Bacteria and Soil Enzymes Supporting the Valorization of Forested Soils
Abstract
:1. Introduction
2. Materials and Methods
2.1. Characteristics of the Study Area
2.2. Chemical and Physicochemical Analyses of Soil
2.3. Microbiological and Enzymatic Analyses of Soil
2.4. Statistical Analysis
- IFT—the index of a tree stand effect;
- AT—the count of microorganisms/the activity of enzymes in the soil from the afforested area;
- A0—the count of microorganisms/the activity of enzymes in the soil from the non-afforested area.
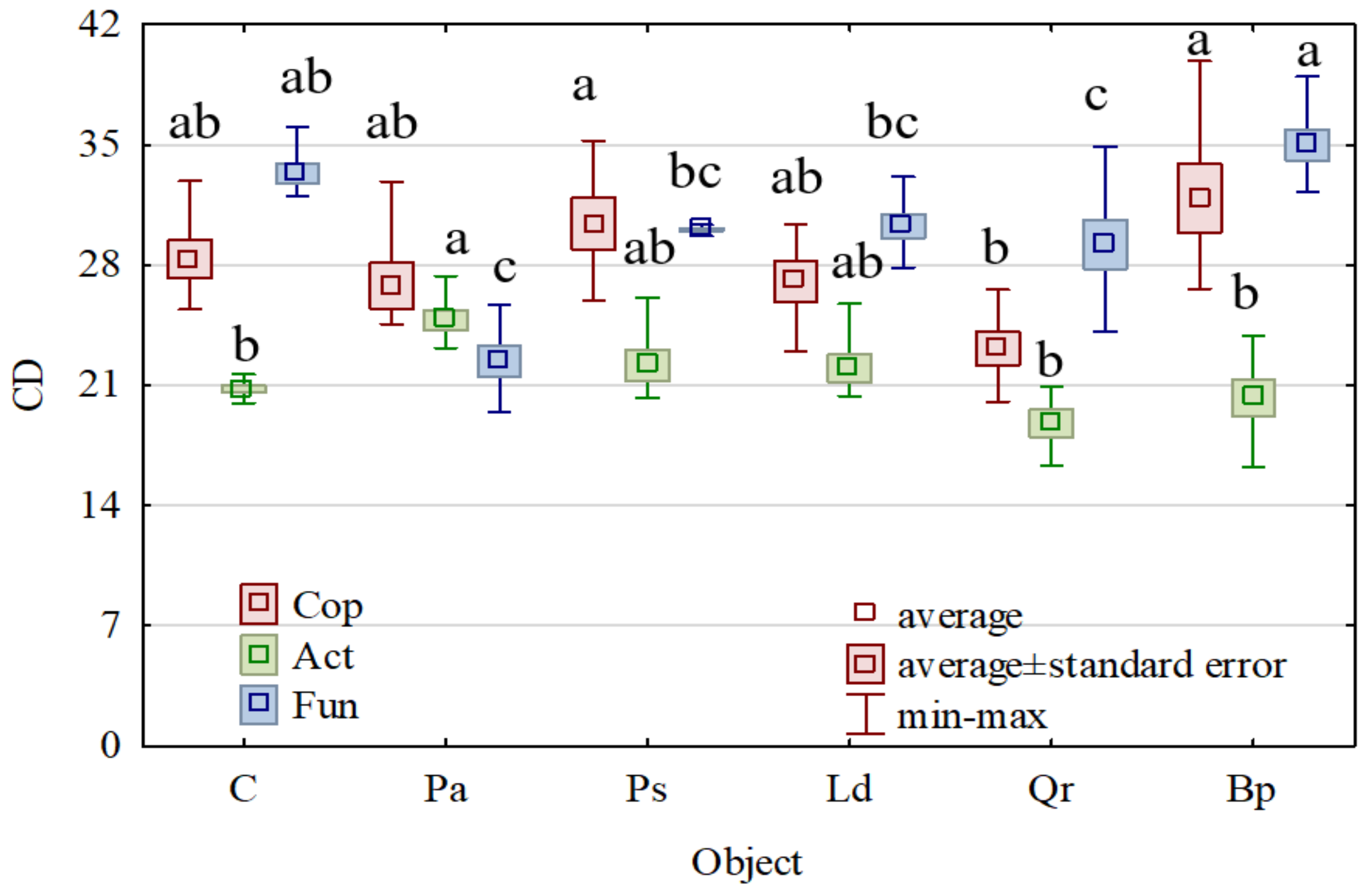
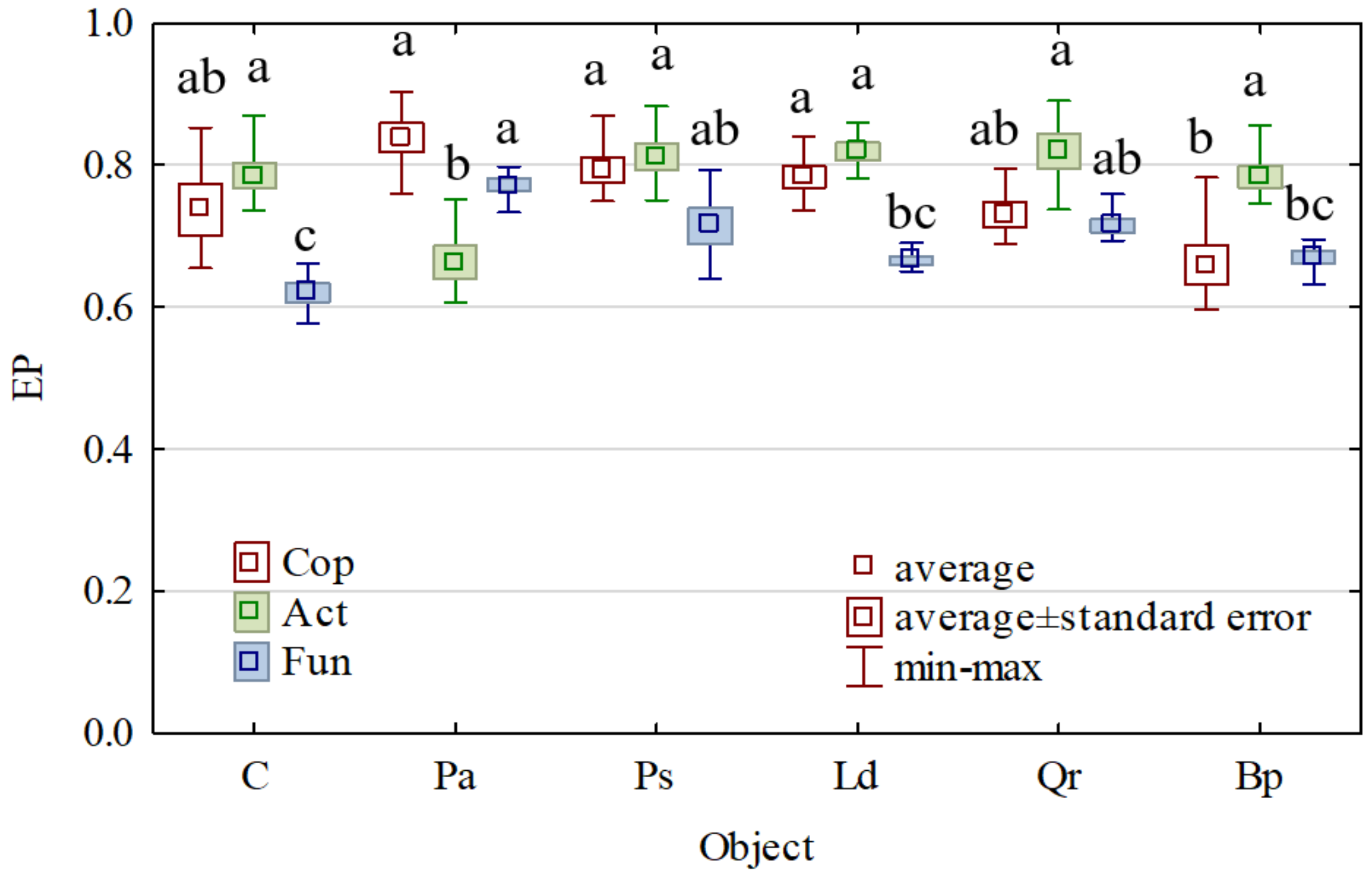
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cukor, J.; Vacek, Z.; Vacek, S.; Linda, R.; Podrázský, V. Biomass productivity, forest stability, carbon balance, and soil transformation of agricultural land afforestation: A case study of suitability of native tree species in the submontane zone in Czechia. CATENA 2022, 210, 105893. [Google Scholar] [CrossRef]
- Smal, H.; Olszewska, M.; Ligęza, S.; Urban, D. The effect of afforestation of post-arable soils with scots pine (Pinus sylvestris L.) on their enzymatic activity. Acta Agrophysica 2017, 24, 509–521. [Google Scholar]
- Mohan, M.; Richardson, G.; Gopan, G.; Aghai, M.M.; Bajaj, S.; Galgamuwa, G.A.P.; Vastaranta, M.; Arachchige, P.S.P.; Amorós, L.; Corte, A.P.D.; et al. UAV-supported forest regeneration: Current trends, challenges and implications. Remote Sens. 2021, 13, 2596. [Google Scholar] [CrossRef]
- Goenster, S.; Gründler, C.; Buerkert, A.; Joergensen, R.G. Soil microbial indicators across land use types in the river oasis Bulgan sum center, Western Mongolia. Ecol. Indic. 2017, 76, 111–118. [Google Scholar] [CrossRef]
- Lacerda-Júnior, G.V.; Noronha, M.F.; Cabral, L.; Delforno, T.P.; de Sousa, S.T.P.; Fernandes-Júnior, P.I.; Melo, I.S.; Oliveira, V.M. Land use and seasonal effects on the soil microbiome of a Brazilian dry forest. Front. Microbiol. 2019, 10, 648. [Google Scholar] [CrossRef] [Green Version]
- Sousa, F.P.; Ferreira, T.; Mendonça, E.; Romero, R.E.; Oliveira, J. Carbon and nitrogen in degraded Brazilian semi-arid soils undergoing desertification. Agric. Ecosyst. Environ. 2012, 148, 11–21. [Google Scholar] [CrossRef]
- Thapa, V.R.; Ghimire, R.; Mikha, M.M.; Idowu, O.J.; Marsalis, M.A. Land use effects on soil health in semiarid drylands. Agric. Environ. Lett. 2018, 3, 180022. [Google Scholar] [CrossRef]
- Meena, A.; Rao, K.S. Assessment of soil microbial and enzyme activity in the rhizosphere zone under different land use/cover of a semiarid region, India. Ecol. Process. 2021, 10, 16. [Google Scholar] [CrossRef]
- Zhao, D.; Li, F.; Yang, Q.; Wang, R.; Song, Y.; Tao, Y. The influence of different types of urban land use on soil microbial biomass and functional diversity in Beijing, China. Soil Use Manag. 2013, 29, 230–239. [Google Scholar] [CrossRef]
- Wang, R.; Cavagnaro, T.R.; Jiang, Y.; Keitel, C.; Dijkstra, F.A. Carbon allocation to the rhizosphere is affected by drought and nitrogen addition. J. Ecol. 2021, 109, 3699–3709. [Google Scholar] [CrossRef]
- Chao, L.; Liu, Y.; Zhang, W.; Wang, Q.; Guan, X.; Yang, Q.; Chen, L.; Zhang, J.; Hu, B.; Liu, Z.; et al. Root Functional Traits, but Not Biomass, Determine the Magnitude of the Rhizosphere Priming Effect among Eight Tree Species; SSRN Scholarly Paper No. ID 3973502; Social Science Research Network: Rochester, NY, USA, 2021. [Google Scholar] [CrossRef]
- Potthoff, M.; Jackson, L.E.; Steenwerth, K.L.; Ramirez, I.; Stromberg, M.R.; Rolston, D.E. Soil biological and chemical properties in restored perennial grassland in California. Restor. Ecol. 2005, 13, 61–73. [Google Scholar] [CrossRef]
- Bhargava, S.; Mitra, S. Elevated atmospheric CO2 and the future of crop plants. Plant Breed. 2021, 140, 1–11. [Google Scholar] [CrossRef]
- Global Climate Change. Vital Signs of the Planet. Available online: https://climate.nasa.gov/vital-signs/carbon-dioxide/ (accessed on 21 February 2021).
- Walkiewicz, A.; Bieganowski, A.; Rafalska, A.; Khalil, M.I.; Osborne, B. Contrasting effects of forest type and stand age on soil microbial activities: An analysis of local scale variability. Biology 2021, 10, 850. [Google Scholar] [CrossRef] [PubMed]
- Dhillion, S.S.; Roy, J.; Abrams, M. Assessing the impact of elevated CO2 on soil microbial activity in a Mediterranean model ecosystem. Plant Soil 1995, 187, 333–342. [Google Scholar] [CrossRef]
- Babur, E.; Dindaroğlu, T. Seasonal changes of soil organic carbon and microbial biomass carbon in different forest ecosystems. In Environmental Factors Affecting Human Health; IntechOpen: London, UK, 2020; pp. 1–22. [Google Scholar] [CrossRef] [Green Version]
- Salazar, S.; Sánchez, L.E.; Alvarez, J.; Valverde, A.; Galindo, P.; Igual, J.M.; Peix, A.; Santa-Regina, I. Correlation among soil enzyme activities under different forest system management practices. Ecol. Eng. 2011, 37, 1123–1131. [Google Scholar] [CrossRef]
- Chakrawal, A.; Herrmann, A.M.; Manzoni, S. Leveraging energy flows to quantify microbial traits in soils. Soil Biol. Biochem. 2021, 155, 108169. [Google Scholar] [CrossRef]
- Luo, P.; Han, X.; Wang, Y.; Han, M.; Shi, H.; Liu, N.; Bai, H. Influence of long-term fertilization on soil microbial biomass, dehydrogenase activity, and bacterial and fungal community structure in a brown soil of northeast China. Ann. Microbiol. 2015, 65, 533–542. [Google Scholar] [CrossRef] [Green Version]
- Drigo, B.; Pijl, A.S.; Duyts, H.; Kielak, A.M.; Gamper, H.A.; Houtekamer, M.J.; Boschker, H.T.S.; Bodelier, P.L.E.; Whiteley, A.S.; van Veen, J.A.; et al. Shifting carbon flow from roots into associated microbial communities in response to elevated atmospheric CO2. Proc. Natl. Acad. Sci. USA 2010, 107, 10938–10942. [Google Scholar] [CrossRef] [Green Version]
- DeAngelis, K.M.; Brodie, E.L.; DeSantis, T.Z.; Andersen, G.L.; Lindow, S.E.; Firestone, M.K. Selective progressive response of soil microbial community to wild oat roots. ISME J. 2009, 3, 168–178. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Wang, Z.; Jiang, P.; He, Y.; Mu, Y.; Lv, X.; Zhuang, L. Bacterial diversity and community structure in the rhizosphere of four Ferula species. Sci. Rep. 2018, 8, 5345. [Google Scholar] [CrossRef] [Green Version]
- Uksa, M.; Buegger, F.; Gschwendtner, S.; Lueders, T.; Kublik, S.; Kautz, T.; Athmann, M.; Köpke, U.; Munch, J.C.; Schloter, M.; et al. Bacteria utilizing plant-derived carbon in the rhizosphere of Triticum aestivum change in different depths of an arable soil. Environ. Microbiol. Rep. 2017, 9, 729–741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kabir, M.E.; Johansen, C.; Bell, R.W. Subsoil rhizosphere modification by chickpea under a dry topsoil: Implications for phosphorus acquisition. J. Plant Nutr. Soil Sci. 2015, 178, 904–913. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.-H.; Crowley, D.E. Rhizosphere microbial community structure in relation to root location and plant iron nutritional status applied and environmental microbiology. Appl. Environ. Microbiol. 2020, 66, 345–351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raiesi, F.; Riahi, M. The influence of grazing exclosure on soil C stocks and dynamics, and ecological indicators in upland arid and semi-arid rangelands. Ecol. Indic. 2014, 41, 145–154. [Google Scholar] [CrossRef]
- Wang, C.; Han, G.; Jia, Y.; Feng, X.; Guo, P.; Tian, X. Response of litter decomposition and related soil enzyme activities to different forms of nitrogen fertilization in a subtropical forest. Ecol. Res. 2011, 26, 505–513. [Google Scholar] [CrossRef]
- Borowik, A.; Wyszkowska, J.; Wyszkowski, M. Resistance of aerobic microorganisms and soil enzyme response to soil contamination with Ekodiesel Ultra fuel. Environ. Sci. Pollut. Res. Int. 2017, 24, 24346–24363. [Google Scholar] [CrossRef] [Green Version]
- Liu, D.; Huang, Y.; An, S.; Sun, H.; Bhople, P.; Chen, Z. Soil physicochemical and microbial characteristics of contrasting land-use types along soil depth gradients. CATENA 2018, 162, 345–353. [Google Scholar] [CrossRef]
- Acosta-Martínez, V.; Mikha, M.M.; Vigil, M.F. Microbial communities and enzyme activities in soils under alternative crop rotations compared to wheat–fallow for the Central Great Plains. Appl. Soil Ecol. 2007, 37, 41–52. [Google Scholar] [CrossRef]
- Pandey, D.; Agrawal, M.; Bohra, J.S. Effects of conventional tillage and no tillage permutations on extracellular soil enzyme activities and microbial biomass under rice cultivation. Soil Tillage Res. 2014, 136, 51–60. [Google Scholar] [CrossRef]
- De Medeiros, K.; de Alcantara, N.; de Barros, J.A.; da Silva Moraes, W.; Silva, A.O.; Moreira, K.A. Absolute and specific enzymatic activities of sandy entisol from tropical dry forest, monoculture and intercropping areas. Soil Tillage Res. 2015, 145, 208–215. [Google Scholar] [CrossRef]
- Solon, J.; Borzyszkowski, J.; Bidłasik, M.; Richling, A.; Badora, K.; Balon, J.; Teresa, B.-W.; Chab, L.; Dobrowolski, R.; Grzegorczyk, I.; et al. Physico-geographical mesoregions of Poland: Verification and adjustment of boundaries on the basis of contemporary spatial data. Geogr. Pol. 2018, 91, 143–170. [Google Scholar] [CrossRef]
- Stan Zdrowotny Lasów W Polsce W 2018 Roku Na Podstawie Badań Monitoringowych. Available online: https://www.bdl.lasy.gov.pl/portal/Media/Default/Publikacje/Stan_zdrowotny_lasow_2018.pdf (accessed on 26 April 2022).
- Nelson, D.W.; Sommers, L.E. Total carbon, organic carbon, and organic matter. In Methods of Soil Analysis; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 1996; pp. 961–1010. [Google Scholar] [CrossRef]
- ISO 11261. Soil Quality—Determination of Total Nitrogen—Modified Kjeldahl Method; International Organization for Standardization: Geneva, Switzerland, 1995. [Google Scholar]
- Egner, H.; Riehm, H.; Domingo, W.R. Untersuchungen über die chemische Bodenanalyse als Grundlage für die Beurteilung des Nährstozustandes der Böden. II. Chemische Extractionsmethoden zur Phospor und Kaliumbestimmung. Ann. R. Agric. Coll. Swed. 1960, 26, 199–215. [Google Scholar]
- Schlichting, E.; Blume, H.P.; Stahr, K. Bodenkundliches Praktikum; Pareys Studientexte 81; Blackwell Wissenschafts-Verlag: Berlin, Germany, 1995. [Google Scholar]
- ISO 11260 Preview. Soil Quality—Determination of Effective Cation Exchange Capacity and Base Saturation Level Using Barium Chloride Solution; International Organization for Standardization: Geneva, Switzerland, 2018. [Google Scholar]
- ISO 10390. Soil Quality—Determination of pH; International Organization for Standardization: Geneva, Switzerland, 2005. [Google Scholar]
- Carter, M.R.; Gregorich, E.G. Soil Sampling and Methods of Analysis, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2008; p. 1224. [Google Scholar]
- De Leij, F.A.A.M.; Whipps, J.M.; Lynch, J.M. The use of colony development for the characterization of bacterial communities in soil and on roots. Microb. Ecol. 1993, 27, 81–97. [Google Scholar] [CrossRef] [PubMed]
- Borowik, A.; Wyszkowska, J.; Kucharski, M.; Kucharski, J. The role of Dactylis glomerata and diesel oil in the formation of microbiome and soil enzyme activity. Sensors 2020, 20, 3362. [Google Scholar] [CrossRef]
- Gomez, E.; Ferreras, L.; Toresani, S. Soil bacterial functional diversity as influenced by organic amendment application. Bioresour. Technol. 2006, 97, 1484–1489. [Google Scholar] [CrossRef]
- Öhlinger, R. Dehydrogenase activity with the substrate TTC. In Methods in Soil Biology; Schinner, F., Öhlinger, R., Kandler, E., Margesin, R., Eds.; Springer: Berlin, Germany, 1996; pp. 241–243. [Google Scholar]
- Johnson, J.L.; Temple, K.L. Some variables affecting the measurement of “catalase activity” in soil. Soil Sci. Soc. Am. J. 1964, 28, 207–209. [Google Scholar] [CrossRef]
- Methods in Applied Soil Microbiology and Biochemistry; Alef, K.; Nannipieri, P. (Eds.) Academic Press: Cambridge, MA, USA, 1998; pp. 316–365. [Google Scholar]
- Tibco Software Inc. Statistica (Data Analysis Software System), version 13; Tibco Software Inc.: Palo Alto, CA, USA, 2017; Available online: https://statistica.io (accessed on 17 November 2021).
- Shapiro, S.S.; Wilk, M.B. An analysis of variance test for normality (complete samples). Biometrika 1965, 52, 591–611. [Google Scholar] [CrossRef]
- Kruskal-Wallis Test. In The Concise Encyclopedia of Statistics; Springer: New York, NY, USA, 2008. [CrossRef]
- Parks, D.H.; Tyson, G.W.; Hugenholtz, P.; Beiko, R.G. STAMP: Statistical analysis of taxonomic and functional profiles. Bioinformatics 2014, 30, 3123–3124. [Google Scholar] [CrossRef] [Green Version]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [Green Version]
- Warnes, G.R.; Bolker, B.; Bonebakker, L.; Gentleman, R.; Huber, W.; Liaw, A.; Lumley, T.; Maechler, M.; Magnusson, M.; Moeller, S.; et al. Various R programming tools for plotting data. R package version 2.17.0. 2020. Available online: https://CRAN.R-project.org/package=gplots (accessed on 23 February 2020).
- Heberle, H.; Meirelles, G.V.; da Silva, F.R.; Telles, G.P.; Minghim, R. InteractiVenn: A web-based tool for the analysis of sets through Venn diagrams. BMC Bioinform. 2015, 16, 169. [Google Scholar] [CrossRef]
- Lladó, S.; López-Mondéjar, R.; Baldrian, P. Drivers of microbial community structure in forest soils. Appl. Microbiol. Biotechnol. 2018, 102, 4331–4338. [Google Scholar] [CrossRef] [PubMed]
- Błońska, E. Enzyme activity in forest peat soils. Folia For. Pol. Ser. A 2010, 52, 20–25. [Google Scholar] [CrossRef]
- Cheng, F.; Peng, X.; Zhao, P.; Yuan, J.; Zhong, C.; Cheng, Y.; Cui, C.; Zhang, S. Soil microbial biomass, basal respiration and enzyme activity of main forest types in the Qinling Mountains. PLoS ONE 2013, 8, e67353. [Google Scholar] [CrossRef] [PubMed]
- Stolnikova, E.V.; Ananyeva, N.D.; Chernova, O.V. The microbial biomass and its activity and structure in the soils of old forests in the European Russia. Eurasian Soil Sci. 2011, 44, 437–452. [Google Scholar] [CrossRef]
- Shannon, V.L.; Vanguelova, E.I.; Morison, J.I.L.; Shaw, L.J.; Clark, J.M. The contribution of deadwood to soil carbon dynamics in contrasting temperate forest ecosystems. Eur. J. For. Res. 2021, 141, 241–252. [Google Scholar] [CrossRef]
- Lu, J.-Z.; Scheu, S. Response of soil microbial communities to mixed beech-conifer forests varies with site conditions. Soil Biol. Biochem. 2021, 155, 108155. [Google Scholar] [CrossRef]
- Fanin, N.; Kardol, P.; Farrell, M.; Nilsson, M.-C.; Gundale, M.J.; Wardle, D.A. The ratio of Gram-positive to Gram-negative bacterial PLFA markers as an indicator of carbon availability in organic soils. Soil Biol. Biochem. 2019, 128, 111–114. [Google Scholar] [CrossRef]
- Thomson, B.C.; Ostle, N.; McNamara, N.; Bailey, M.J.; Whiteley, A.S.; Griffiths, R.I. Vegetation affects the relative abundances of dominant soil bacterial taxa and soil respiration rates in an upland grassland soil. Microb. Ecol. 2010, 59, 335–343. [Google Scholar] [CrossRef]
- Baćmaga, M.; Wyszkowska, J.; Borowik, A.; Kucharski, J.; Paprocki, Ł. Microbiological and biochemical properties in eutric/dystric brunic arenosols, eutric/endocalcaric cambisols, and haplic/albic luvisols soils. J. Soil Sci. Plant Nutr. 2021, 21, 1277–1292. [Google Scholar] [CrossRef]
- Ganault, P.; Nahmani, J.; Hättenschwiler, S.; Gillespie, L.M.; David, J.-F.; Henneron, L.; Iorio, E.; Mazzia, C.; Muys, B.; Pasquet, A.; et al. Relative importance of tree species richness, tree functional type, and microenvironment for soil macrofauna communities in European forests. Oecologia 2021, 196, 455–468. [Google Scholar] [CrossRef]
- Tláskal, V.; Zrůstová, P.; Vrška, T.; Baldrian, P. Bacteria associated with decomposing dead wood in a natural temperate forest. FEMS Microbiol. Ecol. 2017, 93, fix157. [Google Scholar] [CrossRef] [PubMed]
- Gałązka, A.; Marzec-Grządziel, A.; Varsadiya, M.; Niedźwiecki, J.; Gawryjołek, K.; Furtak, K.; Przybyś, M.; Grządziel, J. Biodiversity and metabolic potential of bacteria in bulk soil from the peri-root zone of black alder (Alnus glutinosa), Silver Birch (Betula pendula) and Scots Pine (Pinus sylvestris). Int. J. Mol. Sci. 2022, 23, 2633. [Google Scholar] [CrossRef] [PubMed]
- Norman, J.S.; Barrett, J.E. Substrate availability drives spatial patterns in richness of ammonia-oxidizing bacteria and archaea in temperate forest soils. Soil Biol. Biochem. 2016, 94, 169–172. [Google Scholar] [CrossRef] [Green Version]
- Hansel, C.M.; Fendorf, S.; Jardine, P.M.; Francis, C.A. Changes in bacterial and archaeal community structure and functional diversity along a geochemically variable soil profile. Appl. Environ. Microbiol. 2008, 74, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kielak, A.M.; Barreto, C.C.; Kowalchuk, G.A.; van Veen, J.A.; Kuramae, E.E. The ecology of acidobacteria: Moving beyond genes and genomes. Front. Microbiol. 2016, 7, 744. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fierer, N.; Bradford, M.A.; Jackson, R.B. Toward an ecological classification of soil bacteria. Ecology 2007, 88, 1354–1364. [Google Scholar] [CrossRef]
- Shen, C.; Xiong, J.; Zhang, H.; Feng, Y.; Lin, X.; Li, X.; Liang, W.; Chu, H. Soil pH drives the spatial distribution of bacterial communities along elevation on Changbai Mountain. Soil Biol. Biochem. 2013, 57, 204–211. [Google Scholar] [CrossRef]
- Yun, Y.; Wang, H.; Man, B.; Xiang, X.; Zhou, J.; Qiu, X.; Duan, Y.; Engel, A.S. The relationship between pH and bacterial communities in a single karst ecosystem and its implication for soil acidification. Front. Microbiol. 2016, 7, 1955. [Google Scholar] [CrossRef]
- Baćmaga, M.; Wyszkowska, J.; Borowik, A.; Kucharski, J.; Paprocki, Ł. Role of forest site type in determining bacterial and biochemical properties of soil. Ecol. Indic. 2022, 135, 108557. [Google Scholar] [CrossRef]
- Kim, H.-S.; Lee, S.-H.; Jo, H.Y.; Finneran, K.T.; Kwon, M.J. Diversity and composition of soil Acidobacteria and Proteobacteria communities as a bacterial indicator of past land-use change from forest to farmland. Sci. Total Environ. 2021, 797, 148944. [Google Scholar] [CrossRef]
- Cheng, J.; Zhao, M.; Cong, J.; Qi, Q.; Xiao, Y.; Cong, W.; Deng, Y.; Zhou, J.; Zhang, Y. Soil pH exerts stronger impacts than vegetation type and plant diversity on soil bacterial community composition in subtropical broad-leaved forests. Plant Soil 2020, 450, 273–286. [Google Scholar] [CrossRef]
- Navarrete, A.A.; Tsai, S.M.; Mendes, L.W.; Faust, K.; de Hollander, M.; Cassman, N.A.; Raes, J.; van Veen, J.A.; Kuramae, E.E. Soil microbiome responses to the short-term effects of Amazonian deforestation. Mol. Ecol. 2015, 24, 2433–2448. [Google Scholar] [CrossRef] [PubMed]
- Zhelezova, A.; Chernov, T.; Tkhakakhova, A.; Xenofontova, N.; Semenov, M.; Kutovaya, O. Prokaryotic community shifts during soil formation on sands in the tundra zone. PLoS ONE 2019, 14, e0206777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rughöft, S.; Herrmann, M.; Lazar, C.S.; Cesarz, S.; Levick, S.R.; Trumbore, S.E.; Küsel, K. Community composition and abundance of bacterial, archaeal and nitrifying populations in savanna soils on contrasting bedrock material in Kruger National Park, South Africa. Front. Microbiol. 2016, 7, 1638. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, J.-T.; Hu, H.-W.; Ma, Y.-B.; Zhang, L.-M.; He, J.-Z. Copper pollution decreases the resistance of soil microbial community to subsequent dry-rewetting disturbance. J. Environ. Sci. China 2016, 39, 155–164. [Google Scholar] [CrossRef] [Green Version]
- Lasota, J.; Błońska, E.; Babiak, T.; Piaszczyk, W.; Stępniewska, H.; Jankowiak, R.; Boroń, P.; Lenart-Boroń, A. Effect of charcoal on the properties, enzyme activities and microbial diversity of temperate pine forest soils. Forests 2021, 12, 1488. [Google Scholar] [CrossRef]
- Eaton, W.D.; McGee, K.M.; Larimer, M.; Hoke, E.; Karas, O.; Hernandez, B.; Waylan, N.A. Changes in soil bacterial communities, and carbon and nitrogen metrics as potential indicators of land use effects in a humid tropical forest. Pedobiologia 2021, 85–86, 150730. [Google Scholar] [CrossRef]
- Pratiwi, E.; Satwika, T.; Agus, F. Analysis of peat bacterial diversity in oil palm plantations and a logged forest in Jambi, Indonesia, using PCR-DGGE technique. IOP Conf. Ser. Earth Environ. Sci. 2021, 648, 012200. [Google Scholar] [CrossRef]
- Nacke, H.; Thürmer, A.; Wollherr, A.; Will, C.; Hodac, L.; Herold, N.; Schöning, I.; Schrumpf, M.; Daniel, R. Pyrosequencing-based assessment of bacterial community structure along different management types in German forest and grassland soils. PLoS ONE 2011, 6, e17000. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Wang, S.; Wang, Z.; Zhang, Z.; Qin, H.; Wei, Z.; Feng, K.; Li, S.; Wu, Y.; Yin, H.; et al. Soil microbiome mediated nutrients decline during forest degradation process. Soil Ecol. Lett. 2019, 1, 59–71. [Google Scholar] [CrossRef] [Green Version]
- Błońska, E.; Lasota, J.; Gruba, P. Effect of temperate forest tree species on soil dehydrogenase and urease activities in relation to other properties of soil derived from loess and glaciofluvial sand. Ecol. Res. 2016, 31, 655–664. [Google Scholar] [CrossRef] [Green Version]
- Kaczyńska, G.; Borowik, A.; Wyszkowska, J. Soil dehydrogenases as an indicator of contamination of the environment with petroleum products. Water Air Soil Pollut. 2015, 226, 372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wyszkowska, J.; Borowik, A.; Olszewski, J.; Kucharski, J. Soil bacterial community and soil enzyme activity depending on the cultivation of Triticum aestivum, Brassica napus, and Pisum sativum ssp. arvense. Diversity 2019, 11, 246. [Google Scholar] [CrossRef] [Green Version]
- Silva-Olaya, A.M.; Mora-Motta, D.A.; Cherubin, M.R.; Grados, D.; Somenahally, A.; Ortiz-Morea, F.A. Soil enzyme responses to land use change in the tropical rainforest of the Colombian Amazon region. PLoS ONE 2021, 16, e0255669. [Google Scholar] [CrossRef] [PubMed]
- Hernández, D.; Hobbie, S. The effects of substrate composition, quantity, and diversity on microbial activity. Plant Soil 2010, 335, 397–411. [Google Scholar] [CrossRef]
- Zhan, X.; Wu, W.; Zhou, L.; Liang, J.; Jiang, T. Interactive effect of dissolved organic matter and phenanthrene on soil enzymatic activities. J. Environ. Sci. China 2010, 22, 607–614. [Google Scholar] [CrossRef]
- Brzezińska, M.; Stępniewski, W.; Stępniewska, Z.; Przywara, G.; Włodarczyk, T. Effect of oxygen deficiency on soil dehydrogenase activity in a pot experiment with triticale cv. Jago vegetation. Int. Agrophysics 2001, 15, 145–149. [Google Scholar]
- Andreoni, V.; Cavalca, L.; Rao, M.A.; Nocerino, G.; Bernasconi, S.; Dell’Amico, E.; Colombo, M.; Gianfreda, L. Bacterial communities and enzyme activities of PAHs polluted soils. Chemosphere 2004, 57, 401–412. [Google Scholar] [CrossRef]
- Zaborowska, M.; Wyszkowska, J.; Kucharski, J. Role of Chlorella sp. and rhamnolipid 90 in maintaining homeostasis in soil contaminated with bisphenol A. J. Soils Sediments 2021, 21, 27–41. [Google Scholar] [CrossRef]
- De la Peña, E.; Baeten, L.; Steel, H.; Viaene, N.; De Sutter, N.; De Schrijver, A.; Verheyen, K. Beyond plant–soil feedbacks: Mechanisms driving plant community shifts due to land-use legacies in post-agricultural forests. Funct. Ecol. 2016, 30, 1073–1085. [Google Scholar] [CrossRef] [Green Version]
- Borowik, A.; Wyszkowska, J.; Kucharski, J. Impact of various grass species on soil bacteriobiome. Diversity 2020, 12, 212. [Google Scholar] [CrossRef]
- Dinesh, R.; Chaudhuri, S.; Sheeja, T. Soil biochemical and microbial indices in wet tropical forests: Effects of deforestation and cultivation. J. Plant Nutr. Soil Sci. 2004, 167, 24–32. [Google Scholar] [CrossRef]
- Chen, D.; Wang, X.; Carrión, V.J.; Yin, S.; Yue, Z.; Liao, Y.; Dong, Y.; Li, X. Acidic amelioration of soil amendments improves soil health by impacting rhizosphere microbial assemblies. Soil Biol. Biochem. 2022, 167, 108599. [Google Scholar] [CrossRef]
- Costantini, E.A.C.; Mocali, S. Soil health, soil genetic horizons and biodiversity. J. Plant Nutr. Soil Sci. 2022, 185, 24–34. [Google Scholar] [CrossRef]
- Mocali, S.; Gelsomino, A.; Nannipieri, P.; Pastorelli, R.; Giagnoni, L.; Petrovicova, B.; Renella, G. Short-term resilience of soil microbial communities and functions following severe environmental changes. Agriculture 2022, 12, 268. [Google Scholar] [CrossRef]
 —15,900 seedlings of English oak (Quercus robur L.—Qr) planted on the area of 1.95 ha;
—15,900 seedlings of English oak (Quercus robur L.—Qr) planted on the area of 1.95 ha;  —4500 seedlings of Norway spruce (Picea abies L.—Pa) planted on the area of 0.97 ha;
—4500 seedlings of Norway spruce (Picea abies L.—Pa) planted on the area of 0.97 ha;  —7700 seedlings of Scots pine (Pinus sylvestris L.—Ps) planted on the area of 0.97 ha;
—7700 seedlings of Scots pine (Pinus sylvestris L.—Ps) planted on the area of 0.97 ha;  —700 seedlings of European larch (Larix decidua M.—Ld) planted on the area of 0.43 ha;
—700 seedlings of European larch (Larix decidua M.—Ld) planted on the area of 0.43 ha;  —2400 seedlings of warty birch (Betula pendula L.—Bp) planted on the area of 0.55 ha; and
—2400 seedlings of warty birch (Betula pendula L.—Bp) planted on the area of 0.55 ha; and  —non-afforested land.
—non-afforested land.  —a soil sampling scheme with the use of Egners Riehm’s staff.
—a soil sampling scheme with the use of Egners Riehm’s staff.
 —15,900 seedlings of English oak (Quercus robur L.—Qr) planted on the area of 1.95 ha;
—15,900 seedlings of English oak (Quercus robur L.—Qr) planted on the area of 1.95 ha;  —4500 seedlings of Norway spruce (Picea abies L.—Pa) planted on the area of 0.97 ha;
—4500 seedlings of Norway spruce (Picea abies L.—Pa) planted on the area of 0.97 ha;  —7700 seedlings of Scots pine (Pinus sylvestris L.—Ps) planted on the area of 0.97 ha;
—7700 seedlings of Scots pine (Pinus sylvestris L.—Ps) planted on the area of 0.97 ha;  —700 seedlings of European larch (Larix decidua M.—Ld) planted on the area of 0.43 ha;
—700 seedlings of European larch (Larix decidua M.—Ld) planted on the area of 0.43 ha;  —2400 seedlings of warty birch (Betula pendula L.—Bp) planted on the area of 0.55 ha; and
—2400 seedlings of warty birch (Betula pendula L.—Bp) planted on the area of 0.55 ha; and  —non-afforested land.
—non-afforested land.  —a soil sampling scheme with the use of Egners Riehm’s staff.
—a soil sampling scheme with the use of Egners Riehm’s staff.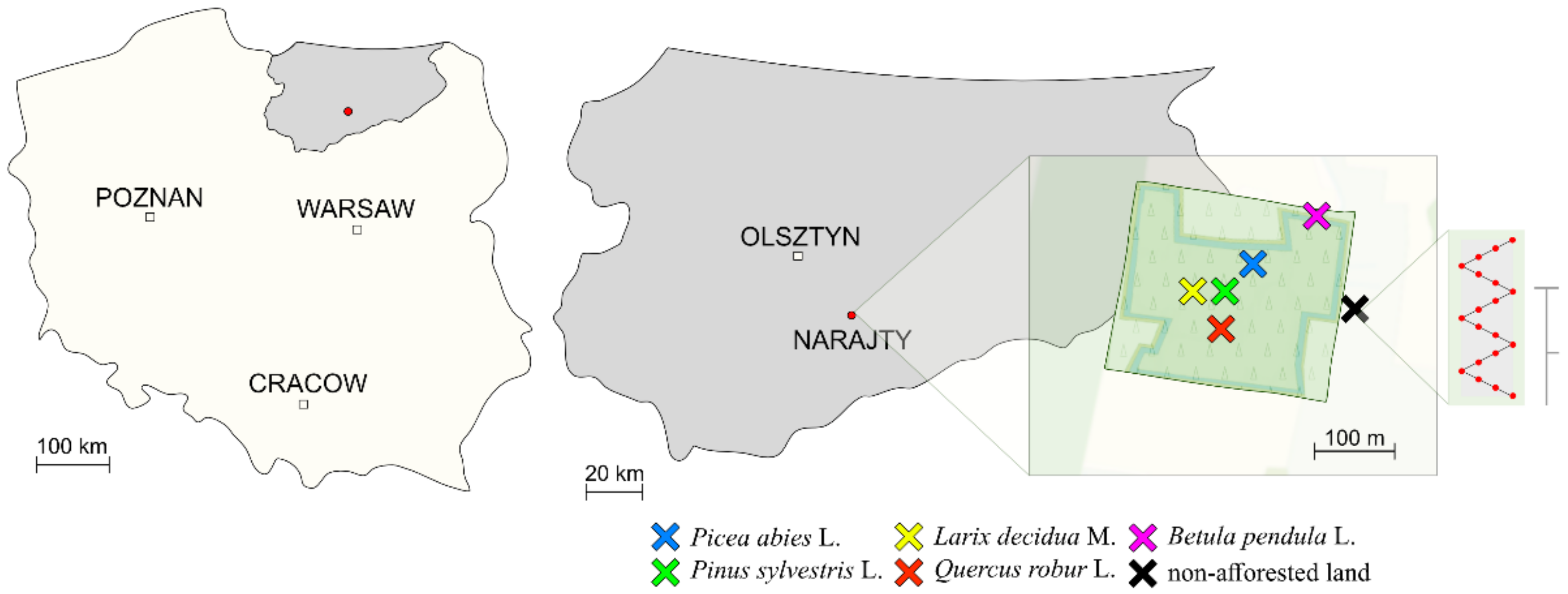
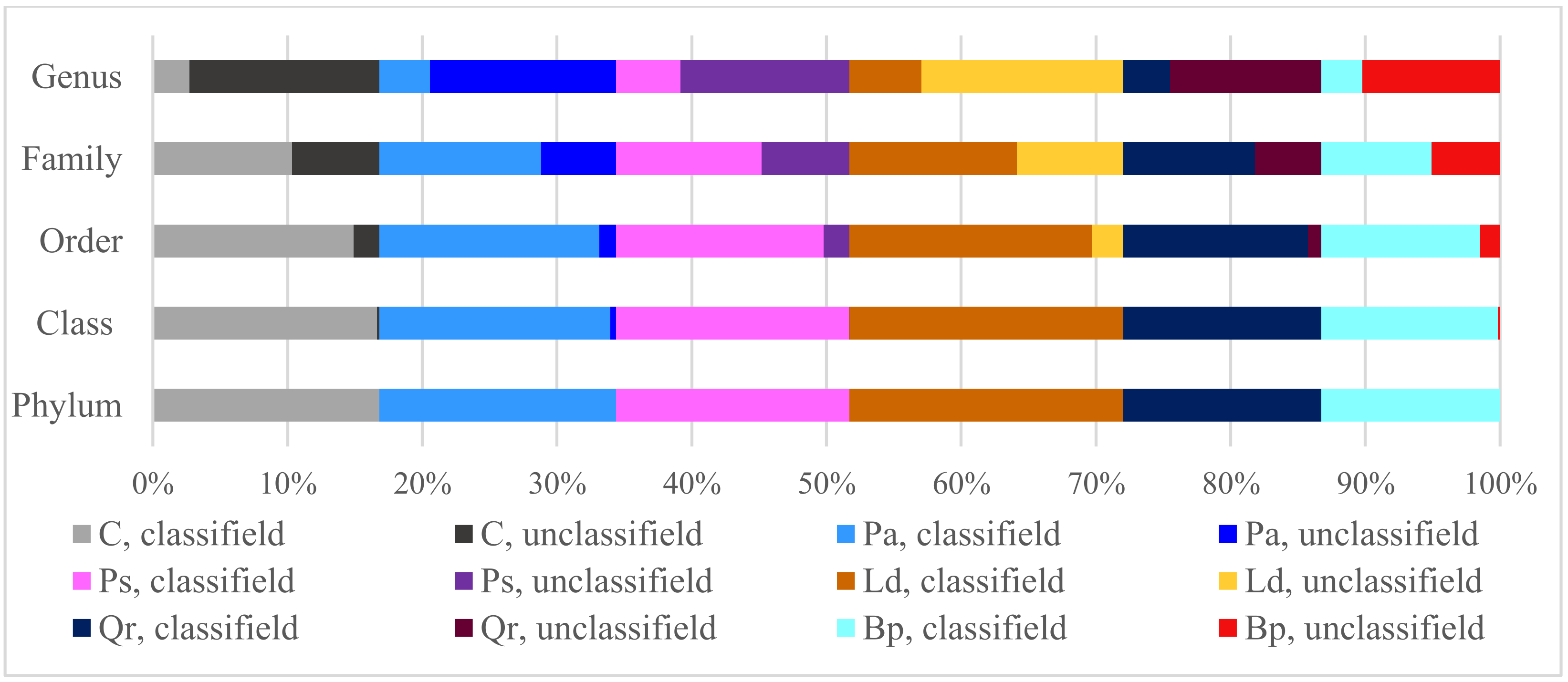

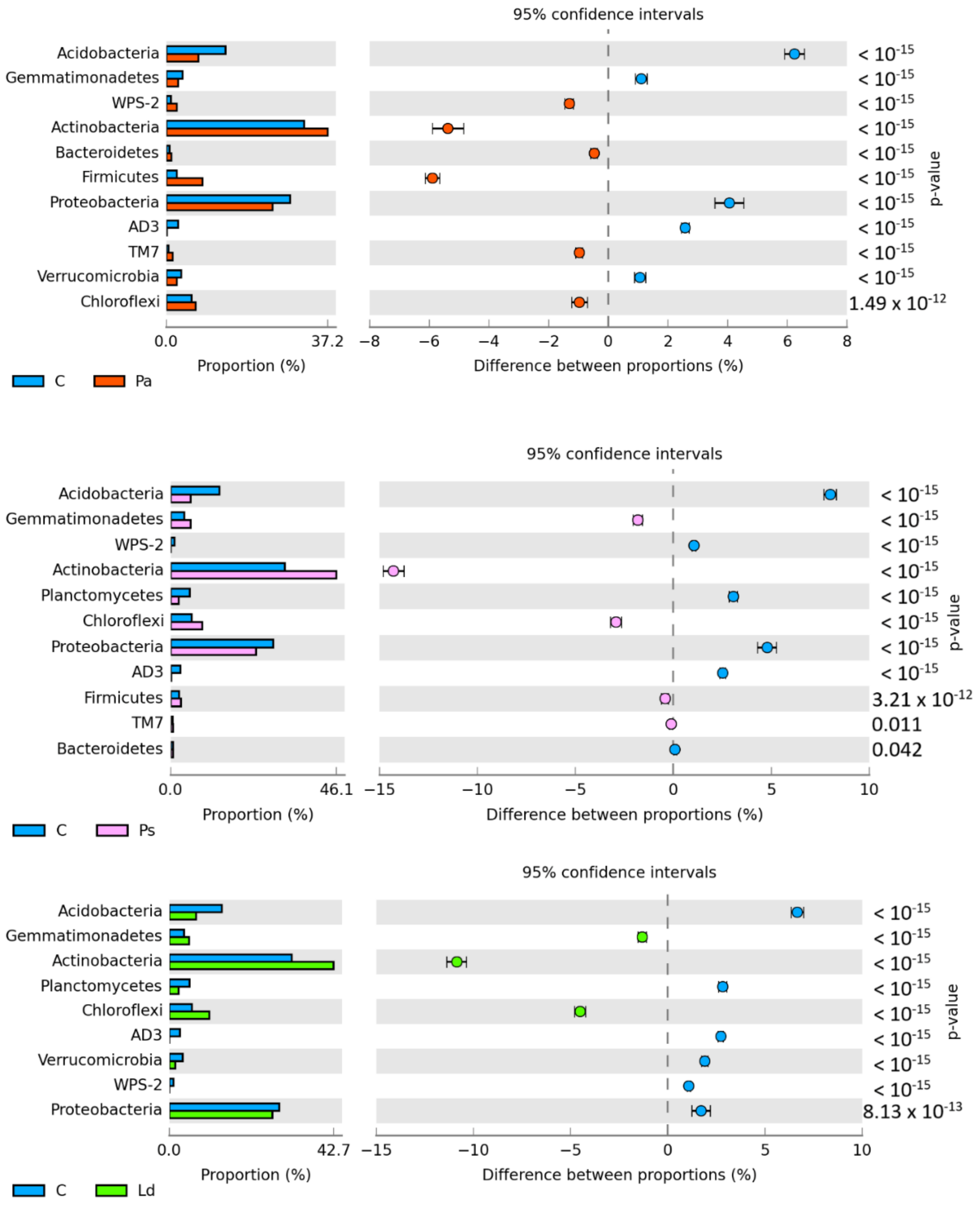
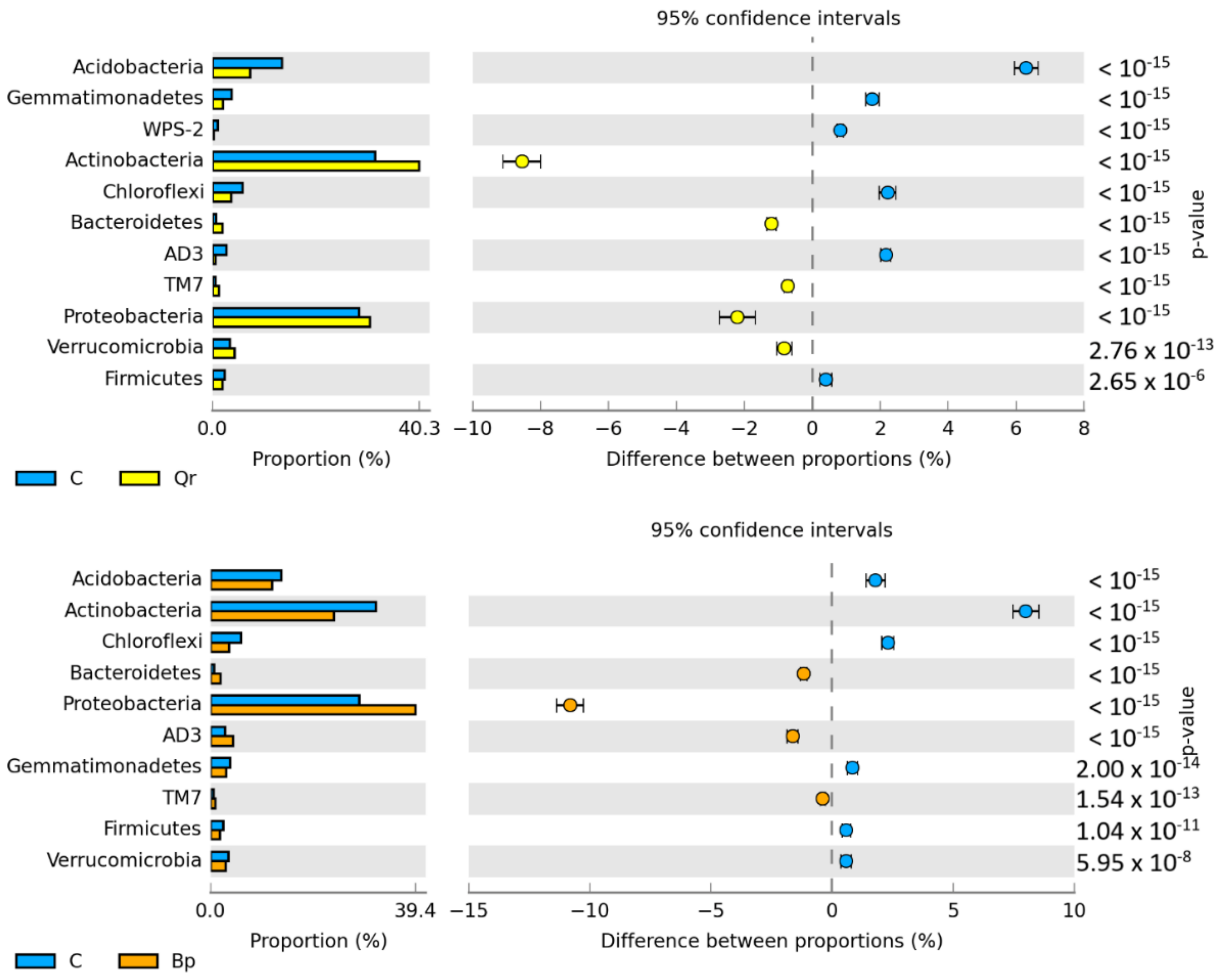
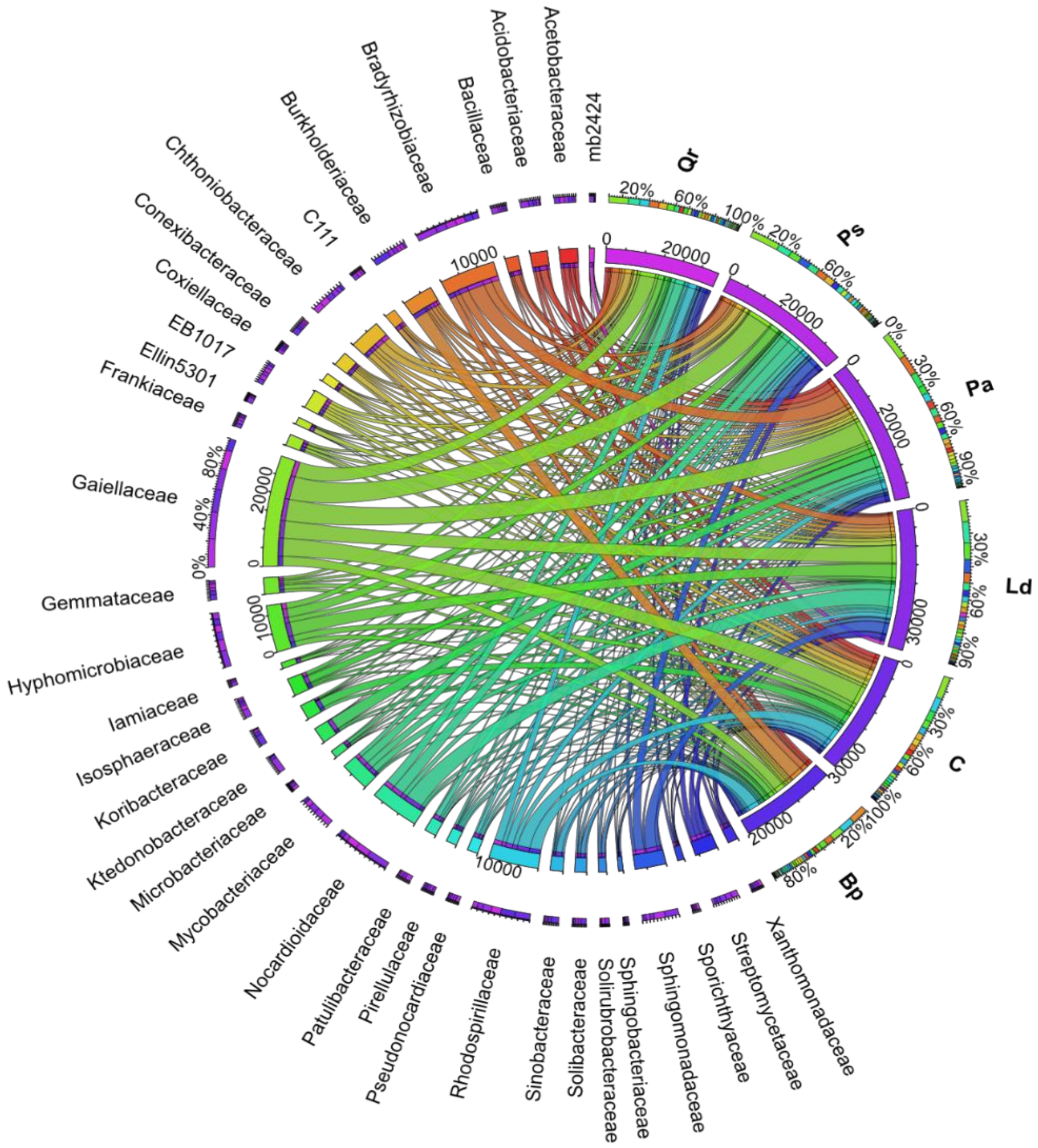
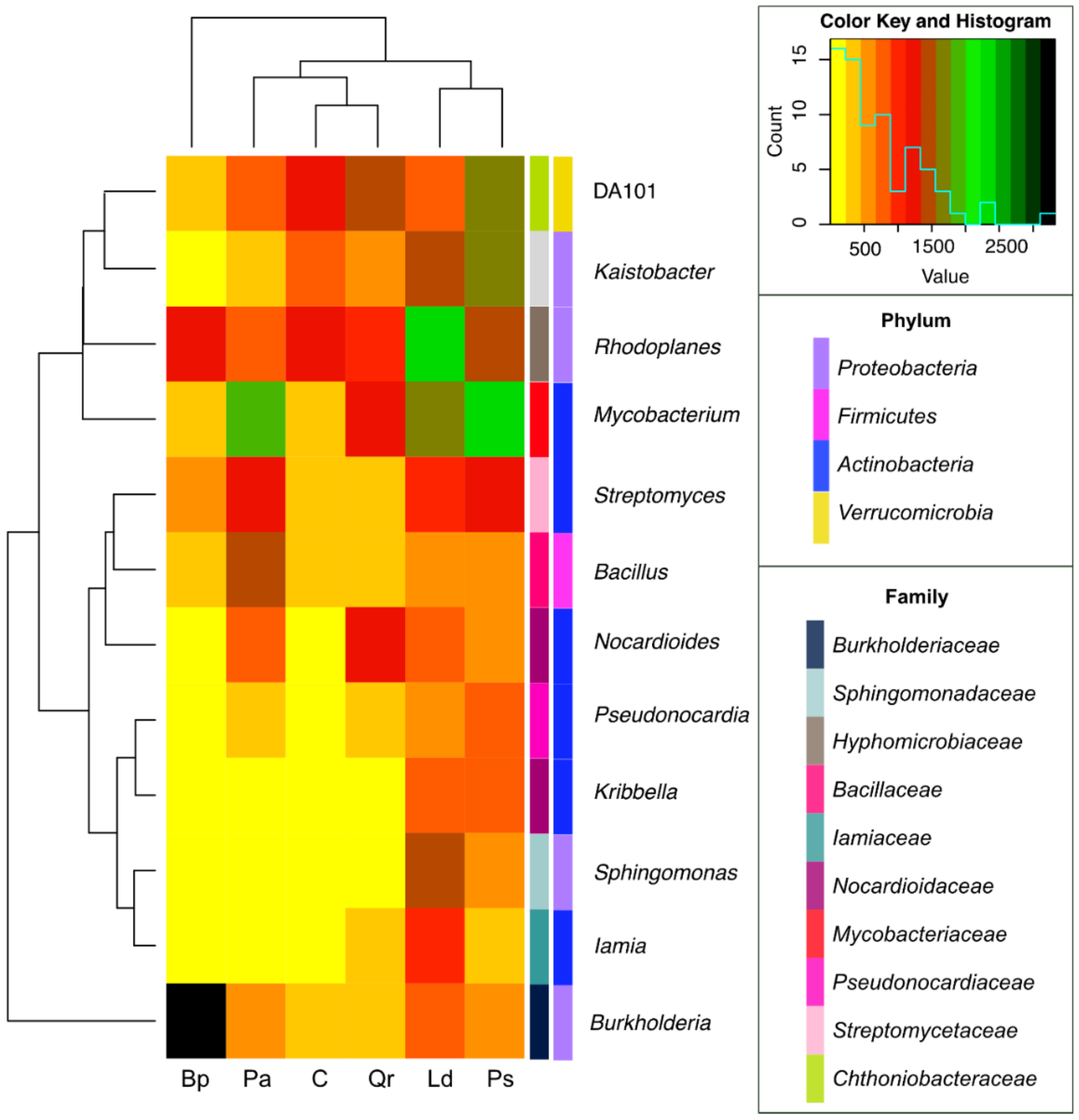
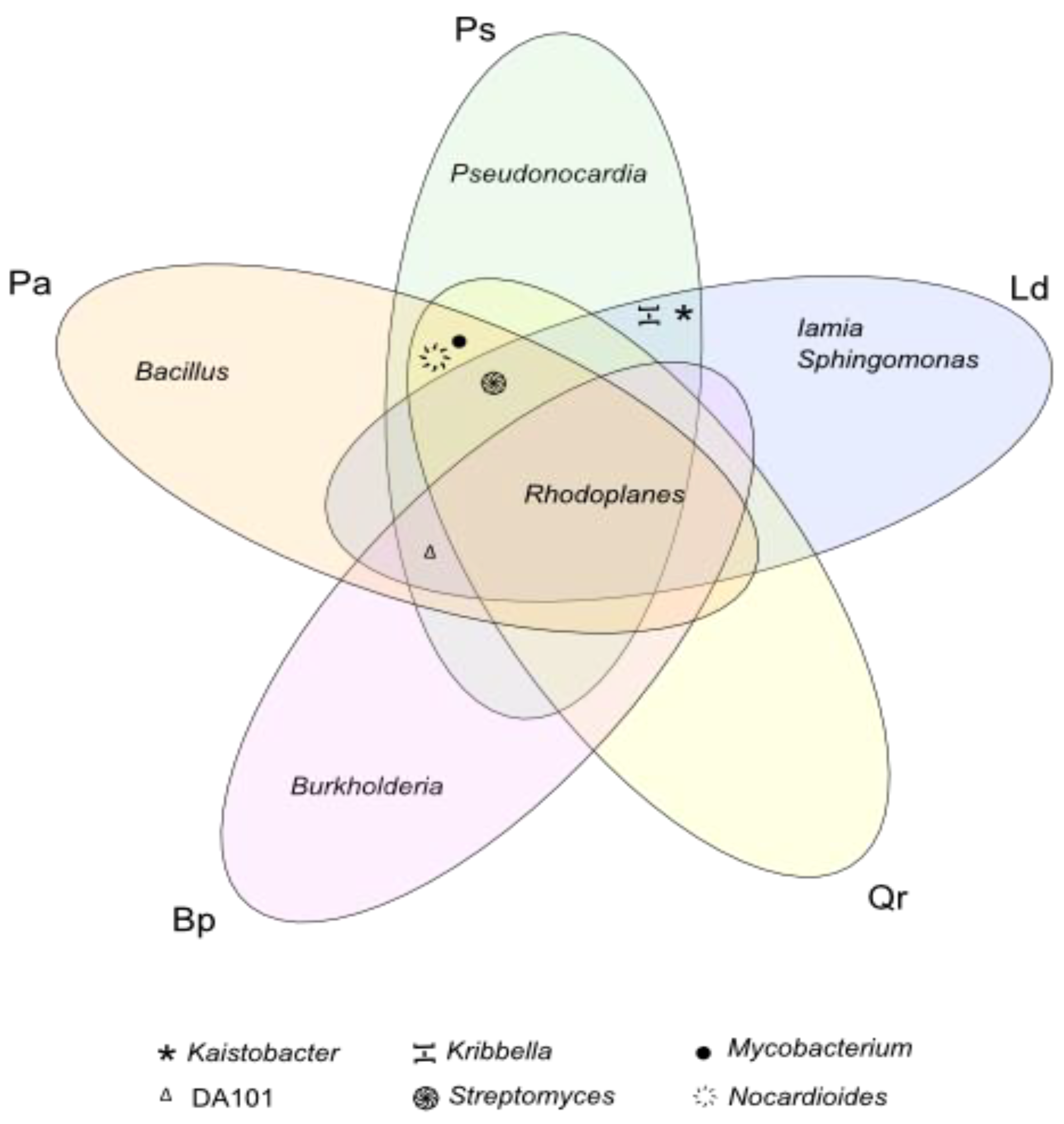
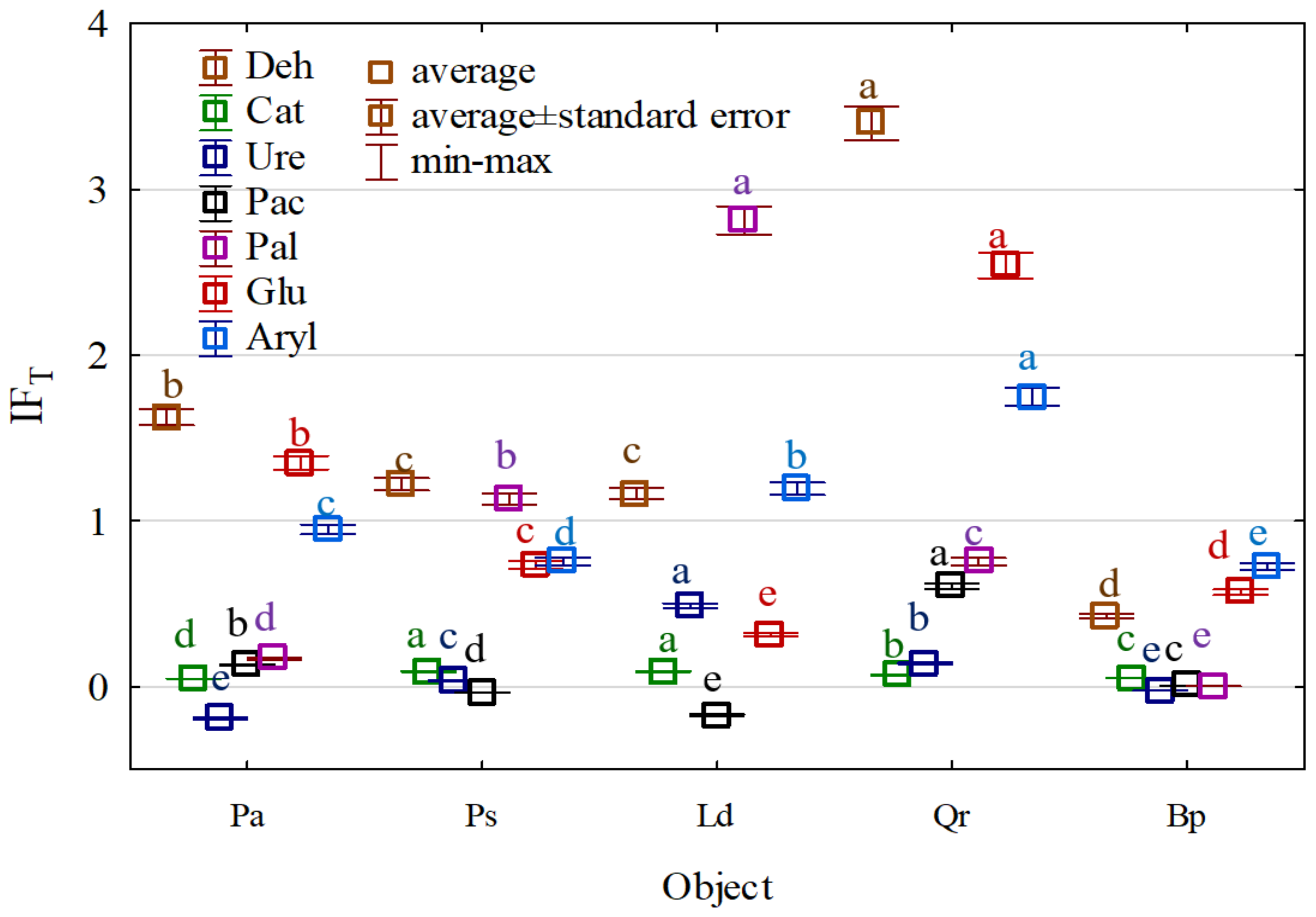
| Taxa | C | Pa | Ps | Ld | Qr | Bp |
|---|---|---|---|---|---|---|
| Phylum | 1.86 a | 1.86 a | 1.62 b | 1.61 b | 1.67 b | 1.82 a |
| Class | 2.72 a | 2.47 b | 2.50 b | 2.50 b | 2.54 b | 2.67 a |
| Order | 2.90 a | 2.70 c | 2.62 c | 2.68 c | 2.78 b | 2.93 a |
| Family | 3.08 b | 3.05 b | 2.99 b | 3.05 b | 3.17 a | 3.02 b |
| Genus | 2.05 d | 2.16 c | 2.32 a | 2.37 a | 2.25 b | 1.68 e |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Borowik, A.; Wyszkowska, J.; Kucharski, J. Bacteria and Soil Enzymes Supporting the Valorization of Forested Soils. Materials 2022, 15, 3287. https://doi.org/10.3390/ma15093287
Borowik A, Wyszkowska J, Kucharski J. Bacteria and Soil Enzymes Supporting the Valorization of Forested Soils. Materials. 2022; 15(9):3287. https://doi.org/10.3390/ma15093287
Chicago/Turabian StyleBorowik, Agata, Jadwiga Wyszkowska, and Jan Kucharski. 2022. "Bacteria and Soil Enzymes Supporting the Valorization of Forested Soils" Materials 15, no. 9: 3287. https://doi.org/10.3390/ma15093287
APA StyleBorowik, A., Wyszkowska, J., & Kucharski, J. (2022). Bacteria and Soil Enzymes Supporting the Valorization of Forested Soils. Materials, 15(9), 3287. https://doi.org/10.3390/ma15093287








