DNA Quantity and Quality Comparisons between Cryopreserved and FFPE Tumors from Matched Pan-Cancer Samples
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Procurement
2.2. DNA Extraction
2.3. DNA Concentration and Purity Measurements
2.4. DNA Fragment Analysis
2.5. Statistics
3. Results
3.1. Matched Tumor Sample Deidentified Patient Demographics
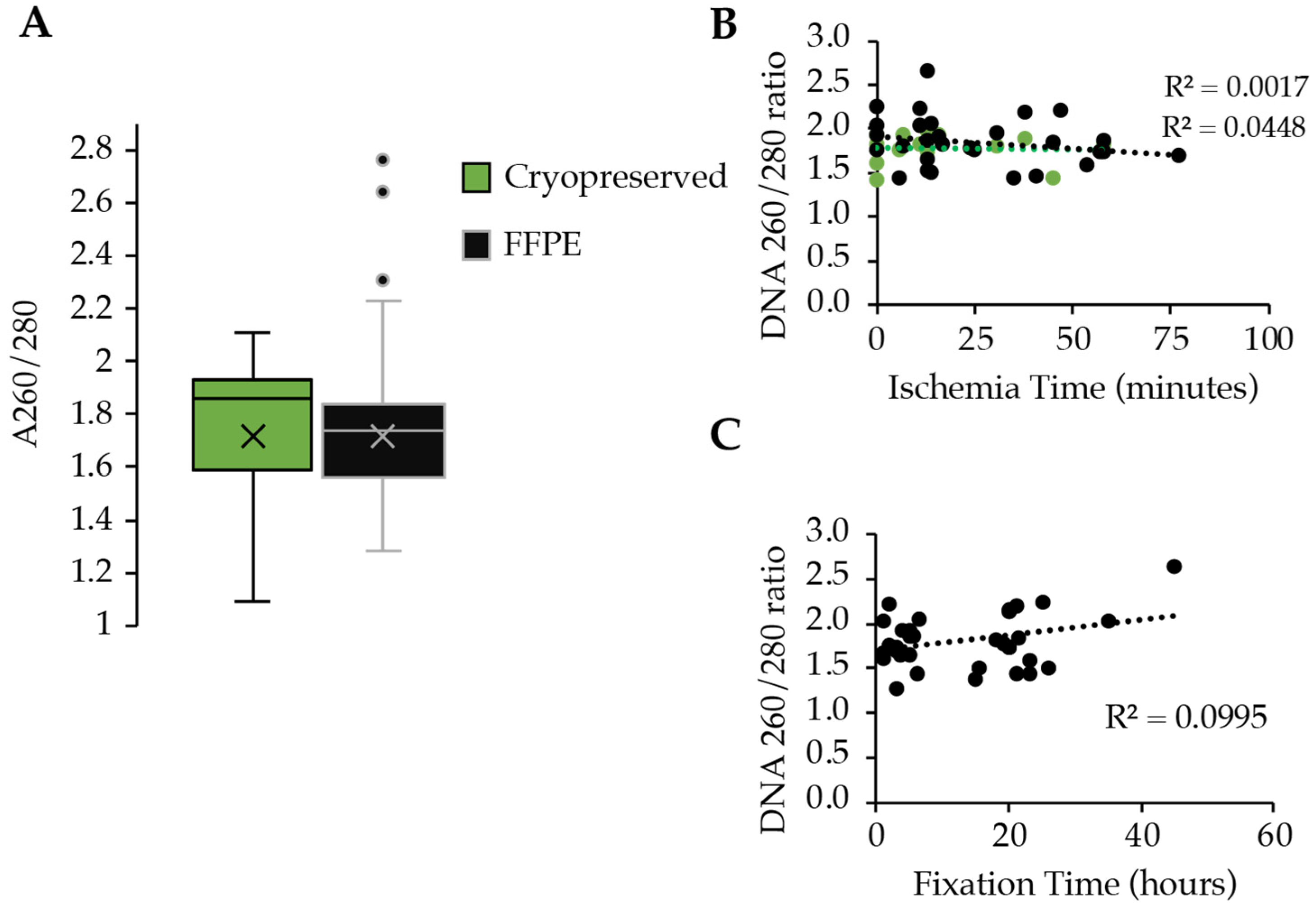
3.2. Sample Preparation and Purity
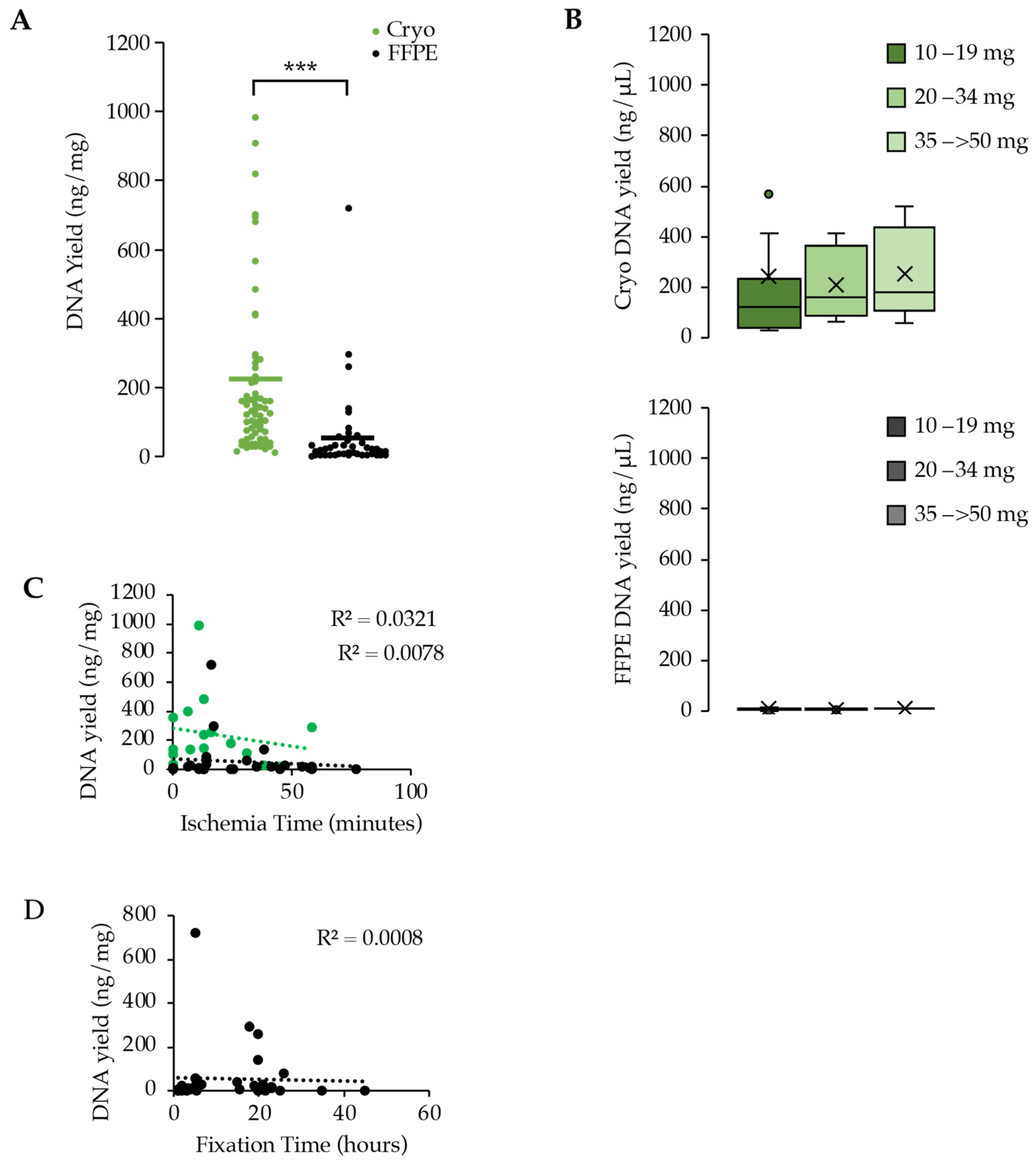
3.3. DNA Extraction and Yield
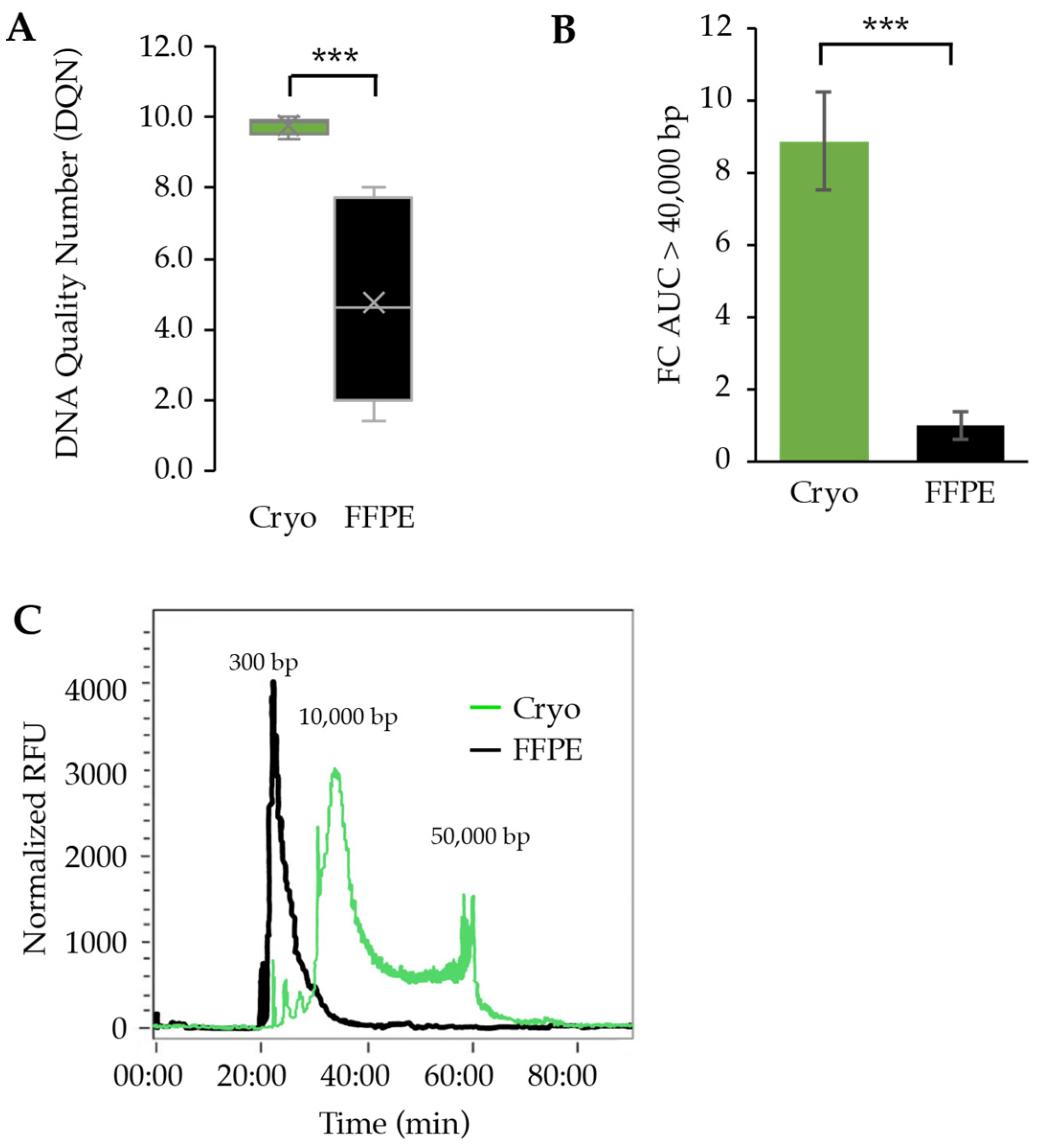
3.4. Assessment of DNA Quality between Cryopreserved and FFPE
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Murciano-Goroff, Y.R.; Drilon, A.; Stadler, Z.K. The NCI-MATCH: A National, Collaborative Precision Oncology Trial for Diverse Tumor Histologies. Cancer Cell 2021, 39, 22–24. [Google Scholar] [CrossRef] [PubMed]
- Meric-Bernstam, F.; Ford, J.M.; O’Dwyer, P.J.; Shapiro, G.I.; McShane, L.M.; Freidlin, B.; O’Cearbhaill, R.E.; George, S.; Glade-Bender, J.; Lyman, G.H.; et al. National Cancer Institute Combination Therapy Platform Trial with Molecular Analysis for Therapy Choice (ComboMATCH). Clin. Cancer Res. 2023, 29, 1412–1422. [Google Scholar] [CrossRef]
- Musiał, A.; Gryglewski, R.W.; Kielczewski, S.; Loukas, M.; Wajda, J. Formalin use in anatomical and histological science in the 19th and 20th centuries. Folia Med. Cracov. 2016, 56, 31–40. [Google Scholar] [PubMed]
- Akhoundova, D.; Rubin, M.A. Clinical application of advanced multi-omics tumor profiling: Shaping precision oncology of the future. Cancer Cell 2022, 40, 920–938. [Google Scholar] [CrossRef]
- Do, H.; Dobrovic, A. Sequence artifacts in DNA from formalin-fixed tissues: Causes and strategies for minimization. Clin. Chem. 2015, 61, 64–71. [Google Scholar] [CrossRef]
- Cazzato, G.; Caporusso, C.; Arezzo, F.; Cimmino, A.; Colagrande, A.; Loizzi, V.; Cormio, G.; Lettini, T.; Maiorano, E.; Scarcella, V.S.; et al. Formalin-Fixed and Paraffin-Embedded Samples for Next Generation Sequencing: Problems and Solutions. Genes 2021, 12, 1472. [Google Scholar] [CrossRef] [PubMed]
- Gallegos Ruiz, M.I.; Floor, K.; Rijmen, F.; Grünberg, K.; Rodriguez, J.A.; Giaccone, G. EGFR and K-ras Mutation Analysis in Non-Small Cell Lung Cancer: Comparison of Paraffin Embedded versus Frozen Specimens. Cell. Oncol. 2007, 29, 568205. [Google Scholar] [CrossRef]
- Haga, Y.; Sakamoto, Y.; Arai, M.; Suzuki, Y.; Suzuki, A. Long-Read Whole-Genome Sequencing Using a Nanopore Sequencer and Detection of Structural Variants in Cancer Genomes. Methods Mol. Biol. 2023, 2632, 177–189. [Google Scholar] [CrossRef]
- Fang, L.T.; Zhu, B.; Zhao, Y.; Chen, W.; Yang, Z.; Kerrigan, L.; Langenbach, K.; de Mars, M.; Lu, C.; Idler, K.; et al. Establishing community reference samples, data and call sets for benchmarking cancer mutation detection using whole-genome sequencing. Nat. Biotechnol. 2021, 39, 1151–1160. [Google Scholar] [CrossRef]
- Fujimoto, A.; Wong, J.H.; Yoshii, Y.; Akiyama, S.; Tanaka, A.; Yagi, H.; Shigemizu, D.; Nakagawa, H.; Mizokami, M.; Shimada, M. Whole-genome sequencing with long reads reveals complex structure and origin of structural variation in human genetic variations and somatic mutations in cancer. Genome Med. 2021, 13, 65. [Google Scholar] [CrossRef]
- Wolff, A.C.; Hammond, M.E.; Hicks, D.G.; Dowsett, M.; McShane, L.M.; Allison, K.H.; Allred, D.C.; Bartlett, J.M.; Bilous, M.; Fitzgibbons, P.; et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. Arch. Pathol. Lab. Med. 2014, 138, 241–256. [Google Scholar] [CrossRef] [PubMed]
- Lucena-Aguilar, G.; Sánchez-López, A.M.; Barberán-Aceituno, C.; Carrillo-Ávila, J.A.; López-Guerrero, J.A.; Aguilar-Quesada, R. DNA Source Selection for Downstream Applications Based on DNA Quality Indicators Analysis. Biopreserv. Biobank. 2016, 14, 264–270. [Google Scholar] [CrossRef] [PubMed]
- Cho, M.; Ahn, S.; Hong, M.; Bang, H.; Van Vrancken, M.; Kim, S.; Lee, J.; Hoon Park, S.; Oh Park, J.; Suk Park, Y.; et al. Tissue recommendations for precision cancer therapy using next generation sequencing: A comprehensive single cancer center’s experiences. Oncotarget 2017, 8, 42478–42486. [Google Scholar] [CrossRef] [PubMed]
- Bhagwate, A.V.; Liu, Y.; Winham, S.J.; McDonough, S.J.; Stallings-Mann, M.L.; Heinzen, E.P.; Davila, J.I.; Vierkant, R.A.; Hoskin, T.L.; Frost, M.; et al. Bioinformatics and DNA-extraction strategies to reliably detect genetic variants from FFPE breast tissue samples. BMC Genom. 2019, 20, 689. [Google Scholar] [CrossRef] [PubMed]
- Lüder Ripoli, F.; Mohr, A.; Conradine Hammer, S.; Willenbrock, S.; Hewicker-Trautwein, M.; Hennecke, S.; Murua Escobar, H.; Nolte, I. A Comparison of Fresh Frozen vs. Formalin-Fixed, Paraffin-Embedded Specimens of Canine Mammary Tumors via Branched-DNA Assay. Int. J. Mol. Sci. 2016, 17, 724. [Google Scholar] [CrossRef] [PubMed]
- Mathieson, W.; Thomas, G.A. Why Formalin-fixed, Paraffin-embedded Biospecimens Must Be Used in Genomic Medicine: An Evidence-based Review and Conclusion. J. Histochem. Cytochem. 2020, 68, 543–552. [Google Scholar] [CrossRef]
- Gao, X.H.; Li, J.; Gong, H.F.; Yu, G.Y.; Liu, P.; Hao, L.Q.; Liu, L.J.; Bai, C.G.; Zhang, W. Comparison of Fresh Frozen Tissue With Formalin-Fixed Paraffin-Embedded Tissue for Mutation Analysis Using a Multi-Gene Panel in Patients With Colorectal Cancer. Front. Oncol. 2020, 10, 310. [Google Scholar] [CrossRef]
- Sakamoto, Y.; Sereewattanawoot, S.; Suzuki, A. A new era of long-read sequencing for cancer genomics. J. Hum. Genet. 2020, 65, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Staňková, H.; Hastie, A.R.; Chan, S.; Vrána, J.; Tulpová, Z.; Kubaláková, M.; Visendi, P.; Hayashi, S.; Luo, M.; Batley, J.; et al. BioNano genome mapping of individual chromosomes supports physical mapping and sequence assembly in complex plant genomes. Plant Biotechnol. J. 2016, 14, 1523–1531. [Google Scholar] [CrossRef]
- Pang, A.W.C.; Kosco, K.; Sahajpal, N.S.; Sridhar, A.; Hauenstein, J.; Clifford, B.; Estabrook, J.; Chitsazan, A.D.; Sahoo, T.; Iqbal, A.; et al. Analytic Validation of Optical Genome Mapping in Hematological Malignancies. Biomedicines 2023, 11, 3263. [Google Scholar] [CrossRef]
- Soler, G.; Ouedraogo, Z.G.; Goumy, C.; Lebecque, B.; Aspas Requena, G.; Ravinet, A.; Kanold, J.; Véronèse, L.; Tchirkov, A. Optical Genome Mapping in Routine Cytogenetic Diagnosis of Acute Leukemia. Cancers 2023, 15, 2131. [Google Scholar] [CrossRef] [PubMed]
- van Belzen, I.A.E.M.; Schönhuth, A.; Kemmeren, P.; Hehir-Kwa, J.Y. Structural variant detection in cancer genomes: Computational challenges and perspectives for precision oncology. NPJ Precis. Oncol. 2021, 5, 15. [Google Scholar] [CrossRef]
- Choo, Z.-N.; Behr, J.M.; Deshpande, A.; Hadi, K.; Yao, X.; Tian, H.; Takai, K.; Zakusilo, G.; Rosiene, J.; Da Cruz Paula, A.; et al. Most large structural variants in cancer genomes can be detected without long reads. Nat. Genet. 2023, 55, 2139–2148. [Google Scholar] [CrossRef] [PubMed]
- Chong, I.Y.; Starling, N.; Rust, A.; Alexander, J.; Aronson, L.; Llorca-Cardenosa, M.; Chauhan, R.; Chaudry, A.; Kumar, S.; Fenwick, K.; et al. The Mutational Concordance of Fixed Formalin Paraffin Embedded and Fresh Frozen Gastro-Oesophageal Tumours Using Whole Exome Sequencing. J. Clin. Med. 2021, 10, 215. [Google Scholar] [CrossRef]
- Robbe, P.; Popitsch, N.; Knight, S.J.L.; Antoniou, P.; Becq, J.; He, M.; Kanapin, A.; Samsonova, A.; Vavoulis, D.V.; Ross, M.T.; et al. Clinical whole-genome sequencing from routine formalin-fixed, paraffin-embedded specimens: Pilot study for the 100,000 Genomes Project. Genet. Med. 2018, 20, 1196–1205. [Google Scholar] [CrossRef] [PubMed]
- Solassol, J.; Ramos, J.; Crapez, E.; Saifi, M.; Mangé, A.; Vianès, E.; Lamy, P.-J.; Costes, V.; Maudelonde, T. KRAS Mutation Detection in Paired Frozen and Formalin-Fixed Paraffin-Embedded (FFPE) Colorectal Cancer Tissues. Int. J. Mol. Sci. 2011, 12, 3191–3204. [Google Scholar] [CrossRef]
- De Paoli-Iseppi, R.; Johansson, P.A.; Menzies, A.M.; Dias, K.R.; Pupo, G.M.; Kakavand, H.; Wilmott, J.S.; Mann, G.J.; Hayward, N.K.; Dinger, M.E.; et al. Comparison of whole-exome sequencing of matched fresh and formalin fixed paraffin embedded melanoma tumours: Implications for clinical decision making. Pathology 2016, 48, 261–266. [Google Scholar] [CrossRef]
- Ondracek, R.P.; Chen, J.; Marosy, B.; Szewczyk, S.; Medico, L.; Mohan, A.S.; Nair, P.; Pratt, R.; Roh, J.M.; Khoury, T.; et al. Results and lessons from dual extraction of DNA and RNA from formalin-fixed paraffin-embedded breast tumor tissues for a large Cancer epidemiologic study. BMC Genom. 2022, 23, 614. [Google Scholar] [CrossRef]
- Ohmomo, H.; Komaki, S.; Ono, K.; Sutoh, Y.; Hachiya, T.; Arai, E.; Fujimoto, H.; Yoshida, T.; Kanai, Y.; Sasaki, M.; et al. Evaluation of clinical formalin-fixed paraffin-embedded tissue quality for targeted-bisulfite sequencing. Pathol. Int. 2021, 71, 135–140. [Google Scholar] [CrossRef]
- Kreula, J.; Bondestam, S.; Virkkunen, P. Sample size in fine needle aspiration biopsy. Br. J. Surg. 1989, 76, 1270–1272. [Google Scholar] [CrossRef]
- Byron, S.A.; Van Keuren-Jensen, K.R.; Engelthaler, D.M.; Carpten, J.D.; Craig, D.W. Translating RNA sequencing into clinical diagnostics: Opportunities and challenges. Nat. Rev. Genet. 2016, 17, 257–271. [Google Scholar] [CrossRef] [PubMed]
- Shohdy, K.S.; Bareja, R.; Sigouros, M.; Wilkes, D.C.; Dorsaint, P.; Manohar, J.; Bockelman, D.; Xiang, J.Z.; Kim, R.; Ohara, K.; et al. Functional comparison of exome capture-based methods for transcriptomic profiling of formalin-fixed paraffin-embedded tumors. NPJ Genom. Med. 2021, 6, 66. [Google Scholar] [CrossRef] [PubMed]
- de Ruijter, T.C.; de Hoon, J.P.J.; Slaats, J.; de Vries, B.; Janssen, M.J.F.W.; van Wezel, T.; Aarts, M.J.B.; van Engeland, M.; Tjan-Heijnen, V.C.G.; Van Neste, L.; et al. Formalin-fixed, paraffin-embedded (FFPE) tissue epigenomics using Infinium HumanMethylation450 BeadChip assays. Lab. Investig. 2015, 95, 833–842. [Google Scholar] [CrossRef] [PubMed]
- Kalmár, A.; Péterfia, B.; Wichmann, B.; Patai, Á.V.; Barták, B.K.; Nagy, Z.B.; Furi, I.; Tulassay, Z.; Molnár, B. Comparison of Automated and Manual DNA Isolation Methods for DNA Methylation Analysis of Biopsy, Fresh Frozen, and Formalin-Fixed, Paraffin-Embedded Colorectal Cancer Samples. J. Lab. Autom. 2015, 20, 642–651. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jovanović, B.; Sheng, Q.; Seitz, R.S.; Lawrence, K.D.; Morris, S.W.; Thomas, L.R.; Hout, D.R.; Schweitzer, B.L.; Guo, Y.; Pietenpol, J.A.; et al. Comparison of triple-negative breast cancer molecular subtyping using RNA from matched fresh-frozen versus formalin-fixed paraffin-embedded tissue. BMC Cancer 2017, 17, 241. [Google Scholar] [CrossRef]
- He, A.; Powell, S.; Kyle, M.; Rose, M.; Masmila, E.; Estrada, V.; Sicklick, J.K.; Molinolo, A.; Kaushal, S. Cryopreservation of Viable Human Tissues: Renewable Resource for Viable Tissue, Cell Lines, and Organoid Development. Biopreserv. Biobank. 2020, 18, 222–227. [Google Scholar] [CrossRef]
- Restivo, G.; Tastanova, A.; Balázs, Z.; Panebianco, F.; Diepenbruck, M.; Ercan, C.; Preca, B.-T.; Hafner, J.; Weber, W.P.; Kurzeder, C.; et al. Live slow-frozen human tumor tissues viable for 2D, 3D, ex vivo cultures and single-cell RNAseq. Commun. Biol. 2022, 5, 1144. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Okojie, J.; O’Neal, N.; Burr, M.; Worley, P.; Packer, I.; Anderson, D.; Davis, J.; Kearns, B.; Fatema, K.; Dixon, K.; et al. DNA Quantity and Quality Comparisons between Cryopreserved and FFPE Tumors from Matched Pan-Cancer Samples. Curr. Oncol. 2024, 31, 2441-2452. https://doi.org/10.3390/curroncol31050183
Okojie J, O’Neal N, Burr M, Worley P, Packer I, Anderson D, Davis J, Kearns B, Fatema K, Dixon K, et al. DNA Quantity and Quality Comparisons between Cryopreserved and FFPE Tumors from Matched Pan-Cancer Samples. Current Oncology. 2024; 31(5):2441-2452. https://doi.org/10.3390/curroncol31050183
Chicago/Turabian StyleOkojie, Jeffrey, Nikole O’Neal, Mackenzie Burr, Peyton Worley, Isaac Packer, DeLaney Anderson, Jack Davis, Bridger Kearns, Kaniz Fatema, Ken Dixon, and et al. 2024. "DNA Quantity and Quality Comparisons between Cryopreserved and FFPE Tumors from Matched Pan-Cancer Samples" Current Oncology 31, no. 5: 2441-2452. https://doi.org/10.3390/curroncol31050183
APA StyleOkojie, J., O’Neal, N., Burr, M., Worley, P., Packer, I., Anderson, D., Davis, J., Kearns, B., Fatema, K., Dixon, K., & Barrott, J. J. (2024). DNA Quantity and Quality Comparisons between Cryopreserved and FFPE Tumors from Matched Pan-Cancer Samples. Current Oncology, 31(5), 2441-2452. https://doi.org/10.3390/curroncol31050183





