fitness calculations.
After generating the trial vector , the bound constraint handling method is applied. Finally, the selection step is performed after calculating the fitness function value in the following way: if the trial vector outperforms or is equal to the parent in terms of fitness, then the target vector in the population is replaced by the trial vector .
3.3.1. L-SHADE Algorithm
The L-SHADE approach is a modification of the DE algorithm, firstly introduced in [
7]. As previously mentioned, DE has three main parameters, and deciding upon which parameter to employ presents a difficult task, as they have an impact on the algorithm’s speed and efficiency. The L-SHADE algorithm uses a set of
historical memory cells containing values
to generate new parameter values
F and
for every mutation and crossover procedure. Mentioned parameter values are sampled using a randomly chosen memory index
as follows:
where
and
are random numbers generated by Cauchy and normal distributions, respectively. The
value is set to 0 or 1 if it spans outside the range
, and the
F value is set to 1 if
or is generated again if
. The values of
F and
, which caused an improvement to an individual, are saved into two arrays,
and
, together with the difference of the fitness value
.
The memory cell with index
h, incrementing from 1 to
H every generation, is updated as follows:
where
S is either
or
. Then, the previous parameter values are used to set the new ones in the following way:
In the last two formulas, c is an update parameter set to , and g is the current iteration number.
The L-SHADE algorithm uses the Linear Population Size Reduction (LPSR) approach to adjust the population size
. To be more specific, it is recalculated at the end of each iteration, and the worst individuals are removed from the population. The new number of individuals depends on the available computational resources:
where
and
are the minimal and initial population sizes, respectively,
is the current number of function evaluations, while
is the maximal number of function evaluations.
Finally, L-SHADE uses an external archive A of inferior solutions. The archive, A, contains parent solutions rejected during selection operation, and is filled until its size reaches the predefined value . Once the archive is full, new solutions replace randomly selected ones in A. The current-to-pbest mutation (5) is changed to use the individuals from the archive so that the l index is taken either from the population or the archive A with a probability of . Additionally, the archive size, , decreases together with the population size in the same manner.
3.3.2. NL-SHADE-RSP Algorithm
The NL-SHADE-RSP algorithm is a modification of the L-SHADE approach, first introduced in [
6]. It is a further development of the L-SHADE-RSP algorithm, presented in [
78], which uses selective pressure for the mutation procedure. The effect of the selective pressure was studied in detail in [
79]. The mutation strategy proposed for the L-SHADE-RSP approach is called current-to-pbest/r, and it is different from the original current-to-pbest strategy only with regard to the choosing of indexes
k and
l. To be more specific, for each individual
i the probability
is calculated in the following way:
where
is the rank of the
i-th individual. In addition, ranks
,
, were set as indexes of individuals in an array sorted by the fitness values, with largest ranks assigned to the best individuals. Finally, indexes
k and
l were chosen with the corresponding probabilities. In NL-SHADE-RSP, the same current-to-pbest/r strategy is used, however, the rank-based selective pressure is applied only to the index
l, and only if it is chosen from the population, while the index
k is chosen uniformly.
In contrast to the L-SHADE and L-SHADE-RSP approaches, in NL-SHADE-RSP the population size is reduced in a non-linear manner:
where
is the ratio of the current number of fitness evaluations.
The external archive
A is used for the index
l in the current-to-pbest/r mutation procedure with probability
, and in the NL-SHADE-RSP algorithm this probability is automatically adjusted. The latter is achieved by implementing the adaptation strategy, originally proposed for the IMODE algorithm [
80]. Firstly, the probability
should be within the range
, and initially it is set to
, unless the archive is empty. Then, the probability
is calculated in the following way:
where
is the amount of archive usage, which is incremented every time an offspring is generated using archive
A,
and
are the fitness improvements achieved with and without archive, respectively. It should be noted that the new value of
is checked to be within the range
by applying the following rule:
In NL-SHADE-RSP, both binomial and exponential crossover operators are used with a probability of
. The description of the exponential crossover is given in [
6]. For exponential crossover, the Success-History Adaptation (SHA) is applied, but at the beginning of the generation, the crossover rates
generated for each individual
i are sorted according to fitness values, so that smaller crossover rate values are assigned to better individuals. For the binomial crossover, value
is calculated in the following way:
Finally, the
value for current-to-pbest/r mutation in NL-SHADE-RSP is controlled in the same way as in the jSO algorithm [
81]:
Note that the following user-defined parameters were used:
and
. Furthermore, the
parameter linearly decreases as it is performed in jSO.Detailed pseudo-code of the NL-SHADE-RSP algorithm is presented in [
6].
3.4. The HFuzzNDA Approach
The HFuzzNDA algorithm, in contrast to the PFuzzND approach, has an additional parameter, namely the maximum possible number of clusters per class, denoted as . This parameter changes over time and is initially set to a minimal value (to be more specific, it is equal to 2). To calculate the new value, the following notations are used:
First, we check how many instances are classified (this number is denoted as ); after the offline stage this number is equal to , and grows incrementally during the online stage (one new instance per iteration).
Two new parameters are introduced, namely and ; controls the speed at which the value grows, and shows how much grows over each iteration.
A given number of considered instances, denoted as , is saved after some number of iterations, right after the offline stage.
Thus, is updated as shown in Algorithm 1 (note that in Algorithm 1 the temporary variable only grows).
| Algorithm 1 update procedure |
- 1:
After the offline stage set , , - 2:
while online stage do - 3:
if then - 4:
- 5:
- 6:
- 7:
end if - 8:
end while
|
Evolutionary and biology-inspired optimization algorithms are popular among researchers and are frequently used for anomaly and/or novelty detection. For example, in [
82] the hybrid algorithm based on Fruit Fly Algorithm (FFA) and Ant Lion Optimizer (ALO), and in [
83] the Farmland Fertility Algorithm were used, respectively, for feature selection to reduce the dimensionality of the data. Data, pre-processed in such a way, were later classified as normal or anomalous by using some well-known machine learning approaches (support vector machines, decision trees,
k nearest neighbours). In this study, we did not apply an evolutionary algorithm as a pre-processing technique, instead it was used for clustering, namely, clusters’ centres were determined by the NL-SHADE-RSP algorithm. To be more specific, NL-SHADE-RSP determined centres of clusters belonging to each class and each individual represented the whole set of clusters’ centres; and no feature selection was applied. Thus, during the offline stage, the NL-SHADE-RSP approach is used to divide each known class into two clusters. For this purpose, function (2) is optimized and each individual represents the centres of the clusters. After that, the PFCM algorithm is used to obtain the membership
U and typicality
T matrices for these classes. The “age” of all clusters, designed during the offline stage, is recorded (they are considered as the oldest clusters).
The online stage starts with Algorithm 1 and then proceeds to check conditions, described for the PFuzzND approach, for each new instance iteratively. However, some changes were implemented to the online procedure. To be more specific, a new merging technique is applied to clusters to check if there is a need to decrease their number, and the NL-SHADE-RSP approach is used to refine clusters belonging to each class. The pseudo-code for the online stage is presented in Algorithm 2. Note that in Algorithm 2 is the current number of instances stored in short memory. Furthermore, the NL-SHADE-RSP approach can change the final number of clusters belonging to the considered class. To be more specific, it is executed for all possible variants of the for a given class from 2 to the current value of , and the best variant is chosen at the end of the optimization process.
| Algorithm 2 One iteration of the online phase |
- 1:
Set , , , , , , , - 2:
for each new j-th instance do - 3:
- 4:
- 5:
Calculate the highest typicality value and determine the corresponding existing class - 6:
For instances, previously labelled as , calculate the mean of the highest typicality values - 7:
if then - 8:
- 9:
end if - 10:
if then - 11:
The j-th instance belongs to class - 12:
Update clusters that belong to the class by using PFCM - 13:
if The number of clusters is greater than 2 then - 14:
Execute the merging procedure - 15:
end if - 16:
else if then - 17:
The j-th instance belongs to class - 18:
if The number of clusters that belong to class is less than then - 19:
Increment the number of clusters that belong to class - 20:
Create new cluster with j-th instance as its center - 21:
Execute NL-SHADE-RSP to refine the clusters - 22:
Calculate the membership and typicality matrices for all clusters - 23:
else - 24:
Determine the oldest cluster that belongs to class - 25:
Create new cluster with j-th instance as its center - 26:
Execute NL-SHADE-RSP to refine the clusters - 27:
Calculate the membership and typicality matrices for all clusters - 28:
end if - 29:
if The number of clusters is greater than 2 then - 30:
Execute the merging procedure - 31:
end if - 32:
else - 33:
if then - 34:
- 35:
Store the j-th instance in the short memory - 36:
else - 37:
Execute the novelty detection procedure - 38:
Store the j-th instance in the updated short memory - 39:
end if - 40:
end if - 41:
end for
|
The original PFuzzND algorithm allows an increase in the number of clusters belonging to each class, but not the other way around. Experiments showed that in some cases the instances belonging to the same class might be divided into an excessive number of clusters which can can lead to bad classification results. Thus, in this study, it is proposed to merge clusters belonging to the same class if they are similar to each other, which allows for a decrease in their number.
In [
9], fuzzy similarity metric was introduced. It can be described in the following way: firstly, the dispersions of two considered clusters are calculated, then the dissimilarity between these clusters is determined and, finally, the sum of dispersions is divided by the dissimilarity value [
9]. Each cluster’s dispersion is the weighted sum of distances between instances belonging to this cluster and its centre, averaged by the number of considered instances. Note that the membership values are used as weights. The dissimilarity between two clusters is the Euclidean distance between their centres.
In our study, the typicality values are used as the weight coefficients for calculating the similarity metric. We find two of the most similar clusters belonging to a given class, and determine if they should be merged by using the generalized soft
C index metric [
10] (here, we denote it as
). To use the latter, we have to conduct calculations for all instances that belong to the considered class. However, this can significantly slow the algorithm, so only a part
(here,
c is the class number and
B is the batch size) of instances participate in calculating the
values. The pseudo-code of the proposed merging procedure is demonstrated in Algorithm 3.
| Algorithm 3 The merging procedure |
- 1:
Denote the current set of clusters belonging to the considered class as - 2:
for p-th cluster belonging to the considered class, do - 3:
for q-th cluster belonging to the considered class, do - 4:
if then - 5:
Calculate fuzzy similarities between p-th and q-th clusters - 6:
end if - 7:
end for - 8:
end for - 9:
Find the - 10:
Determine centres of the most similar clusters and corresponding to the value - 11:
ifthen - 12:
- 13:
Create a new (empty) set , which can contain clusters - 14:
Create new cluster with centre by merging two the most similar clusters - 15:
- 16:
Fill set with newly created cluster and all clusters from except two the most similar - 17:
Execute the PFCM algorithm with new clusters from to update them - 18:
Choose instances belonging to the considered class - 19:
For chosen instances and both sets and calculate the values - 20:
Choose the set of clusters with better value - 21:
end if
|
Finally, during the novelty detection phase, instances stored in the short memory are divided into clusters using the NL-SHADE-RSP algorithm. The maximum possible number of short-memory clusters is set to the current value of . After that, membership and typicality matrices are determined. Then, the standard steps of the novelty detection procedure introduced for the PFuzzND approach are conducted.
The general scheme of the proposed approach is demonstrated in
Figure 1.