Drug Sensitivity Testing in Osteosarcoma: A Case Report
Abstract
1. Introduction
2. Materials and Methods
Drug Sensitivity Testing
3. Results
3.1. Case Presentation
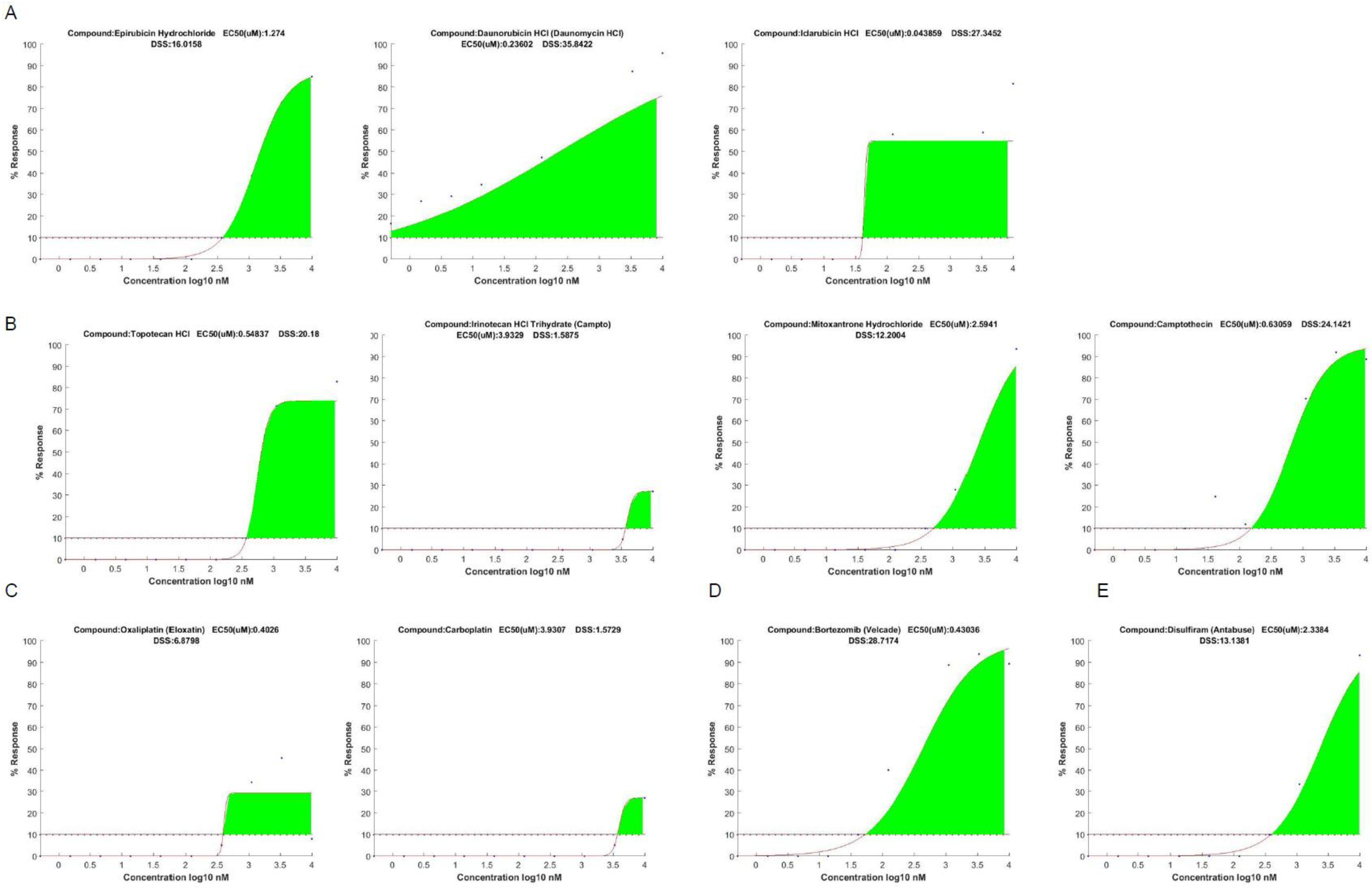
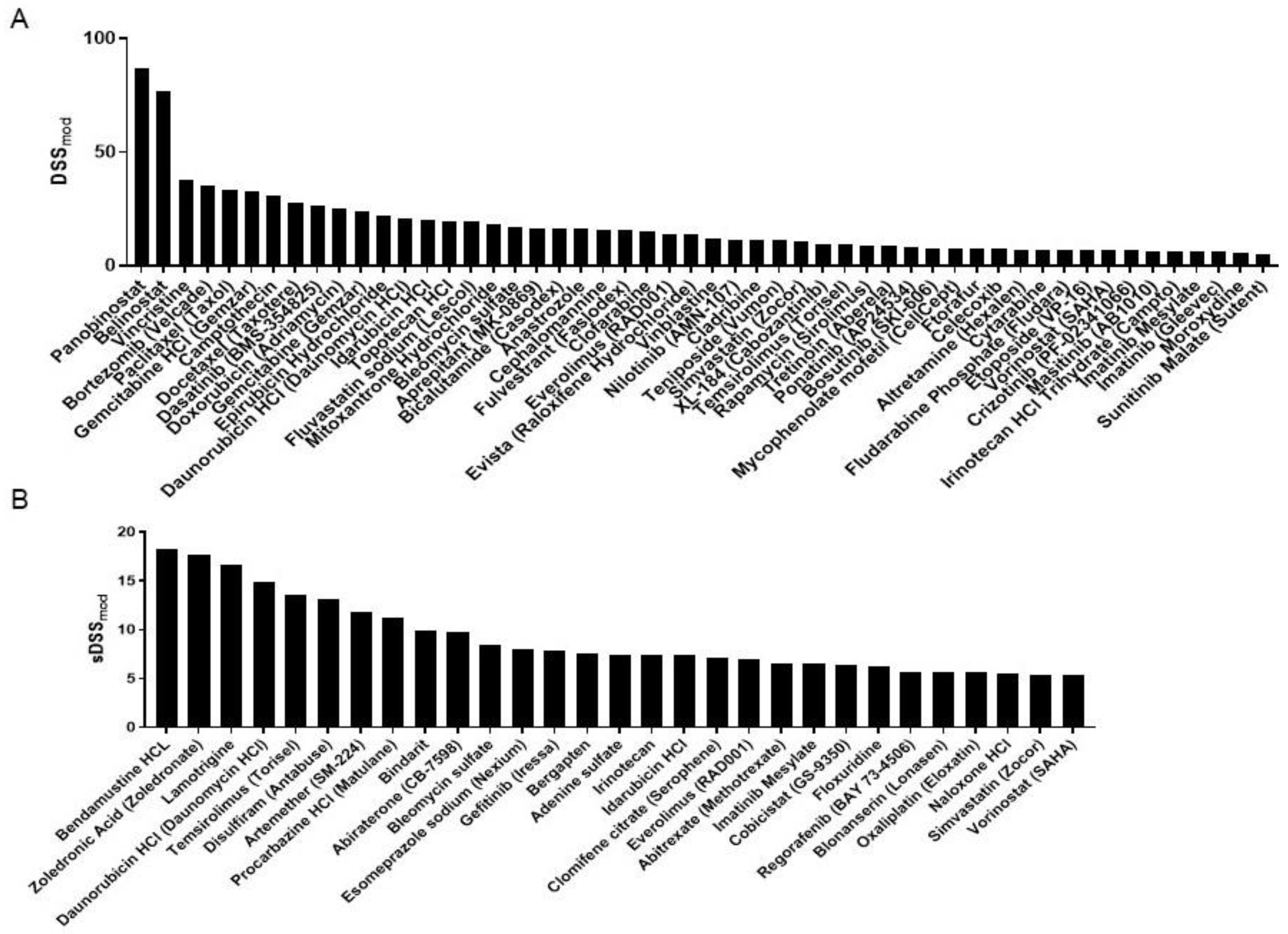
3.2. DST Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bielack, S.S.; Kempf-Bielack, B.; Delling, G.; Exner, G.U.; Flege, S.; Helmke, K.; Kotz, R.; Salzer-Kuntschik, M.; Werner, M.; Winkelmann, W. Prognostic factors in high-grade osteosarcoma of the extremities or trunk: An analysis of 1,702 patients treated on neoadjuvant cooperative osteosarcoma study group protocols. J. Clin. Oncol. 2002, 20, 776–790. [Google Scholar] [CrossRef] [PubMed]
- Sheng, G.; Gao, Y.; Yang, Y.; Wu, H. Osteosarcoma and Metastasis. Front. Oncol. 2021, 11, 780264. [Google Scholar] [CrossRef] [PubMed]
- Anderson, M.E. Update on Survival in Osteosarcoma. Orthop. Clin. N. Am. 2016, 47, 283–292. [Google Scholar] [CrossRef] [PubMed]
- Lohse, I.; Azzam, D.J.; Al-Ali, H.; Volmar, C.-H.; Brothers, S.P.; Ince, T.A.; Wahlestedt, C. Ovarian Cancer Treatment Stratification Using Ex Vivo Drug Sensitivity Testing. Anticancer Res. 2019, 39, 4023–4030. [Google Scholar] [CrossRef]
- Swords, R.T.; Azzam, D.; Al-Ali, H.; Lohse, I.; Volmar, C.-H.; Watts, J.M.; Perez, A.; Rodriguez, A.; Vargas, F.; Elias, R.; et al. Ex-vivo sensitivity profiling to guide clinical decision making in acute myeloid leukemia: A pilot study. Leuk Res. 2018, 64, 34–41. [Google Scholar] [CrossRef]
- Mandell, J.B.; Douglas, N.; Ukani, V.; Beumer, J.H.; Guo, J.; Payne, J.; Newman, R.; Mancinelli, L.; Intini, G.; Anderson, C.J.; et al. ALDH1A1 Gene Expression and Cellular Copper Levels between Low and Highly Metastatic Osteosarcoma Provide a Case for Novel Repurposing with Disulfiram and Copper. Sarcoma 2022, 2022, 7157507. [Google Scholar] [CrossRef]
- Cho, H.-J.; Lee, T.-S.; Park, J.-B.; Park, K.-K.; Choe, J.-Y.; Sin, D.-I.; Park, Y.-Y.; Moon, Y.-S.; Lee, K.-G.; Yeo, J.-H.; et al. Disulfiram suppresses invasive ability of osteosarcoma cells via the inhibition of MMP-2 and MMP-9 expression. J. Biochem. Mol. Biol. 2007, 40, 1069–1076. [Google Scholar] [CrossRef]
- Armenian, S.; Bhatia, S. Predicting and Preventing Anthracycline-Related Cardiotoxicity. Am. Soc. Clin. Oncol. Educ. Book 2018, 38, 3–12. [Google Scholar] [CrossRef]
- Carvalho, C.; Santos, R.X.; Cardoso, S.; Correia, S.; Oliveira, P.J.; Santos, M.S.; Moreira, P.I. Doxorubicin: The good, the bad and the ugly effect. Curr. Med. Chem. 2009, 16, 3267–3285. [Google Scholar] [CrossRef]
- van Dalen, E.C.; Raphaël, M.F.; Caron, H.N.; Kremer, L.C.M. Treatment including anthracyclines versus treatment not including anthracyclines for childhood cancer. Cochrane Database Syst. Rev. 2014, CD006647. [Google Scholar] [CrossRef]
- Anninga, J.K.; Gelderblom, H.; Fiocco, M.; Kroep, J.R.; Taminiau, A.H.M.; Hogendoorn, P.C.W.; Egeler, R.M. Chemotherapeutic adjuvant treatment for osteosarcoma: Where do we stand? Eur. J. Cancer 2011, 47, 2431–2445. [Google Scholar] [CrossRef] [PubMed]
- Hogendoorn, P.C.W.; Athanasou, N.; Bielack, S.; De Alava, E.; Tos, A.P.D.; Ferrari, S.; Gelderblom, H.; Grimer, R.; Hall, K.S.; Hassan, B.; et al. Bone sarcomas: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2010, 21 (Suppl. S5), v204–v213. [Google Scholar] [CrossRef] [PubMed]
- Lagmay, J.P.; Krailo, M.D.; Dang, H.; Kim, A.; Hawkins, D.S.; Beaty, O.; Widemann, B.C.; Zwerdling, T.; Bomgaars, L.; Langevin, A.-M.; et al. Outcome of Patients With Recurrent Osteosarcoma Enrolled in Seven Phase II Trials Through Children’s Cancer Group, Pediatric Oncology Group, and Children’s Oncology Group: Learning From the Past to Move Forward. J. Clin. Oncol. 2016, 34, 3031–3038. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Bahrami, A.; Pappo, A.; Easton, J.; Dalton, J.; Hedlund, E.; Ellison, D.; Shurtleff, S.; Wu, G.; Wei, L.; et al. Recurrent somatic structural variations contribute to tumorigenesis in pediatric osteosarcoma. Cell Rep. 2014, 7, 104–112. [Google Scholar] [CrossRef]
- Bishop, M.W.; Janeway, K.A. Emerging concepts for PI3K/mTOR inhibition as a potential treatment for osteosarcoma. F1000Research 2016, 5, F1000 Faculty Rev-1590. [Google Scholar] [CrossRef]
- Perry, J.A.; Kiezun, A.; Tonzi, P.; Van Allen, E.M.; Carter, S.L.; Baca, S.C.; Cowley, G.S.; Bhatt, A.S.; Rheinbay, E.; Pedamallu, C.S.; et al. Complementary genomic approaches highlight the PI3K/mTOR pathway as a common vulnerability in osteosarcoma. Proc. Natl. Acad. Sci. USA 2014, 111, E5564–E5573. [Google Scholar] [CrossRef]
- Cabrera-Andrade, A.; López-Cortés, A.; Jaramillo-Koupermann, G.; González-Díaz, H.; Pazos, A.; Munteanu, C.R.; Pérez-Castillo, Y.; Tejera, E. A Multi-Objective Approach for Anti-Osteosarcoma Cancer Agents Discovery through Drug Repurposing. Pharmaceuticals 2020, 13, 409. [Google Scholar] [CrossRef]
- Triscott, J.; Pambid, M.R.; Dunn, S.E. Concise review: Bullseye: Targeting cancer stem cells to improve the treatment of gliomas by repurposing disulfiram. Stem Cells 2015, 33, 1042–1046. [Google Scholar] [CrossRef]
- Mandell, J.B.; Lu, F.; Fisch, M.; Beumer, J.H.; Guo, J.; Watters, R.J.; Weiss, K.R. Combination Therapy with Disulfiram, Copper, and Doxorubicin for Osteosarcoma: In Vitro Support for a Novel Drug Repurposing Strategy. Sarcoma 2019, 2019, 1320201. [Google Scholar] [CrossRef]
- Jiao, Y.; Hannafon, B.N.; Ding, W.-Q. Disulfiram’s Anticancer Activity: Evidence and Mechanisms. Anticancer Agents Med. Chem. 2016, 16, 1378–1384. [Google Scholar] [CrossRef]
- Cong, J.; Wang, Y.; Zhang, X.; Zhang, N.; Liu, L.; Soukup, K.; Michelakos, T.; Hong, T.; DeLeo, A.; Cai, L.; et al. A novel chemoradiation targeting stem and nonstem pancreatic cancer cells by repurposing disulfiram. Cancer Lett. 2017, 409, 9–19. [Google Scholar] [CrossRef]
- Schweizer, M.T.; Lin, J.; Blackford, A.; Bardia, A.; King, S.; Armstrong, A.J.; A Rudek, M.; Yegnasubramanian, S.; A Carducci, M. Pharmacodynamic study of disulfiram in men with non-metastatic recurrent prostate cancer. Prostate Cancer Prostatic Dis. 2013, 16, 357–361. [Google Scholar] [CrossRef] [PubMed]
- Nechushtan, H.; Hamamreh, Y.; Nidal, S.; Gotfried, M.; Baron, A.; Shalev, Y.I.; Nisman, B.; Peretz, T.; Peylan-Ramu, N. A phase IIb trial assessing the addition of disulfiram to chemotherapy for the treatment of metastatic non-small cell lung cancer. Oncologist 2015, 20, 366–367. [Google Scholar] [CrossRef] [PubMed]
- Gorlick, R.; Janeway, K.; Lessnick, S.; Randall, R.L.; Marina, N.; COG Bone Tumor Committee. Children’s Oncology Group’s 2013 blueprint for research: Bone tumors. Pediatr. Blood Cancer 2013, 60, 1009–1015. [Google Scholar] [CrossRef] [PubMed]
- Grohar, P.J.; Janeway, K.A.; Mase, L.D.; Schiffman, J.D. Advances in the Treatment of Pediatric Bone Sarcomas. Am. Soc. Clin. Oncol. Educ. Book 2017, 37, 725–735. [Google Scholar] [CrossRef]
- Lohse, I.; Al-Ali, H.; Volmar, C.-H.; Trotta, A.D.A.; Brothers, S.P.; Capobianco, A.J.; Wahlestedt, C. Ex vivo drug sensitivity testing as a means for drug repurposing in esophageal adenocarcinoma. PLoS ONE 2018, 13, e0203173. [Google Scholar] [CrossRef]
- Ren, Y.; Lin, Y.; Chen, J.; Jin, Y. Disulfiram Chelated with Copper Promotes Apoptosis in Osteosarcoma via ROS/Mitochondria Pathway. Biol. Pharm. Bull. 2021, 44, 1557–1564. [Google Scholar] [CrossRef]

| Class | Compound |
|---|---|
| Alkylating agents | Bendamustine, Busulfan, Carboplatin, Cisplatin, Cyclophosphamide, Dacarbazine, lfosfamide, lomustine, Methazolastone, Oxaliplatin, Procarbazine, Streptozotocin |
| Antimetabolites | Azacitidine, Azaguanine-8, Capecitabine, Carmofur, Cladrabine, Clofarabine, Cytarabine, Decitabine, Febuxostat, Floxuridine, Fludarabine, Fluorouracil, Ftorafur, Gemcitabine, lonidamine, Mercaptopurine, Methotrexate, Nelarabine, Pemetrexed, Thioguanine |
| Antimitotics | 10-Deacetylbaccatin, Cephalomannine, Docetaxel, Paclitaxel, Vinblastine, Vincristine |
| Antitumor antibiotics | Artemether, Azithromycin, Bacitracin, Bleomycin, Hygromycin B, Lincomycin, Methacycline, Ofloxacin |
| HDAC inhibitors | Belinostat, Panobinostat, Sodium Butyrate, Vorinostat |
| Hormone inhibitors | 2-Methoxyestradiol, Abiraterone, Aminoglutethimide, Anastrozole, Bicalutamide, Clomifene Citrate, Diethylstilbestrol, Doxercalciferol, Enzalutamide, Exemestane, Flutamide, Fulvestrant, ltraconazole, Letrozole, Megestrol, Mifepristone, Paeoniflorin, Raloxifene, Tamoxifen, Toremifene, Triamcinolone |
| lmmunomodulators | Aspirin, Azathioprine, Bindarit, Cortisone, Celecoxib, Dexamethasone, Hydrocortisone, lmiquimod, Maraviroc, Meprednisone, Mizoribine, Mycophenolate, Phenylbutazone, Pimecrolimus, Pomalidomide, Prednisone, Sulindac, Tacrolimus, Thalidomide, Vinpocetine, Zileuton |
| Kinase inhibitors | Afatinib, Apatinib, Axitinib, Bosutinib, Cabozantinib, Crizotinib, Dasatinib, Erlotinib, lbrutinib, lmatinib, Lapatinib, Masitinib, Nilotinib, Pazopanib, Ponatinib, Regorafenib, Ruxolitinib, Sorafenib, Sunitinib, Tofacitinib, Vandetanib, Vemurafenib |
| Proteasome inhibitors | Bortezomib, Carfilzomib, Ubenimex |
| Rapalogs | Everolimus, Sirolimus, Temsirolimus |
| Topoisomerase 1/2 inhibitors | Camptothecin, Doxorubicin, Daunorubicin, Epirubicin, Etoposide, ldarubicin, lrinotecan, Mitoxantrone, Teniposide, Topotecan |
| Miscellaneous antineoplastics | Altretamine, Anagrelide, Bexarotene, Eltrombopag, Geniposide, Hydroxyurea, Mitotane, MLN4924, lsotretinoin, Tretinoin |
| Other | Adenine, Aprepitant, Atazanavir, Bepotastine, Bergapten, Blonanserin, Carbazochrome, Clorsulon, DAPT (GSI-IX), Disulfram, Dorzolamide, Ellagic acid, Epinephrine bitartrate, Esomeprazole, Ezetimibe, Flunarizine, Fluvastatin, Gadodiamide, Genistein, L-Arginine, Lamotrigine, Leucovorin, Linagliptin, Mesna, Mirabegron, Naloxone, Noscapine, Pamidronate, Pioglitazone, Ranolazine, Rosiglitazone, Orthovanadate, Temocapril, Tolbutamide, Valproic acid, Zoledronic acid, Vismodegib |
| Compound | DSSmod | Compound | DSSmod | Compound | DSSmod |
|---|---|---|---|---|---|
| Daunorubicin | 35.84 | Vorinostat | 12.19 | Bepotastine besilate | 6.76 |
| Doxorubicin | 29.08 | Artemether | 11.82 | Abitrexate | 6.59 |
| Bortezomib | 28.71 | Procarbazine | 11.16 | Ftorafur | 6.58 |
| Idarubicin | 27.34 | Irinotecan | 11.04 | Nilotinib | 6.55 |
| Bleomycin | 25.25 | Cobicistat | 10.83 | Cabozantinib | 6.48 |
| Camptothecin | 24.14 | Fluvastatin | 10.74 | Hygromycin b | 6.17 |
| Temsirolimus | 22.89 | Anastrozole | 10.73 | Naloxone hcl | 5.80 |
| Everolimus | 20.86 | Celecoxib | 10.58 | Regorafenib | 5.67 |
| Topotecan | 20.18 | Bindarit | 9.88 | Blonanserin | 5.66 |
| Zoledronic acid | 19.31 | Cladribine | 9.83 | Epinephrine bitartrate | 5.50 |
| Paclitaxel | 18.75 | Abiraterone | 9.79 | Vinblastine | 5.27 |
| Bendamustine | 18.21 | Clomifene citrate | 9.63 | Methacycline | 5.02 |
| Lamotrigine | 16.70 | Docetaxel | 9.03 | ||
| Simvastatin | 16.30 | Bergapten | 8.41 | ||
| Epirubicin | 16.01 | Streptozotocin | 8.03 | ||
| Vincristine | 14.01 | Esomeprazole | 7.92 | ||
| Dasatinib | 13.98 | Gefitinib | 7.86 | ||
| Disulfiram | 13.13 | Adenine sulfate | 7.45 | ||
| Imatinib mesylate | 12.70 | Oxaliplatin | 6.87 | ||
| Bicalutamide | 12.47 | Floxuridine | 6.85 | ||
| Mitoxantrone | 12.20 | Gemcitabine | 6.76 |
| Compound | sDSSmod | Compound | sDSSmod |
|---|---|---|---|
| Bendamustine HCL | 18.21 | Cobicistat | 6.43 |
| Zoledronic acid | 17.66 | Floxuridine | 6.17 |
| Lamotrigine | 16.70 | Regorafenib | 5.67 |
| Daunorubicin HCl | 14.92 | Blonanserin | 5.66 |
| Temsirolimus | 13.58 | Oxaliplatin | 5.66 |
| Disulfiram | 13.13 | Naloxone HCl | 5.44 |
| Artemether | 11.82 | Simvastatin | 5.35 |
| Procarbazine HCl | 11.16 | Vorinostat | 5.35 |
| Bindarit | 9.88 | ||
| Abiraterone | 9.79 | ||
| Bleomycin sulfate | 8.49 | ||
| Esomeprazole sodium | 7.92 | ||
| Gefitinib | 7.86 | ||
| Bergapten | 7.55 | ||
| Adenine sulfate | 7.45 | ||
| Irinotecan | 7.36 | ||
| Idarubicin HCl | 7.33 | ||
| Clomifene citrate | 7.08 | ||
| Everolimus | 6.94 | ||
| Abitrexate | 6.59 | ||
| Imatinib mesylate | 6.48 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lohse, I.; Dutcher, G.; Al-Ali, H.; Alperstein, W.; Coulter, D.W.; Trucco, M.; Trent, J.C.; Wahlestedt, C. Drug Sensitivity Testing in Osteosarcoma: A Case Report. Curr. Oncol. 2025, 32, 271. https://doi.org/10.3390/curroncol32050271
Lohse I, Dutcher G, Al-Ali H, Alperstein W, Coulter DW, Trucco M, Trent JC, Wahlestedt C. Drug Sensitivity Testing in Osteosarcoma: A Case Report. Current Oncology. 2025; 32(5):271. https://doi.org/10.3390/curroncol32050271
Chicago/Turabian StyleLohse, Ines, Giselle Dutcher, Hassan Al-Ali, Warren Alperstein, Donald W. Coulter, Matteo Trucco, Jonathan C. Trent, and Claes Wahlestedt. 2025. "Drug Sensitivity Testing in Osteosarcoma: A Case Report" Current Oncology 32, no. 5: 271. https://doi.org/10.3390/curroncol32050271
APA StyleLohse, I., Dutcher, G., Al-Ali, H., Alperstein, W., Coulter, D. W., Trucco, M., Trent, J. C., & Wahlestedt, C. (2025). Drug Sensitivity Testing in Osteosarcoma: A Case Report. Current Oncology, 32(5), 271. https://doi.org/10.3390/curroncol32050271









