Proteomics Disclose the Potential of Gingival Crevicular Fluid (GCF) as a Source of Biomarkers for Severe Periodontitis
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents, Chemicals, and Solvents
2.2. Patient Selection
2.3. GCF Samples Collection
2.4. GCF Protein Extraction and Quantification
2.5. SDS-PAGE Separation
2.6. LC-MS/MS Analysis
2.7. Data Processing and Statistics
3. Results
3.1. SDS-PAGE
3.2. LC-MS/MS Identifications
3.3. STRING Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| A2M | Alpha-2-macroglobulin |
| C3 | Complement C3 |
| CALR | Calreticulin |
| D-GCF | Diseased-Gingival Crevicular Fluid |
| EIF4G3 | Eukaryotic translation initiation factor 4 gamma 3 |
| ENO1 | Alpha-enolase |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase |
| GC | Vitamin D-binding protein |
| H-GCF | Healthy-Gingival Crevicular Fluid |
| HP | Haptoglobin |
| HPX | Hemopexin |
| IGHA1 | Immunoglobulin heavy constant alpha 1 |
| IGHG1 | Immunoglobulin heavy constant gamma 1 |
| IGHG2 | Immunoglobulin heavy constant gamma 2 |
| IGHV3-74 | Immunoglobulin heavy variable 3-74 |
| KRT1 | Keratin, type II cytoskeletal 1 isoform Iso 1 |
| KRT13 | Keratin, type I cytoskeletal 13 isoform Iso 1 |
| KRT14 | Keratin, type I cytoskeletal 14 isoform Iso 1 |
| KRT16 | Keratin, type I cytoskeletal 16 isoform Iso 1 |
| KRT19 | Keratin, type I cytoskeletal 19 isoform Iso 1 |
| KRT4 | Keratin, type II cytoskeletal 4 isoform Iso 1 |
| KRT6A | Keratin, type II cytoskeletal 6A isoform Iso 1 |
| KRT76 | Keratin, type II cytoskeletal 2 oral isoform Iso 1 |
| LC-MS/MS | Liquid Chromatography/Tandem Mass Spectrometry |
| PBS | Phosphate Buffer Saline |
| PDs | Periodontal Diseases |
| SDS-PAGE | Sodium Dodecyl Sulphate-PolyAcrylamide Gel Electrophoresis |
| SERPINA1 | Alpha-1-antitrypsin |
| SERPINA3 | Alpha-1-antichymotrypsin |
| SERPINB1 | Leukocyte elastase inhibitor |
| SPTLC3 | Serine palmitoyltransferase 3 |
| TMEM201 | Transmembrane protein 201 |
| TSCM | Tooth Surface Collected Material |
References
- Pihlstrom, B.L.; Michalowicz, B.S.; Johnson, N.W. Periodontal diseases. Lancet 2005, 366, 1809–1820. [Google Scholar] [CrossRef]
- Preianò, M.; Savino, R.; Villella, C.; Pelaia, C.; Terracciano, R. Gingival crevicular fluid peptidome profiling in healthy and in periodontal diseases. Int. J. Mol. Sci. 2020, 21, 5270. [Google Scholar] [CrossRef] [PubMed]
- Papapanou, P.N.; Sanz, M.; Buduneli, N.; Dietrich, T.; Feres, M.; Fine, D.H.; Flemmig, T.F.; Garcia, R.; Giannobile, W.V.; Graziani, F.; et al. Periodontitis: Consensus report of workgroup 2 of the 2017 World Workshop on the Classification of Periodontal and Peri-Implant Diseases and Conditions. J. Periodontol. 2018, 89, S173–S182. [Google Scholar] [CrossRef] [PubMed]
- Khurshid, Z.; Khan, E. Future of oral proteomics. J. Oral Res. 2018, 7, 42–43. [Google Scholar] [CrossRef][Green Version]
- Nisha, K.J.; Annie, K.G. Proteomics–The future of periodontal diagnostics. Biomed. J. Sci. Tech. Res. 2017, 1, 1402–1406. [Google Scholar] [CrossRef][Green Version]
- Grover, H.S.; Kapoor, S.; Saksena, N. Periodontal proteomics: Wonders never cease! Int. J. Proteom. 2013, 2013, 850235. [Google Scholar] [CrossRef] [PubMed]
- AlRowis, R.; AlMoharib, H.S.; AlMubarak, A.; Bhaskardoss, J.; Preethanath, R.S.; Anil, S. Oral fluid-based biomarkers in periodontal disease–Part 2. Gingival crevicular fluid. J. Int. Oral Health 2014, 6, 126–135. [Google Scholar]
- Baliban, R.C.; Sakellari, D.; Li, Z.; Guzman, Y.A.; Garcia, B.A.; Floudas, C.A. Discovery of biomarker combinations that predict periodontal health or disease with high accuracy from GCF samples based on high-throughput proteomic analysis and mixed-integer linear optimization. J. Clin. Periodontol. 2013, 40, 131–139. [Google Scholar] [CrossRef]
- Khurshid, Z.; Mali, M.; Naseem, M.; Najeeb, S.; Zafar, M.S. Human gingival crevicular fluids (GCF) proteomics: An overview. Dent. J. 2017, 5, 12. [Google Scholar] [CrossRef]
- Tsuchida, S.; Satoh, M.; Takiwaki, M.; Nomura, F. Current status of proteomic technologies for discovering and identifying gingival crevicular fluid biomarkers for periodontal disease. Int. J. Mol. Sci. 2019, 20, 86. [Google Scholar] [CrossRef]
- Barros, S.P.; Williams, R.; Offenbacher, S.; Morelli, T. Gingival crevicular fluid as a source of biomarkers for periodontitis. Periodontology 2000 2016, 70, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Heboyan, A.; Syed, A.U.Y.; Rokaya, D.; Cooper, P.R.; Manrikyan, M.; Markaryan, M. Cytomorphometric analysis of inflammation dynamics in the periodontium following the use of fixed dental prostheses. Molecules 2020, 25, 4650. [Google Scholar] [CrossRef] [PubMed]
- Papathanasiou, E.; Teles, F.; Griffin, T.; Arguello, E.; Finkelman, M.; Hanley, J.; Theoharides, T.C. Gingival crevicular fluid levels of interferon-g, but not interleukin-4 or -33 or thymic stromal lymphopoietin, are increased in inflamed sites in patients with periodontal disease. J. Periodontol. Res. 2014, 49, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Caton, J.G.; Armitage, G.; Berglundh, T.; Chapple, I.L.C.; Jepsen, S.; Kornman, K.S.; Mealey, B.L.; Papapanou, P.N.; Sanz, M.; Tonetti, M.S. A new classification scheme for periodontal and peri-implant diseases and conditions-Introduction and key changes from the 1999 classification. J. Periodontol. 2018, 89 (Suppl. 1), S1–S8. [Google Scholar] [CrossRef]
- Tonetti, M.S.; Greenwell, H.; Kornman, K.S. Staging and grading of periodontitis: Framework and proposal of a new classification and case definition. J. Periodontol. 2018, 89 (Suppl. 1), S159–S172. [Google Scholar] [CrossRef] [PubMed]
- Carnevale, G.; Cairo, F.; Nieri, M.; Tonetti, M.S. Fibre retention osseous resective surgery: How deep is the infrabony component of the osseous-resected defects? J. Clin. Periodontol. 2008, 35, 133–138. [Google Scholar] [CrossRef] [PubMed]
- Cortellini, P.; Tonetti, M.S. Improved wound stability with a modified minimally invasive surgical technique in the regenerative treatment of isolated interdental intrabony defects. J. Clin. Periodontol. 2009, 36, 157–163. [Google Scholar] [CrossRef]
- Bertoldi, C.; Generali, L.; Cortellini, P.; Lalla, M.; Luppi, S.; Tomasi, A.; Zaffe, D.; Salvatori, R.; Bergamini, S. Influence of Tooth-Brushing on Early Healing after Access Flap Surgery: A Randomized Controlled Preliminary Study. Materials 2021, 14, 2933. [Google Scholar] [CrossRef]
- Duvina, M.; Barbato, L.; Brancato, L.; Rose, G.D.; Amunni, F.; Tonelli, P. Biochemical markers as predictors of bone remodelling in dental disorders: A narrative description of literature. Clin. Cases Miner. Bone Metab. 2012, 9, 100–106. [Google Scholar]
- Bertoldi, C.; Monari, E.; Cortellini, P.; Generali, L.; Lucchi, A.; Spinato, S.; Zaffe, D. Clinical and histological reaction of periodontal tissues to subgingival resin composite restorations. Clin. Oral Investig. 2020, 24, 1001–1011. [Google Scholar] [CrossRef]
- Eickholz, P.; Kaltschmitt, J.; Berbig, J.; Reitmeir, P.; Pretzl, B. Tooth loss after active periodontal therapy. 1: Patient-related factors for risk, prognosis, and quality of outcome. J. Clin. Periodontol. 2008, 35, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Bertoldi, C.; Venuta, M.; Guaraldi, G.; Lalla, M.; Guaitolini, S.; Generali, L.; Monzani, D.; Cortellini, P.; Zaffe, D. Are periodontal outcomes affected by personality patterns? A 18-month follow-up study. Acta Odontol. Scand. 2018, 76, 48–57. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.; Song, T.; Xiao, X.; Liu, Y.; Sun, H.; Guo, Z.; Liu, X.; Shao, C.; Li, Q.; Sun, W. A qualitative and quantitative analysis of the human gingival crevicular fluid proteome and metaproteome. Proteomics 2021, 21, 2000321. [Google Scholar] [CrossRef]
- Mohammed-Salih, H.S.; Saloom, H.F. Collection, storage and protein extraction method of gingival crevicular fluid for proteomic analysis. Baghdad Sci. J. 2021, 19, 368–377. [Google Scholar] [CrossRef]
- Preianò, M.; Maggisano, G.; Murfuni, M.S.; Villella, C.; Pelaia, C.; Montalcini, T.; Lombardo, N.; Pelaia, G.; Savino, R.; Terracciano, R. An analytical method for assessing optimal storage conditions of gingival crevicular fluid and disclosing a peptide biomarker signature of gingivitis by MALDI-TOF MS. Proteom. Clin. Appl. 2018, 12, e1800005. [Google Scholar] [CrossRef]
- Bellei, E.; Cuoghi, A.; Monari, E.; Bergamini, S.; Fantoni, L.I.; Zappaterra, M.; Guerzoni, S.; Bazzocchi, A.; Tomasi, A.; Pini, L.A. Proteomic analysis of urine in medication-overuse headache patients: Possible relation with renal damages. J. Headache Pain 2012, 13, 45–52. [Google Scholar] [CrossRef][Green Version]
- Bellei, E.; Rustichelli, C.; Bergamini, S.; Monari, E.; Baraldi, C.; Lo Castro, F.; Tomasi, A.; Ferrari, A. Proteomic serum profile in menstrual-related and post menopause migraine. J. Pharm. Biomed. Anal. 2020, 184, 113165. [Google Scholar] [CrossRef]
- Ishihama, Y.; Oda, Y.; Tabata, T.; Sato, T.; Nagasu, T.; Rappsilber, J.; Mann, M. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol. Cell. Proteom. 2005, 4, 1265–1272. [Google Scholar] [CrossRef]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef]
- Bertoldi, C.; Bellei, E.; Pellacani, C.; Ferrari, D.; Lucchi, A.; Cuoghi, A.; Bergamini, S.; Cortellini, P.; Tomasi, A.; Zaffe, D.; et al. Non-bacterial protein expression in periodontal pockets by proteome analysis. J. Clin. Periodontol. 2013, 40, 573–582. [Google Scholar] [CrossRef]
- Monari, E.; Cuoghi, A.; Bellei, E.; Bergamini, S.; Lucchi, A.; Tomasi, A.; Cortellini, P.; Zaffe, D.; Bertoldi, C. Analysis of protein expression in periodontal pocket tissue: A preliminary study. Proteome Sci. 2015, 13, 33. [Google Scholar] [CrossRef] [PubMed]
- Bergamini, S.; Bellei, E.; Generali, L.; Tomasi, A.; Bertoldi, C. A proteomic analysis of discolored tooth surfaces after the use of 0.12% Chlorhexidine (CHX) mouthwash and CHX provided with an anti-discoloration system (ADS). Materials 2021, 14, 4338. [Google Scholar] [CrossRef] [PubMed]
- Savage, A.; Eaton, K.A.; Moles, D.R.; Needleman, I. A systematic review of definitions of periodontitis and methods that have been used to identify this disease. J. Clin. Periodontol. 2009, 36, 458–467. [Google Scholar] [CrossRef]
- Giacomelli, L.; Covani, U. Bioinformatics and data mining studies in oral genomics and proteomics: New trends and challenges. Open Dent. J. 2010, 4, 67–71. [Google Scholar] [CrossRef] [PubMed]
- Khurshid, Z.; Zohaib, S.; Najeeb, S.; Zafar, M.S. Proteomics advancements in dentistry. J. Dent. Health Oral Disord. Ther. 2016, 4, 54–55. [Google Scholar] [CrossRef][Green Version]
- Zhu, M.; Zhao, B.; Wei, L.; Wang, S. Alpha-2-macroglobulin, a native and powerful proteinase inhibitor, prevents cartilage degeneration disease by inhibiting majority of catabolic enzymes and cytokines. Curr. Mol. Biol. Rep. 2021, 7, 1–7. [Google Scholar] [CrossRef]
- Gavish, H.; Bab, I.; Tartakovsky, A.; Chorev, M.; Mansur, N.; Greenberg, Z.; Namdar-Attar, M.; Muhlrad, A. Human-2-macroglobulin is an osteogenic growth peptide-binding protein. Biochemistry 1997, 36, 14883–14888. [Google Scholar] [CrossRef]
- Choi, Y.J.; Kim, S.; Choi, Y.; Nielsen, T.B.; Yan, J.; Lu, A.; Ruan, J.; Lee, H.; Wu, H.; Spellberg, B.; et al. SERPINB1-mediated checkpoint of inflammatory caspase activation. Nat. Immunol. 2019, 20, 276–287. [Google Scholar] [CrossRef]
- Choi, Y.J.; Heo, S.H.; Lee, J.M.; Cho, J.Y. Identification of azurocidin as a potential periodontitis biomarker by a proteomic analysis of gingival crevicular fluid. Proteome Sci. 2011, 9, 42. [Google Scholar] [CrossRef]
- Silva-Boghossian, C.M.; Colombo, A.P.V.; Tanaka, M.; Rayo, C.; Xiao, Y.; Siqueira, W.L. Quantitative proteomic analysis of gingival crevicular fluid in different periodontal conditions. PLoS ONE 2013, 8, e75898. [Google Scholar] [CrossRef]
- Kalsheker, N.; Morgan, K.; Chappell, S. Proteinase inhibitors/Antichymotrypsin. In Encyclopedia of Respiratory Medicine; Elsevier BV: Amsterdam, The Netherlands, 2006; pp. 507–511. [Google Scholar] [CrossRef]
- Eaton, J.W.; Brandt, P.; Mahoney, J.R.; Lee, J.T., Jr. Haptoglobin: A natural bacteriostat. Science 1982, 215, 691–693. [Google Scholar] [CrossRef] [PubMed]
- Tsuchida, S.; Satoh, M.; Umemura, H.; Sogawa, K.; Kawashima, Y.; Kado, S.; Sawai, S.; Nishimura, M.; Kodera, Y.; Matsushita, K.; et al. Proteomic analysis of gingival crevicular fluid for discovery of novel periodontal disease markers. Proteomics 2012, 12, 2190–2202. [Google Scholar] [CrossRef] [PubMed]
- Bostanci, N.; Heywood, W.; Mills, K.; Parkar, M.; Nibali, L.; Donos, N. Application of label-free absolute quantitative proteomics in human gingival crevicular fluid by LC/MSE (gingival exudatome). J. Protoeme Res. 2010, 9, 2191–2199. [Google Scholar] [CrossRef] [PubMed]
- Tolosano, E.; Altruda, F. Hemopexin: Structure, function, and regulation. DNA Cell. Biol. 2002, 21, 297–306. [Google Scholar] [CrossRef]
- Karnaukhova, E.; Owczarek, C.; Schmidt, P.; Schaer, D.J.; Buehler, P.W. Human plasma and recombinant hemopexins: Heme binding revisited. Int. J. Mol. Sci. 2021, 22, 1199. [Google Scholar] [CrossRef]
- D’Aiuto, F.; Nibali, L.; Parkar, M.; Patel, K.; Suvan, J.; Donos, N. Oxidative stress, systemic inflammation, and severe periodontitis. J. Dent. Res. 2010, 89, 1241–1246. [Google Scholar] [CrossRef]
- Yucel-Lindberg, T.; Bage, T. Inflammatory mediators in the pathogenesis of periodontitis. Expert Rev. Mol. Med. 2013, 15, e7. [Google Scholar] [CrossRef]
- Baliban, R.C.; Sakellari, D.; Li, Z.; DiMaggio, P.A.; Garcia, B.A.; Floudas, C.A. Novel protein identification methods for biomarker discovery via a proteomic analysis of periodontally healthy and diseased gingival crevicular fluid samples. J. Clin. Periodontol. 2012, 39, 203–212. [Google Scholar] [CrossRef]
- Nauseef, W.M.; McCormick, S.J.; Clark, R.A. Calreticulin functions as a molecular chaperone in the biosynthesis of myeloperoxidase. J. Biol. Chem. 1995, 270, 4741–4747. [Google Scholar] [CrossRef]
- Mackenzie, I.C.; Gao, Z. Patterns of cytokeratin expression in the epithelia of inflamed human gingiva and periodontal pockets. J. Periodontal Res. 1993, 28, 49–59. [Google Scholar] [CrossRef]
- Bertoldi, C.; Bergamini, S.; Ferrari, M.; Lalla, M.; Bellei, E.; Spinato, S.; Tomasi, A.; Monari, E. Comparative proteomic analysis between the gingival crevicular fluid and the corresponding periodontal pocket: A preliminary study. J. Biol. Regul. Homeost. Agents 2019, 33, 983–986. [Google Scholar] [PubMed]
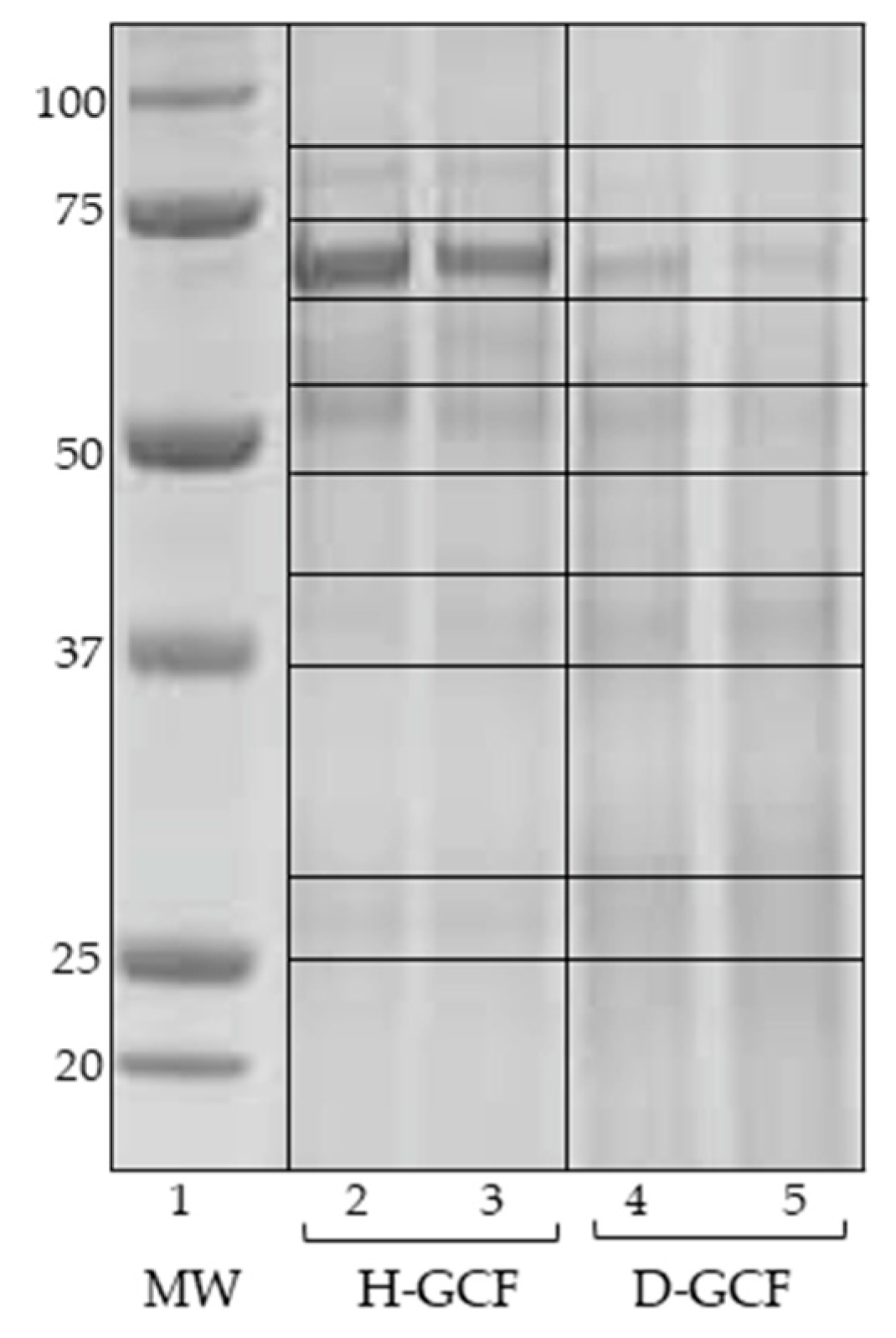
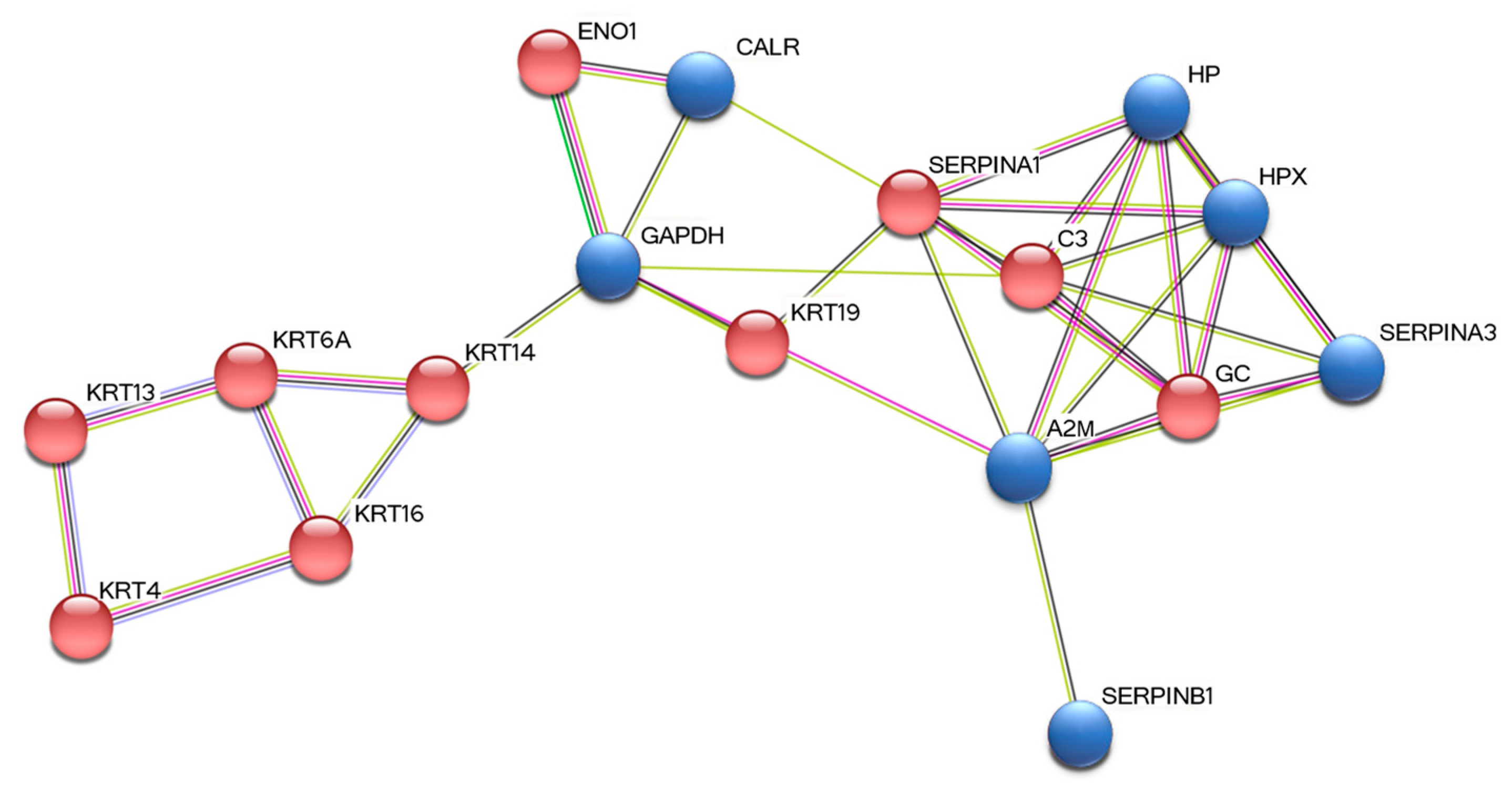
| Protein Full Name a | Gene b | Acc. Number c | Score d | Mass e | emPAI f | Change g |
|---|---|---|---|---|---|---|
| Up-regulated proteins in D-GCF | ||||||
| Alpha-2-macroglobulin (fragment) | A2M | NX_P01023-1 | 18 | 164,613 | 0.05 | +3.00 |
| Leukocyte elastase inhibitor | SERPINB1 | NX_30740-1 | 111 | 42,829 | 0.56 | +1.75 |
| Alpha-1-antichymotrypsin | SERPINA3 | NX_01011-1 | 49 | 47,792 | 0.37 | +1.94 |
| Haptoglobin | HP | NX_P00738-1 | 42 | 45,861 | 0.09 | +2.66 |
| Hemopexin | HPX | NX_P02790-1 | 36 | 52,385 | 0.15 | +1.86 |
| Immunoglobulin heavy constant gamma 1 | IGHG1 | NX_P01857-1 | 143 | 36,596 | 0.86 | +2.33 |
| Immunoglobulin heavy constant gamma 2 | IGHG2 | NX_P01859-1 | 44 | 36,505 | 0.23 | +2.29 |
| Immunoglobulin heavy constant alpha 1 | IGHA1 | NX_P01876-1 | 15 | 38,486 | 0.10 | +1.83 |
| Immunoglobulin heavy variable 3-74 | IGHV3-74 | NX_A0A0B4J1X5-1 | 16 | 13,002 | 0.76 | +1.67 |
| Glyceraldehyde-3-phosphate dehydrogenase | GAPDH | NX_P044406-1 | 46 | 36,201 | 0.37 | +2.00 |
| Calreticulin | CALR | NX_P27797-1 | 21 | 48,283 | 0.08 | +1.61 |
| Eukaryotic translation initiation factor 4 gamma 3 (fragment) | EIF4G3 | NX_O43432-1 | 24 | 177,682 | 0.02 | +1.80 |
| Transmembrane protein 201 | TMEM201 | NX_Q5SNT2-1 | 14 | 73,444 | 0.05 | +1.80 |
| Serine palmitoyltransferase 3 | SPTLC3 | NX_Q9NUV7-1 | 26 | 62,352 | 0.06 | +1.75 |
| Down-regulated proteins in D-GCF | ||||||
| Keratin, type I cytoskeletal 13 isoform Iso 1 | KRT13 | NX_P13646-1 | 580 | 49,900 | 3.59 | 0.60 |
| Keratin, type I cytoskeletal 14 isoform Iso 1 | KRT14 | NX_P02533-1 | 713 | 51,872 | 4.41 | 0.60 |
| Keratin, type I cytoskeletal 16 isoform Iso 1 | KRT16 | NX_P08779-1 | 598 | 51,578 | 4.06 | 0.60 |
| Keratin, type I cytoskeletal 19 isoform Iso 1 | KRT19 | NX_P08727-1 | 87 | 44,079 | 0.54 | 0.80 |
| Keratin, type II cytoskeletal 1 isoform Iso 1 | KRT1 | NX_P04264-1 | 130 | 66,170 | 0.59 | 0.67 |
| Keratin, type II cytoskeletal 4 isoform Iso 1 | KRT4 | NX_P19013-1 | 53 | 57,649 | 0.30 | 0.80 |
| Keratin, type II cytoskeletal 2 oral isoform Iso 1 | KRT76 | NX_Q01546-1 | 150 | 66,370 | 0.78 | 0.43 |
| Keratin, type II cytoskeletal 6A isoform Iso 1 | KRT6A | NX_P02538-1 | 40 | 60,293 | 0.37 | 0.33 |
| Alpha-enolase | ENO1 | NX_P06733-1 | 108 | 47,481 | 0.62 | 0.80 |
| Vitamin D-binding protein | GC | NX_P02774-1 | 28 | 54,480 | 0.15 | 0.66 |
| Complement C3 (fragment) | C3 | NX_P01024-1 | 205 | 188,569 | 0.30 | 0.55 |
| Alpha-1-antitrypsin | SERPINA1 | NX_P01009-1 | 76 | 46,878 | 0.50 | 0.50 |
| Prot. Abbr. a | Principal Functions b | Biological Process c |
|---|---|---|
| Up-regulated proteins | ||
| A2M | Calcium-dependent protein binding; endopeptidase inhibitor activity | Acute-phase response |
| SERPINB1 | Protease inhibitor; regulation of immune response and inflammation | Inflammatory and immune response |
| SERPINA3 | Protease inhibitor; acute-phase response | Inflammatory response |
| HP | Acute inflammatory response and defense response; antibacterial activity | Acute-phase response modulation |
| HPX | Acute-phase reactant; intracellular antioxidant; iron homeostasis | Acute-phase response |
| IGHG1 | Antigen binding; immunoglobulin receptor binding | Immune response |
| IGHG2 | Antigen binding; immunoglobulin receptor binding | Immune response |
| IGHA1 | Antigen binding; antibacterial humoral response | Immune response |
| IGHV3-74 | Participates in the antigen recognition and binding; immune response | Adaptive immunity |
| GAPDH (*,§) | Key enzyme in glycolysis; also plays a role in innate immunity | Energy metabolism |
| CALR | Calcium-binding chaperone; positive regulation of cell cycle | Protein folding |
| EIF4G3 | Regulation of translational initiation; RNA binding | Protein biosynthesis |
| TMEM201 | Actin filament and lamin binding; nuclear envelope organization | Transmembrane protein |
| SPTLC3 | Acyltransferase; sphingolipid pathway; lipid metabolism | Biosynthetic processes |
| Down-regulated proteins | ||
| KRT13 (*) | Structural molecule activity | Cytoskeleton organization |
| KRT14 | Keratin filament binding; structural constituent of cytoskeleton | Epidermis development |
| KRT16 | Epidermis-specific type I keratin; structural constituent of cytoskeleton | Cornification, keratinization |
| KRT19 (*) | Organization of myofibers; structural constituent of cytoskeleton | Cornification, keratinization |
| KRT1 | Structural constituent | Fibrinolysis, keratinization |
| KRT4 (*) | Epithelial cell differentiation | Cornification, keratinization |
| KRT76 | Contributes to terminal cornification | Keratinization |
| KRT6A | Structural constituent of cytoskeleton | Cornification |
| ENO1 (#,*) | Glycolytic enzyme; also involved in hypoxia tolerance | Glycolytic process |
| GC | Vitamin D binding, transport, and storage | Vitamin transport |
| C3 | Central role in complement system activation | Complement activation |
| SERPINA1 | Inhibitor of serine proteases and elastase | Enzymatic inhibitor |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bellei, E.; Bertoldi, C.; Monari, E.; Bergamini, S. Proteomics Disclose the Potential of Gingival Crevicular Fluid (GCF) as a Source of Biomarkers for Severe Periodontitis. Materials 2022, 15, 2161. https://doi.org/10.3390/ma15062161
Bellei E, Bertoldi C, Monari E, Bergamini S. Proteomics Disclose the Potential of Gingival Crevicular Fluid (GCF) as a Source of Biomarkers for Severe Periodontitis. Materials. 2022; 15(6):2161. https://doi.org/10.3390/ma15062161
Chicago/Turabian StyleBellei, Elisa, Carlo Bertoldi, Emanuela Monari, and Stefania Bergamini. 2022. "Proteomics Disclose the Potential of Gingival Crevicular Fluid (GCF) as a Source of Biomarkers for Severe Periodontitis" Materials 15, no. 6: 2161. https://doi.org/10.3390/ma15062161
APA StyleBellei, E., Bertoldi, C., Monari, E., & Bergamini, S. (2022). Proteomics Disclose the Potential of Gingival Crevicular Fluid (GCF) as a Source of Biomarkers for Severe Periodontitis. Materials, 15(6), 2161. https://doi.org/10.3390/ma15062161









