Abstract
In machine learning, ensembles of models based on Multi-Layer Perceptrons (MLPs) or decision trees are considered successful models. However, explaining their responses is a complex problem that requires the creation of new methods of interpretation. A natural way to explain the classifications of the models is to transform them into propositional rules. In this work, we focus on random forests and gradient-boosted trees. Specifically, these models are converted into an ensemble of interpretable MLPs from which propositional rules are produced. The rule extraction method presented here allows one to precisely locate the discriminating hyperplanes that constitute the antecedents of the rules. In experiments based on eight classification problems, we compared our rule extraction technique to “Skope-Rules” and other state-of-the-art techniques. Experiments were performed with ten-fold cross-validation trials, with propositional rules that were also generated from ensembles of interpretable MLPs. By evaluating the characteristics of the extracted rules in terms of complexity, fidelity, and accuracy, the results obtained showed that our rule extraction technique is competitive. To the best of our knowledge, this is one of the few works showing a rule extraction technique that has been applied to both ensembles of decision trees and neural networks.
Keywords:
ensembles; bagging; boosting; model explanation; decision trees; perceptrons; rule extraction 1. Introduction
Deep learning has been very successful in the last decade. Particularly, in domains such as computer vision, deep neural network models have improved their performance significantly over that of Multi-Layer Perceptrons (MLPs), Support Vector Machines (SVMs), Decision Trees (DTs), and ensembles. However, for structured data, deep models do not offer a significant advantage over well-established “classical” models, which therefore remain indispensable. A major problem with MLPs is that it is difficult to interpret their responses; MLPs are therefore very often considered as black boxes. However, a number of works aimed at transforming the knowledge stored in connections and activations into propositional rules. Andrews et al. introduced a nomenclature encompassing all methods for explaining neural network responses with symbolic rules [1]. For SVMs that are functionally equivalent to MLPs, rule extraction methods were also proposed [2].
Ensembles of models often provide better accuracy than a single model. Many learning methods for ensembles were introduced, such as bagging [3] and boosting [4]. They have been applied to decision trees and neural networks. However, even decision trees that are models that are directly translatable into propositional rules lose their interpretability when combined in an ensemble. Solving the interpretability problem is crucial for machine learning models to be well accepted by society. For instance, in Europe, with the General Data Protection Regulation (GDPR), an individual has the right to an explanation when an algorithm makes a decision about her or him, such as denying credit. The purpose of this work is to demonstrate that an existing rule extraction technique that is applied to ensembles of neural networks can also be applied to ensembles of decision trees. In addition, our second purpose is to describe the characteristics of the rulesets generated in terms of fidelity, accuracy, and complexity.
With ensembles of DTs, two main strategies are applied to generate propositional rules. The first tries to reduce the number of DTs by increasing their diversity. Thus, with a reduced number of trees, all of the rules generated by each tree are taken into account. Examples of algorithms for the optimization of diversity are reported in [5]. In the second group of methods, the basic strategy is to remove as many rules as possible. Regarding neural network ensembles, few works have been performed for rule extraction, including the one applied to the DIMLP model (Discretized Interpretable Multi-Layer Perceptron) [6]. Unlike MLPs, in DIMLPs, the discriminative hyperplanes are precisely localized. This makes it possible to define the antecedents of propositional rules. As shown in this work, a DT is transformed into a DIMLP network; hence, the rule extraction technique used for DIMLP ensembles can also be applied to DT ensembles. Specifically, here, this method is applied to Random Forests (RFs) [7] and DT ensembles trained by gradient boosting [8].
We apply rule extraction to both DT and DIMLP ensembles on eight classification problems. The characteristics of the generated rules are compared to those of “Skope-Rules” [9] and other rule extraction techniques. The results obtained by our method were found to be competitive. In the following sections, we briefly present the state of the art of rule extraction from ensembles, the models used, and the experiments, followed by a discussion and the conclusion.
2. Rule Extraction from Ensembles
Since Saito and Nakano’s early work on single MLPs [10], only a few papers have addressed rule extraction from neural network ensembles. As an historical example of a rule extraction technique, Saito and Nakano presented a method that generated rules from changes in levels of input and output neurons. Specifically, their algorithm checked whether a rule could be generated or not. To avoid combinatorial explosion, meaningless combinations of inputs were excluded, and only a limited number of inputs in the antecedents were taken into account.
The author proposed DIMLP networks to generate unordered propositional rules from ensembles [11,12,13,14]. With the DIMLP model, rule extraction is performed by determining the precise location of axis-parallel discriminative hyperplanes. Zhou et al. introduced the REFNE algorithm (Rule Extraction from a Neural Network Ensemble) [15]. In REFNE, a trained ensemble generates additional samples and then extracts propositional rules. Furthermore, attributes are discretized during rule extraction, and it also uses particular fidelity evaluation mechanisms. Finally, rules are limited to only three antecedents. Johansson used a genetic programming technique to produce rules from ensembles of 20 neural networks [16]. Here, rule extraction from ensembles was viewed as an optimization problem in which a trade-off between accuracy and comprehensibility had to be taken into account. Hara and Hayashi introduced a rule extraction technique for a limited number of MLPs in an ensemble [17,18]. In [19], Sendi et al. trained DIMLP ensembles by optimizing their diversity. Then, rule extraction was carried out for each single network, and for each sample, the rule that was chosen was the one with the highest confidence score.
A well-known representative technique for an ensemble of DTs is RuleFit [20]. Here, trees are trained on random subsets of the learning set, the main idea being to define a linear function that includes rules and features that approximate the whole ensemble of DTs. At the end of the process, this linear function represents a regularized regression of the ensemble responses with a large number of coefficients that are equal to zero. Node Harvest (NH) is another rule-based representative technique [21]. Its purpose is to find suitable weights for rules by performing a minimization on a quadratic program with linear inequality constraints. In [22], the rule extraction problem was viewed as a regression problem using the sparse group lasso method [23], such that each rule was assumed to be a feature and the aim was to predict the response. Subsequently, most of the rules were removed by trying to keep the accuracy and fidelity as high as possible. “Skope-Rules” is a recent technique [9], the main objective being to provide propositional rules that verify precision and recall conditions. Similar or duplicated rules are removed based on a similarity threshold of their supports. The final rules are associated to weights that are simply proportional to their out-of-bag precision. For all of the previous rule extraction techniques, questions remain about the interpretability of the coefficients that are different from zero with respect to the rules. In a different approach, Sagi and Rokach proposed a method of transforming a decision forest into a single decision tree, aiming at approximating the predictive performance of the original decision forest [24]. Finally, in [25], rules extracted from a tree ensemble were summarized into a rule-based learner in which all of the original rules were selected in a compact set of relevant and non-redundant rules and then pruned and ranked.
3. Materials and Methods
The key idea in this work is to transform ensembles of DTs into ensembles of interpretable MLPs. Therefore, by being able to generate propositional rules from neural networks, we are also in a position to generate rules for ensembles of DTs. Figure 1 illustrates an example of transformation of an ensemble of three DTs into an ensemble of neural networks. First, from each DT, a number of rules are generated (R11, R12, etc.); second, each rule is inserted into a single neural network (NN11, NN12, etc.); third, the final classification is the result of all of the neural networks’ classifications. Hence, rule extraction is performed at the ensemble level, which can be considered as a unique neural network with an additional layer represented by the two green neurons on the right.
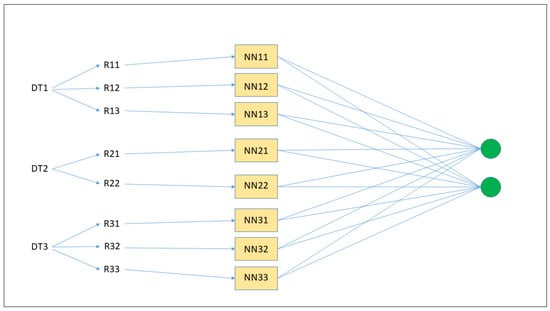
Figure 1.
Transformation of an ensemble of three DTs into an ensemble of neural networks. First, from each DT, a number of rules are generated (R11, R12, etc.); second, each rule is inserted into a single neural network (NN11, NN12, etc.); third, the final classification is the result of all of the neural networks’ classifications.
3.1. Ensembles of Decision Trees
A binary decision tree is a recursive structure containing nodes and edges. Each node represents a predicate with respect to an attribute. Depending on its value, the path taken to classify a sample continues to the left or right branch until a terminal node is reached. Every path from the root to a terminal node defines a propositional rule. Specifically, the format of a symbolic rule is given as: “If tests on antecedents are true, then class C”, where “tests on antecedents” are in the form:
- ; or
- ;
with as the input variable (or attribute) and as a real number. Class C designates a class among several possible classes. Figure 2 illustrates an example of a decision tree. The learning phase of such a model consists in determining at each node of the tree the best attribute for accurately dividing the learning samples. Many possible criteria can be used to determine the best splitting attribute; for more details, see [26,27].
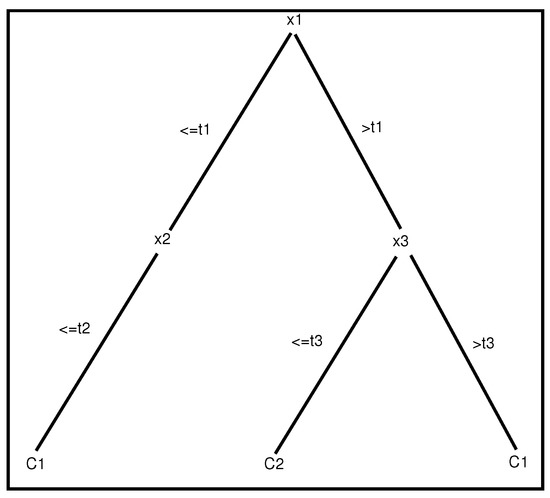
Figure 2.
An example of a tree with two depth levels. Each path from the root to a leaf represents a propositional rule of two different classes ( and ).
The number of nodes in a shallow tree is very limited. It represents a “weak” learner with limited power of expression. For instance, a tree with a unique node performs a test only on an attribute. This type of DT is also called a decision stump. With the use of boosting techniques [4], ensembles of weak learners become strong classifiers [28] that are able to learn complex classification problems. In this work, we train ensembles of Gradient-Boosted Trees (GB) [8].
Random Forests (RF) are ensembles of DTs [7] trained by bagging [3]. Specifically, bagging selects for each classifier included in an ensemble a number of samples drawn with a replacement from the original training set. Since many of the generated samples may be repeated while others may be left out, a certain diversity of each single predictor proves to be beneficial with respect to the whole ensemble of combined classifiers. In addition, each tree can be constrained at each induction stage to select a small proportion of the available attributes.
3.2. Transformation of Decision Trees into Interpretable MLPs
Since the key idea behind extracting rules from ensembles of DTs is their transformation into ensembles of interpretable MLPs, we first describe how to transform a rule with a unique antecedent into an MLP. Then, we generalize to rules with many antecedents and coefficient-weighted rules.
3.2.1. An Example with a Unique Antecedent
Figure 3 shows an MLP that represents a propositional rule with a unique antecedent. Any neuron in the middle or output layer receives a signal, which is the result of a weighted sum of inputs and weight values; here, this sum is .
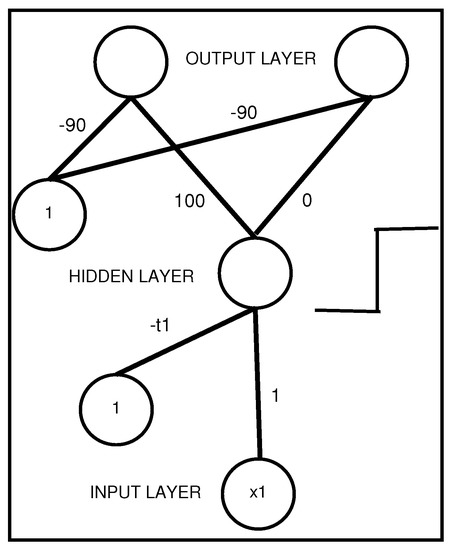
Figure 3.
An interpretable MLP coding a propositional rule with a unique antecedent .
Then, an activation function is applied; in the middle layer, it is a step function that is given as:
In the output layer, we have a sigmoid function that is given as:
Therefore, the MLP presented in Figure 3 represents the following propositional rule:
- ; with designating the first class coded by vector .
3.2.2. An Example with Two Antecedents
Figure 4 shows an MLP that represents a propositional rule with two antecedents:
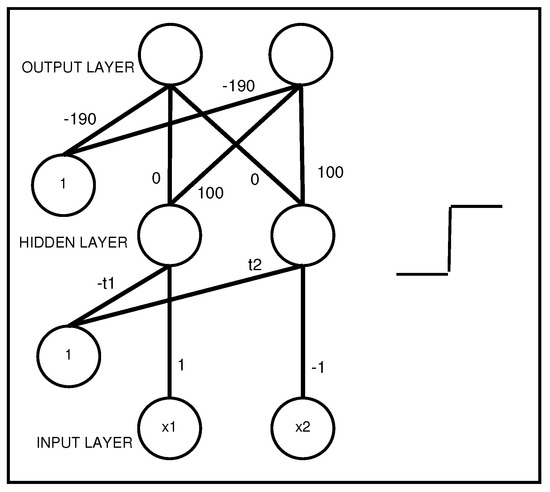
Figure 4.
An interpretable MLP coding a propositional rule with two antecedents, AND .
- AND →, with designating the second class coded by vector .
It should be noted that, between the hidden layer and the output layer, a logical “AND” is performed. Therefore, when all of the rule antecedents are true (e.g., all of the hidden neurons have an activation equal to one), then an output neuron related to a given class is activated to a value very close to one.
3.2.3. Generalization to an Arbitrary Number of Antecedents
Generally, with a propositional rule involving A antecedents, to perform a logical “AND” between the hidden layer and the output layer, we define bias values equal to , with . Moreover, the values of the weights between the hidden neurons and the output neuron encoding the class are equal to K; all other weight values are equal to zero. Each rule generated from the root to a leaf of a DT is inserted into an MLP with the coding described above.
3.2.4. Coefficient-Weighted Rules
Rules extracted from the DTs trained by GB present a coefficient, which somehow represents the importance of the rules. This coefficient is inserted into an MLP by means of an additional layer, as depicted in Figure 5. Specifically, at the top right, symbol w between the second hidden layer and the output layer encodes a rule coefficient.
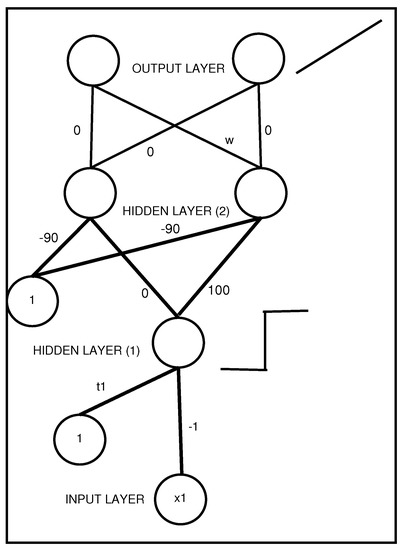
Figure 5.
An MLP with two hidden layers that represents a propositional rule with a unique antecedent . The activation function of the output layer is the identity, with coefficient w representing the rule weight.
3.3. Rule Extraction from Neural Network Ensembles
Particular MLPs, such as those shown in Figure 4, were reported in [29]. Their precise name is Interpretable Multi-Layer Perceptrons (IMLPs). IMLPs often include two hidden layers, with the feature that any neuron in the first hidden layer receives only one connection from an input neuron and the bias neuron. Furthermore, the activation function in the first hidden layer is a step function. Above the first hidden layer, neurons are fully connected.
With IMLPs, the training process was performed with a simulated annealing algorithm, while rule extraction was carried out with the ESPRESSO logic minimizer [30]. More precisely, the binary activations of the first hidden layer put into play a Boolean minimization problem with respect to the IMLP network responses. Later, the step activation function in the first hidden layer was generalized by the staircase activation function with the DIMLP architecture [6]. The staircase function approximates the sigmoid function; thus, it provides quantized values of the sigmoid. With DIMLPs, the training algorithm is a modified back-propagation algorithm [6,12].
The rule extraction algorithm for a single DIMLP is briefly described here; it includes three main steps, the details of which are reported in [6,12]. In the first step, the relevance of axis-parallel hyperplanes is calculated. Specifically, this relevance measure corresponds to the number of samples viewing a hyperplane as the transition to a different class. In the second step, a decision tree is built with all of the training samples. The class represented at the leaves is the one provided by the neural model. In the third step, all of the paths between the root and the leaves are represented as propositional rules. At this point, rules are disjointed, and generally, their number is large, as is their number of antecedents. Then, a pruning strategy is applied; it removes as many antecedents and as many rules as possible.
Rule extraction from ensembles of DIMLPs is achieved with the rule extraction algorithm that was briefly described above, as it can be applied to any DIMLP with any number of hidden layers. It is worth noting that an ensemble of DIMLP networks can be viewed as a single DIMLP with an additional hidden layer [12]. Hence, rule extraction can also be performed for a DIMLP ensemble.
4. Experimental Results
In the experiments, we first describe the characteristics of the eight selected classification problems; secondly, we explain the assessment measures and the values of the learning parameters, and then we show the results obtained through cross-validation. Finally, we illustrate an example of a generated ruleset. In this part, a key research question is whether the proposed rule extraction technique provides good results with respect to other known algorithms.
4.1. Classification Problems
We applied a number of models to classification problems of two classes. We retrieved eight datasets from the Machine Learning Repository at the University of California, Irvine (https://archive.ics.uci.edu/ml/index.php) [31] (accessed on: 14 September 2021). Table 1 describes their main characteristics.

Table 1.
Datasets used in the experiments. From left to right, the columns designate: number of samples; number of input features; types of features (Boolean, categorical, integer, real); proportion of samples in the majority class; references.
4.2. Learning Parameters and Assessment Measures
Training sets were normalized through Gaussian normalization. The following models were trained with the eight classification problems used in this work:
- DIMLP ensembles trained by bagging (DIMLP);
- Random Forests (RF);
- Shallow decision trees trained by Gradient Boosting (GB);
- Skope-Rules (SR) [9].
All DIMLP ensembles were trained through back-propagation with the default learning parameters:
- Learning parameter = 0.1;
- Momentum = 0.6;
- Flat spot elimination = 0.01;
- Number of stairs in the staircase activation function = 50.
The DIMLP architectures were defined with two hidden layers. The default number of neurons in the first hidden layer was equal to the number of input neurons. For the second hidden layer, this number was empirically defined in order to obtain a number of connections less than or similar to the number of training samples. For each classification problem, the number of neurons in the second hidden layer was:
- Australian Credit Appr.: 20;
- Breast Cancer: 20;
- Divorce Prediction: 2;
- Heart Disease: 10;
- Ionosphere: 10;
- Mammographic Mass: 50;
- Student Performance (Math): 6;
- Voting Records: 10.
Finally, out-of-bag samples were used to avoid the overtraining phenomenon by applying an early-stopping technique. Specifically, the out-of-bag set constituted a subset of the training dataset that was not used to fit the weight values of the neural networks.
Ensembles of DTs were trained with the Scikit Python Library [39]. For GB, the depth of the trees was varied from one to three, while for RF, the depth parameter was unconstrained or fixed to three (RF-3). Furthermore, for all of the random forests, the number of attributes that were taken into account at each induction step was, by default, the square root of the total number of attributes.
For comparison purposes, the “Skope-Rules” rule extractor [9] was also included in this work. Specifically, its propositional rules were generated from ensembles of RFs. Three important parameters were used:
- Minimal recall of rules;
- Minimal precision of rules;
- Maximal depth of the trees.
The minimal recall parameter influences the number of rules generated. Specifically, recall is defined as , with designating the number of true positives and denoting the number of false negatives. Precision is defined as , with representing the number of false positives. In order to approach the performance of the other models used in this work, we carried out several preliminary tests. We noticed that when the maximum tree depth was not limited, the average number of antecedents per rule was too high. We found that a maximal depth of three was a good value. Furthermore, with a low value of the minimal rule accuracy, the predictive accuracy tended to be too weak. After several preliminary experiments, we determined that 95% was an appropriate value for the eight selected datasets. Finally, too high of a value of the minimal recall parameter meant that many samples were not covered by the rules, and too low of a value generated a high number of rules. After several trials, we found that a value equal to 5% was a good compromise. In all of the experiments, the number of classifiers in an ensemble varied from 25 to 150.
With predictive accuracy being defined as the ratio of correctly classified samples on the total number of samples of a testing set, from left to right, the columns in the following tables designate:
- Average predictive accuracy of the model (correctly classified samples in the testing set divided by the number of samples);
- Average fidelity on the testing set, which is the degree of matching between the generated rules and the model. Specifically, with P samples in the testing set and Q samples for which the classification of the rules corresponds to the classification of the model, the fidelity is .
- Average predictive accuracy of the rules;
- Average predictive accuracy of the rules when the rules and model agree. Specifically, it is the proportion of correctly classified samples among the Q samples defined above.
- Average number of rules extracted;
- Average number of rule antecedents.
4.3. Cross-Validation Experiments
We conducted experiments based on ten repetitions of stratified 10-fold cross-validation trials. Table 2 illustrates the results for the “Australian” dataset; between brackets are shown the standard deviations. For the DIMLP ensembles, we observed that the numbers in each column were quite stable, although the number of predictors in an ensemble increased from 25 to 150. This was also true for RFs to a lesser extent. For GB and SR, the average complexity of the rules increased with an increasing number of predictors (last two columns).

Table 2.
Average results obtained on the “Australian” dataset. From left to right are presented the average results on predictive accuracy, fidelity on the testing sets, predictive accuracy of the rules, predictive accuracy of the rules when ensembles and rules agreed, number of rules, and number of antecedents per rule. Standard deviations are given between brackets. For DIMLP, RF, RF-3, and RS, the number of predictors is given between brackets (first column). For GB, the first number in brackets is the depth of the trees and the second is the number of predictors. For each column, the highest average accuracy or average fidelity is represented in bold, along with the lowest average number of rules or average number of antecedents.
Note also that the highest average fidelity was obtained with the simplest ensembles; e.g., decision stumps trained by gradient boosting with 25 and 50 predictors. RF ensembles with 50 predictors provided the highest average predictive accuracy (87.2%), but the generated rulesets were among the most complex (76.8 rules on average). Interestingly, the average predictive accuracy obtained by the decision stumps was very close, with an average of 87.1% (see GB (1, 100)), but with less complex rulesets (13.2, on average). As a general observation, the average predictive accuracy of the ensembles tended to be a bit higher than that obtained by the extracted rules. Nevertheless, the average predictive accuracy of the extracted rules, when rules and ensembles agreed (fifth column) tended to be a bit higher than that obtained by the models (first column).
Table 3 presents the results for the “Breast Cancer” classification problem. As in the previous table, the results provided by DIMLPs and RFs were quite stable with the increasing number of predictors in an ensemble. For GB and SR, the complexity of the extracted rulesets tended to be augmented as the number of predictors increased, but to a lesser extent with the decision stumps (trees with only one node). Again, the highest average predictive accuracy of the model and the extracted rules was provided by RF (97.4% and 96.9%). The average highest fidelity was reached by the decision stumps (99.0%). For this model, the average predictive accuracy attained by the rules was equal to 96.7%, and the complexity of the rules was less than half that obtained by RF (11.2 versus 24.3). Finally, the average predictive accuracy obtained by SR decreased as the number of trees in an ensemble increased. Moreover, with SR, the rules were, on average, the most complex.

Table 3.
Average results obtained on the “Breast Cancer” dataset. For each column, the highest average accuracy or average fidelity is represented in bold, along with the lowest average number of rules or average number of antecedents.
Table 4 depicts the results for the “Divorce” dataset. The highest predictive accuracy was obtained by the DIMLP ensembles (98.1%) with 5.5 rules, on average. GB provided the highest fidelity with 99.5% and 3.8 rules, on average. Finally, the highest average predictive accuracy of the rules was reached by ensembles of DTs trained by GB (3, 50) and RF-3 (97.3%). Note that for the latter model, a few more rules were produced (3.8 versus 5.1).

Table 4.
Average results obtained on the “Divorce” dataset. For each column, the highest average accuracy or average fidelity is represented in bold, along with the lowest average number of rules or average number of antecedents.
As shown in Table 5, on the “Heart Disease” classification problem, the DIMLP ensembles achieved the highest predictive accuracy (85.8%), the highest predictive accuracy of the rules (84.4%), and the highest predictive accuracy of the rules when ensembles and rules agreed (86.8%), on average. The decision stumps provided the highest average fidelity (99.0%) and the lowest average complexity of the rules (11.0 rules with 2.6 antecedents). It is worth noting that the average predictive accuracy obtained by the DIMLPs and GB (1, 50) was very close (84.4 versus 84.3), but GB produced fewer rules (15.2 versus 20.4). The average predictive accuracy attained by SR was the lowest; moreover, it decreased as the number of trees increased.

Table 5.
Average results obtained on the “Heart Disease” dataset. For each column, the highest average accuracy or average fidelity is represented in bold, along with the lowest average number of rules or average number of antecedents.
With the “Ionosphere” dataset, the results illustrated in Table 6 show that the highest average predictive accuracy was obtained by both the DIMLP ensembles and RFs (93.4%). Moreover, the same models provided the highest average predictive accuracy of the rules when ensembles and rulesets agreed (94.5%). However, rulesets generated from DIMLPs were less complex than those extracted from RFs, on average (18.3 versus 32.6). The highest average fidelity was attained by GB (99.6%), with the least complex rulesets (6.5 rules with 1.9 antecedents).

Table 6.
Average results obtained on the “Ionosphere” dataset. For each column, the highest average accuracy or average fidelity is represented in bold, along with the lowest average number of rules or average number of antecedents.
Table 7 depicts the results for the “Mammographic” classification problem. The highest average predictive accuracies were achieved by the DIMLPs for both ensembles and their generated rulesets. Moreover, the lowest predictive accuracy averages were provided by RF. The reason could be that the RFs were overtrained in this particular case. Indeed, for the average predictive accuracy, the difference from the DIMLPs was greater than five points, with almost eight times more rules generated. Finally, the rules extracted from the decision stumps provided the highest average fidelity (99.9%) and the lowest complexity (5.4 rules with 2.0 antecedents).

Table 7.
Average results obtained on the “Mammographic” dataset. For each column, the highest average accuracy or average fidelity is represented in bold, along with the lowest average number of rules or average number of antecedents.
Table 8 illustrates the results for the “Students-on-Math” prediction problem. The highest average predictive accuracy was provided by the DIMLP ensembles (92.2%). Again, the most complex rulesets were provided by RFs, on average. With decision stumps trained by GB, we obtained the highest average fidelity (100%) with the simplest extracted rulesets (two rules, on average). Finally, it is worth noting that the rulesets with an average predictive accuracy of 91.8% generated from the DIMLPs had 18.5 rules, on average.

Table 8.
Average results obtained on the “Students-on-Math” dataset. For each column, the highest average accuracy or average fidelity is represented in bold, along with the lowest average number of rules or average number of antecedents.
For the “Voting” dataset, the results are illustrated in Table 9. The highest average predictive accuracy for both the model and the rules was obtained by GB (96.6% and 96.2%, respectively).

Table 9.
Average results obtained on the “Voting” dataset. For each column, the highest average accuracy or average fidelity is represented in bold, along with the lowest average number of rules or average number of antecedents.
Table 10 presents a comparison between the best average accuracies of the rulesets produced by two groups of models. In the first group, we had ensembles of DIMLPs or DTs, while the second included Skope-Rules. A Welch t-test was performed to compare the average accuracies obtained by the two groups, with p-values in the last column. Numbers in bold represent significantly better average predictive accuracies. Overall, in seven of the eight classification problems, the average predictive accuracy obtained by the rulesets generated from the approach proposed in this work was significantly better.

Table 10.
Summary of the best average accuracies of rulesets produced by ensembles of DIMLPs or DTs. A comparison with SR (fourth column) was achieved through a Welch t-test; p-values are illustrated in the last column. A bold number represents a significantly better average predictive accuracy.
The average number of rules generated from the most accurate rulesets provided by ensembles of DIMLPs or DTs is presented in Table 11. Again, a Welch t-test was performed to compare our ensembles to SR. It turned out that on only one classification problem, the average number of rules was significantly lower with SR.

Table 11.
Summary of the average number of rules extracted from the most accurate rulesets produced by DIMLPs or DTs. A comparison with SR was carried out with a Welch t-test, with p-values in the last column. A bold number represents a significantly lower number of rules.
4.4. Related Work
We compare our results with those of other state-of-the-art approaches. For the “Australian” classification problem, Table 12 illustrates the results provided by several rule extraction techniques applied to ensembles of DTs (ET-FBT, RF-FBT, AFBT, and InTrees). Moreover, G-REX is a rule extraction method that is applied to ensembles of MLPs. Finally, the last row indicates our results with respect to Gradient-Boosted Trees (GB-DIMLP). The second column describes the evaluation method. Specifically, “10 × RS” signifies that the random sampling of the training and testing sets was applied ten times, while “1 × 10-fold-CV” indicates one repetition of ten-fold cross-validation. Here, the highest average predictive accuracy was given by our rule extraction method. However, a strict comparison of results is difficult because not all evaluation procedures are completely the same.

Table 12.
Other results on the “Australian” dataset. A bold number represents a highest average accuracy of the rules.
Table 13 depicts the results for the “Breast Cancer” classification problem. The first five rows show a number of rule extraction techniques applied to an ensemble of DTs (NH, RuleFit, RF-DHC, RF-SGL, and RF-mSGL). The last row indicates our results obtained with ensembles of RFs transformed into ensembles of DIMLPs. The highest average predictive accuracies were provided by RuleFit and RF-DIMLP (97% and 96.9%, respectively), with fewer rules generated by RF-DIMLP, on average (24.3 versus 38). It is also important to keep in mind that each rule generated by RuleFit, RF-SGL, or RF-mSGL has a coefficient that makes rule interpretability more difficult, which is not the case for RF-DIMLP.

Table 13.
Other results on the “Breast Cancer” dataset. A bold number represents a highest average accuracy of the rules.
We carried out a Welch t-test on the average predictive accuracies provided by RF-DIMLP and RuleFit. The number of samples for the former model was equal to 100, while for the latter, it was ten, corresponding to only one repetition of 10-fold cross-validation. The difference in their average predictive accuracies was not big enough to be statistically significant (p-value ). Then, for the same models, we conducted a Welch t-test with respect to the average number of rules. The difference between the values provided by RF-DIMLP and RuleFit was big enough to be statistically significant (p-value ). Statistical comparisons between random sampling evaluations and cross-validations were not performed, since they were not fully comparable.
The results on the “Ionosphere” classification problem are compared in Table 14. Rulefit gave the highest average predictive accuracy (93%), with the ensembles of DIMLP networks providing slightly lower values (92.3%), but with the lowest number of extracted rules (18.3 versus 25), on average. Note also that the rule extractors that produced rules without multiplicative coefficients were NH, RF-DHC, and G-Rex. They provided average predictive accuracies equal to 89%, 89%, and 91.4%, which were below that yielded by the DIMLPs.

Table 14.
Other results on the “Ionosphere” dataset. A bold number represents a highest average accuracy of the rules.
We performed a Welch t-test for the average predictive accuracies provided by the DIMLP ensembles and RuleFit. The difference in their average predictive accuracies was not big enough to be statistically significant (p-value ). Then, for the same models, we carried out a Welch t-test with respect to the average number of rules. The difference between the values of the DIMLPs and RuleFit was big enough to be statistically significant (p-value ).
For the “Mammographic” dataset, Table 15 depicts some results obtained by other rule extractors from ensembles of DTs. The DIMLP ensembles obtained the highest average accuracy, with that of AFBT being a bit lower (84.6% versus 83.4%). Nevertheless, the evaluation protocol was different. Hence, these results are rather indicative.

Table 15.
Other results on the “Mammographic” dataset. A bold number represents a highest average accuracy of the rules.
4.5. An Example of a Ruleset Generated from the “Divorce” Dataset
Listing 1 illustrates a ruleset generated from the “Divorce” classification problem for gradient-boosted DTs (GB (3, 50)). It was produced during the cross-validation trials. Specifically, the classification problem consisted in determining whether, based on 54 attributes, a divorce had been declared for a married couple. Each attribute corresponds to a sentence with a certain degree of truth on a scale of zero to four, with the maximal value indicating “true”.

Listing 1.
A ruleset generated from the “Divorce” dataset.
Listing 1.
A ruleset generated from the “Divorce” dataset.
| Rule 1: DIVORCE () |
| Rule 2: NO DIVORCE () |
| Rule 3: NO DIVORCE () |
| Rule 4: NO DIVORCE () |
The accuracy of the rules was 100% for both the training set and on the testing set, respectively. In parentheses at the end of each line is given the number of training/testing samples covered. We enumerate the following specified attributes for the extracted ruleset:
- : “My spouse and I have similar ideas about how marriage should be”.
- : “I know my spouse’s basic anxieties”.
- : “We are just starting a discussion before I know what is going on”.
5. Discussion
The rule extraction method proposed in this work can be applied to any decision tree. In this work, we applied it to ensembles of MLPs and DTs. Although agnostic rule extraction algorithms can be applied to any model, we are not aware of any other scholars that have applied a rule extraction method to both ensembles of neural networks and decision trees. The average fidelity during our cross-validation trials was often well above 95%, with the lowest value being 89.3%. In addition, it can be qualitatively observed that, very often, the less complex the propositional rules are, the higher the average fidelity will be.
During cross-validation experiments, the highest average predictive accuracy of the rules was provided four times by GBs, three times by DIMLP ensembles, and once by RFs. To compare the generated rules with those of another rule extraction technique under the same cross-validation settings, we used Skope-Rules. The average predictive accuracy of the rules produced by Skope-Rules was always lower than that obtained by our rule extraction technique. Finally, the results were compared to those obtained in the state of the art, although it was difficult to actually find results from similar evaluation procedures. On three out of four datasets, we obtained rules with similar or higher accuracy, on average. On only the “Ionosphere” classification problem, the average predictive accuracy of our generated rules was a bit lower than that of another rule extraction technique. Nevertheless, the difference was not statistically significant. On this same dataset, on average, we generated a significantly lower number of rules; we thus obtained rulesets with better comprehensibility. Finally, unlike those of many other algorithms, our extracted rules do not have any coefficients that make their interpretation difficult.
Each learning model learns in a different way. Hence, the extracted rules very often present different characteristics in terms of complexity. For instance, in the “Breast Cancer” problem, the highest predictive accuracy of the rules was achieved by RFs with a value equal to 96.9%. That of GB was a bit lower with a value equal to 96.7%, but GB generated more than half as many rules (11.2 versus 24.3), on average. If one is looking for simplicity in explanations, GB could be considered more interesting than RF in this case. A similar example was given with the “Heart Disease” dataset. Here, the highest average predictive accuracy reached by the rules produced by the ensembles of DIMLPs was 84.4%, and the average number of rules was 20.4. With GB (1,50), the same measures were equal to 84.3% and 15.2, respectively. Therefore, GB generated fewer rules with a very similar average predictive accuracy.
For the eight classification problems related to this work, the average predictive accuracy provided by the rules when the models agreed with the extracted rules was the highest with DIMLPs five times, twice with RFs, and once with GBs. These average values were higher than the highest average predictive accuracies provided by the models. An intuitive reason is that test samples for which the extracted rules and the model disagreed are the most difficult samples to classify. Hence, when these more difficult cases are left out (which is the case when we calculate this type of accuracy), the average predictive accuracy increases. By excluding difficult cases, we also lose the ability to explain them, but with fidelity above 95%, the loss of explanation would occur in less than 5% of cases.
A question arising is whether it is possible to find a way to increase the predictive accuracy of the rules and the fidelity. By combining the symbolic rules of five models among SVMs, ensembles of DTs with several parameterizations, and ensembles of DIMLPs, the author obtained 99.9% average fidelity and 87.5% average predictive accuracy for the “Australian” dataset [13]. For the “Breast” classification problem, the average fidelity was 100%, with an average predictive accuracy of 97.4%. Finally, with the “Ionosphere” dataset, an average fidelity of 99.9% and an average predictive accuracy of 94.2% were reached. Hence, the average predictive accuracy and average fidelity improved; however, the number of rules increased by a factor of between five and ten times the number of rules produced by a DIMLP ensemble, making interpretation more complicated.
In the future, we will apply our rule extraction technique to Convolutional Neural Networks (CNNs). Specifically, a CNN includes kernels that calculate distinct feature maps. Since each feature map represents a new version of the original dataset, we could imagine an ensemble of MLPs that learn all of these maps. Very accurate results have been obtained in this respect with support vector machines [40], but without the possibility of interpretation. With the use of DIMLPs, propositional rules will be able to be produced.
6. Conclusions
A rule extraction technique that is normally used for DIMLP neural networks was applied to random forests and gradient-boosted decision trees. This was made possible by considering that a DT represents a special case of a DIMLP. Through cross-validation trials on eight classification problems, the experiments revealed competitive results with respect to the characteristics of the generated rules. To the best of our knowledge, this is one of the rare works showing a rule extraction technique that is applied to ensembles of both DTs and MLPs. In future work, we will aim to increase the predictive accuracy by combining rulesets generated by models with a high diversity.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All datasets were retrieved from https://archive.ics.uci.edu/ml/index.php (accessed on: 14 September 2021).
Conflicts of Interest
The author declares no conflict of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| GDPR | General Data Protection Regulation |
| MLP | Multi-Layer Perceptron |
| IMLP | Interpretable Multi-Layer Perceptron |
| DIMLP | Discretized Interpretable Multi-Layer Perceptron |
| SVM | Support Vector Machine |
| CNN | Convolutional Neural Network |
| DT | Decision Tree |
| RF | Random Forest |
| RF-3 | Random Forest with a tree depth of three |
| GB | Gradient Boosting |
| SR | Skope-Rules |
| NH | Node Harvest |
| 10 × RS | Ten repetitions of Random Sampling |
| 100 × RS | 100 repetitions of Random Sampling |
References
- Andrews, R.; Diederich, J.; Tickle, A.B. Survey and critique of techniques for extracting rules from trained artificial neural networks. Knowl. Based Syst. 1995, 8, 373–389. [Google Scholar] [CrossRef]
- Diederich, J. Rule Extraction from Support Vector Machines; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2008; Volume 80. [Google Scholar] [CrossRef]
- Breiman, L. Bagging predictors. Mach. Learn. 1996, 24, 123–140. [Google Scholar] [CrossRef]
- Freund, Y.; Schapire, R.E. A desicion-theoretic generalization of on-line learning and an application to boosting. In Proceedings of the European Conference on Computational Learning Theory, Barcelona, Spain, 13–15 March 1995; Springer: Berlin/Heidelberg, Germany, 1995; pp. 23–37. [Google Scholar] [CrossRef]
- Brown, G.; Wyatt, J.; Harris, R.; Yao, X. Diversity creation methods: A survey and categorisation. Inf. Fusion 2005, 6, 5–20. [Google Scholar] [CrossRef]
- Bologna, G. Symbolic rule extraction from the DIMLP neural network. In International Workshop on Hybrid Neural Systems; Springer: Berlin/Heidelberg, Germany, 1998; pp. 240–254. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Hastie, T.; Tibshirani, R.; Friedman, J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2009. [Google Scholar]
- Arrieta, A.B.; Díaz-Rodríguez, N.; Del Ser, J.; Bennetot, A.; Tabik, S.; Barbado, A.; García, S.; Gil-López, S.; Molina, D.; Benjamins, R.; et al. Explainable Artificial Intelligence (XAI): Concepts, taxonomies, opportunities and challenges toward responsible AI. Inf. Fusion 2020, 58, 82–115. [Google Scholar] [CrossRef]
- Saito, K.; Nakano, R. Medical diagnostic expert system based on PDP model. In Proceedings of the IEEE 1988 International Conference on Neural Networks, San Diego, CA, USA, 24–27 July 1988; pp. 255–262. [Google Scholar] [CrossRef]
- Bologna, G. A study on rule extraction from several combined neural networks. Int. J. Neural Syst. 2001, 11, 247–255. [Google Scholar] [CrossRef]
- Bologna, G. Is it worth generating rules from neural network ensembles? J. Appl. Log. 2004, 2, 325–348. [Google Scholar] [CrossRef][Green Version]
- Bologna, G.; Hayashi, Y. A comparison study on rule extraction from neural network ensembles, boosted shallow trees, and SVMs. Appl. Comput. Intell. Soft Comput. 2018, 2018, 4084850. [Google Scholar] [CrossRef]
- Bologna, G. Transparent Ensembles for COVID-19 Prognosis. In Proceedings of the International Cross-Domain Conference for Machine Learning and Knowledge Extraction, Online, 17–20 August 2021; Springer: Berlin/Heidelberg, Germany, 2021; pp. 351–364. [Google Scholar] [CrossRef]
- Zhou, Z.H.; Jiang, Y.; Chen, S.F. Extracting symbolic rules from trained neural network ensembles. Artif. Intell. Commun. 2003, 16, 3–16. [Google Scholar]
- Johansson, U. Obtaining Accurate and Comprehensible Data Mining Models: An Evolutionary Approach; Department of Computer and Information Science, Linköping University: Linköping, Sweden, 2007. [Google Scholar]
- Hara, A.; Hayashi, Y. Ensemble neural network rule extraction using Re-RX algorithm. In Proceedings of the 2012 International Joint Conference on Neural Networks(IJCNN), Brisbane, QLD, Australia, 10–15 June 2012; pp. 1–6. [Google Scholar] [CrossRef]
- Hayashi, Y.; Sato, R.; Mitra, S. A new approach to three ensemble neural network rule extraction using recursive-rule extraction algorithm. In Proceedings of the 2013 International Joint Conference on Neural Networks (IJCNN), Dallas, TX, USA, 4–9 August 2013; pp. 1–7. [Google Scholar] [CrossRef]
- Sendi, N.; Abchiche-Mimouni, N.; Zehraoui, F. A new transparent ensemble method based on deep learning. Procedia Comput. Sci. 2019, 159, 271–280. [Google Scholar] [CrossRef]
- Friedman, J.H.; Popescu, B.E. Predictive learning via rule ensembles. Ann. Appl. Stat. 2008, 2, 916–954. [Google Scholar] [CrossRef]
- Meinshausen, N. Node harvest. Ann. Appl. Stat. 2010, 4, 2049–2072. [Google Scholar] [CrossRef]
- Mashayekhi, M.; Gras, R. Rule Extraction from Decision Trees Ensembles: New Algorithms Based on Heuristic Search and Sparse Group Lasso Methods. Int. J. Inf. Technol. Decis. Mak. 2017, 16, 1707–1727. [Google Scholar] [CrossRef]
- Friedman, J.; Hastie, T.; Tibshirani, R. A note on the group Lasso and a sparse group Lasso. arXiv 2010, arXiv:1001.0736. [Google Scholar]
- Sagi, O.; Rokach, L. Explainable decision forest: Transforming a decision forest into an interpretable tree. Inf. Fusion 2020, 61, 124–138. [Google Scholar] [CrossRef]
- Deng, H. Interpreting tree ensembles with intrees. Int. J. Data Sci. Anal. 2019, 7, 277–287. [Google Scholar] [CrossRef]
- Salzberg, S.L. C4.5: Programs for Machine Learning by J. Ross Quinlan. Morgan Kaufmann Publishers, Inc., 1993. Mach. Learn. 1994, 16, 235–240. [Google Scholar] [CrossRef]
- Breiman, L.; Friedman, J.; Stone, C.J.; Olshen, R.A. Classification and Regression Trees; CRC Press: Boca Raton, FL, USA, 1984. [Google Scholar] [CrossRef]
- Schapire, R.E. A brief introduction to boosting. Ijcai 1999, 99, 1401–1406. [Google Scholar]
- Bologna, G.; Pellegrini, C. Constraining the MLP power of expression to facilitate symbolic rule extraction. In Proceedings of the 1998 IEEE International Joint Conference on Neural Networks Proceedings, IEEE World Congress on Computational Intelligence (Cat. No.98CH36227), Anchorage, AK, USA, 4–9 May 1998; Volume 1, pp. 146–151. [Google Scholar] [CrossRef]
- Brayton, R.; Hachtel, G.; Hemachandra, L.; Newton, A.; Sangiovanni-Vincentelli, A. A comparison of logic minimization strategies using ESPRESSO: An APL program package for partitioned logic minimization. In Proceedings of the International Symposium on Circuits and Systems, Rome, Italy, 10–12 May 1982; pp. 42–48. [Google Scholar]
- Lichman, M. UCI Machine Learning Repository; University of California, School of Information and Computer Sciences: Irvine, CA, USA, 2013. [Google Scholar]
- Quinlan, J.R. Simplifying decision trees. Int. J. Man-Mach. Stud. 1987, 27, 221–234. [Google Scholar] [CrossRef]
- Wolberg, W.H.; Mangasarian, O.L. Multisurface method of pattern separation for medical diagnosis applied to breast cytology. Proc. Natl. Acad. Sci. USA 1990, 87, 9193–9196. [Google Scholar] [CrossRef]
- Yöntem, M.K.; Kemal, A.; Ilhan, T.; KILIÇARSLAN, S. Divorce prediction using correlation based feature selection and artificial neural networks. Nevşehir Hacı Bektaş Veli Üniversitesi SBE Dergisi 2019, 9, 259–273. [Google Scholar]
- Sigillito, V.G.; Wing, S.P.; Hutton, L.V.; Baker, K.B. Classification of radar returns from the ionosphere using neural networks. Johns Hopkins APL Tech. Dig. 1989, 10, 262–266. [Google Scholar]
- Elter, M.; Schulz-Wendtland, R.; Wittenberg, T. The prediction of breast cancer biopsy outcomes using two CAD approaches that both emphasize an intelligible decision process. Med. Phys. 2007, 34, 4164–4172. [Google Scholar] [CrossRef]
- Cortez, P.; Silva, A.M.G. Using data mining to predict secondary school student performance. In Proceedings of the 5th Annual Future Business Technology Conference, Porto, Portugal, 9–11 April 2008; pp. 5–12. [Google Scholar]
- Schlimmer, J.C. Concept Acquisition through Representational Adjustment. Ph.D. Thesis, University of California, Irvine, CA, USA, 1987. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Nanni, L.; Ghidoni, S.; Brahnam, S. Handcrafted vs. non-handcrafted features for computer vision classification. Pattern Recognit. 2017, 71, 158–172. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).