Survival Outcome Prediction in Glioblastoma: Insights from MRI Radiomics
Abstract
1. Introduction
2. Materials and Methods
2.1. Subjects
2.2. Imaging
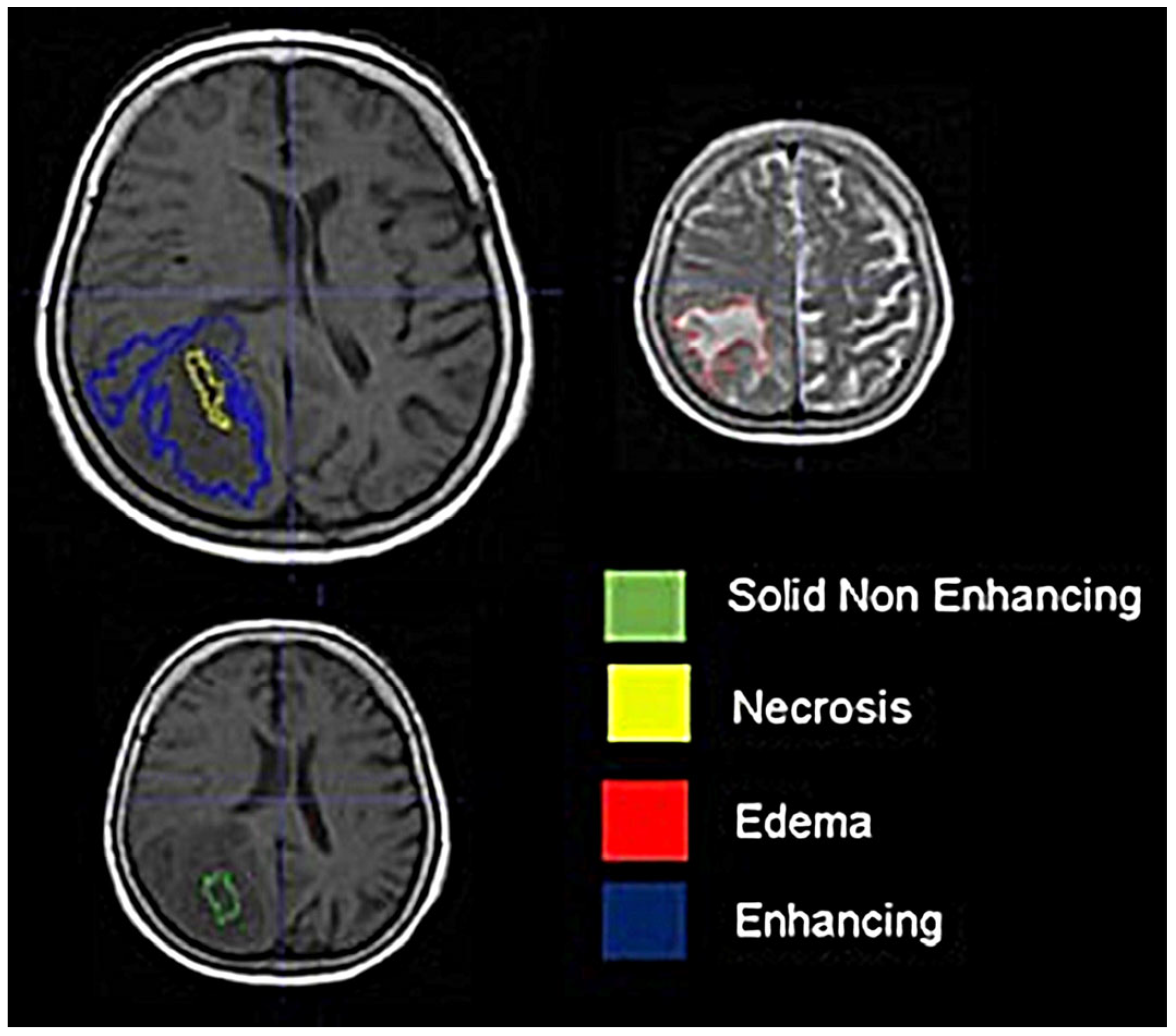
2.3. Image Processing
2.4. Radiomics
2.5. Statistical Analysis
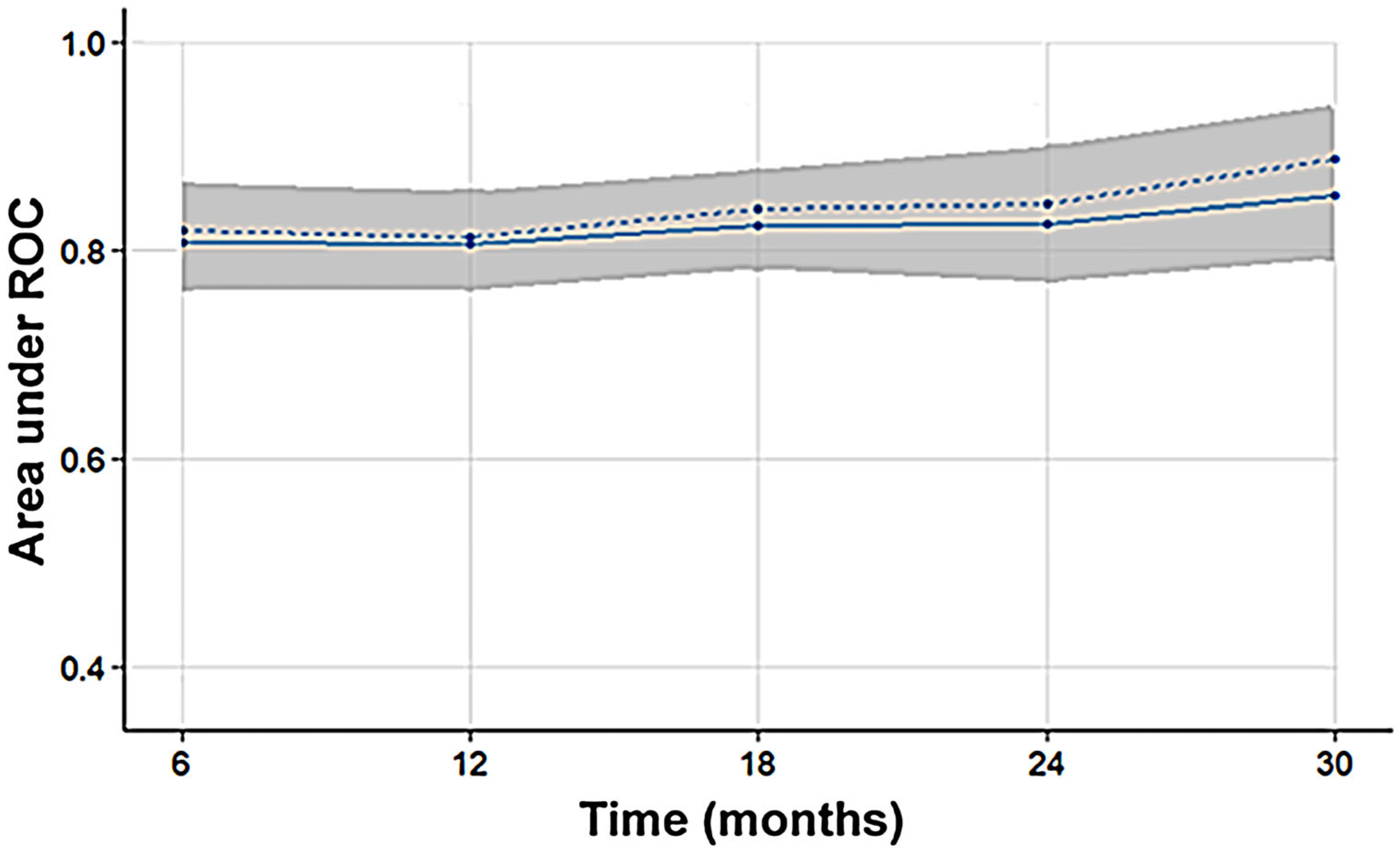
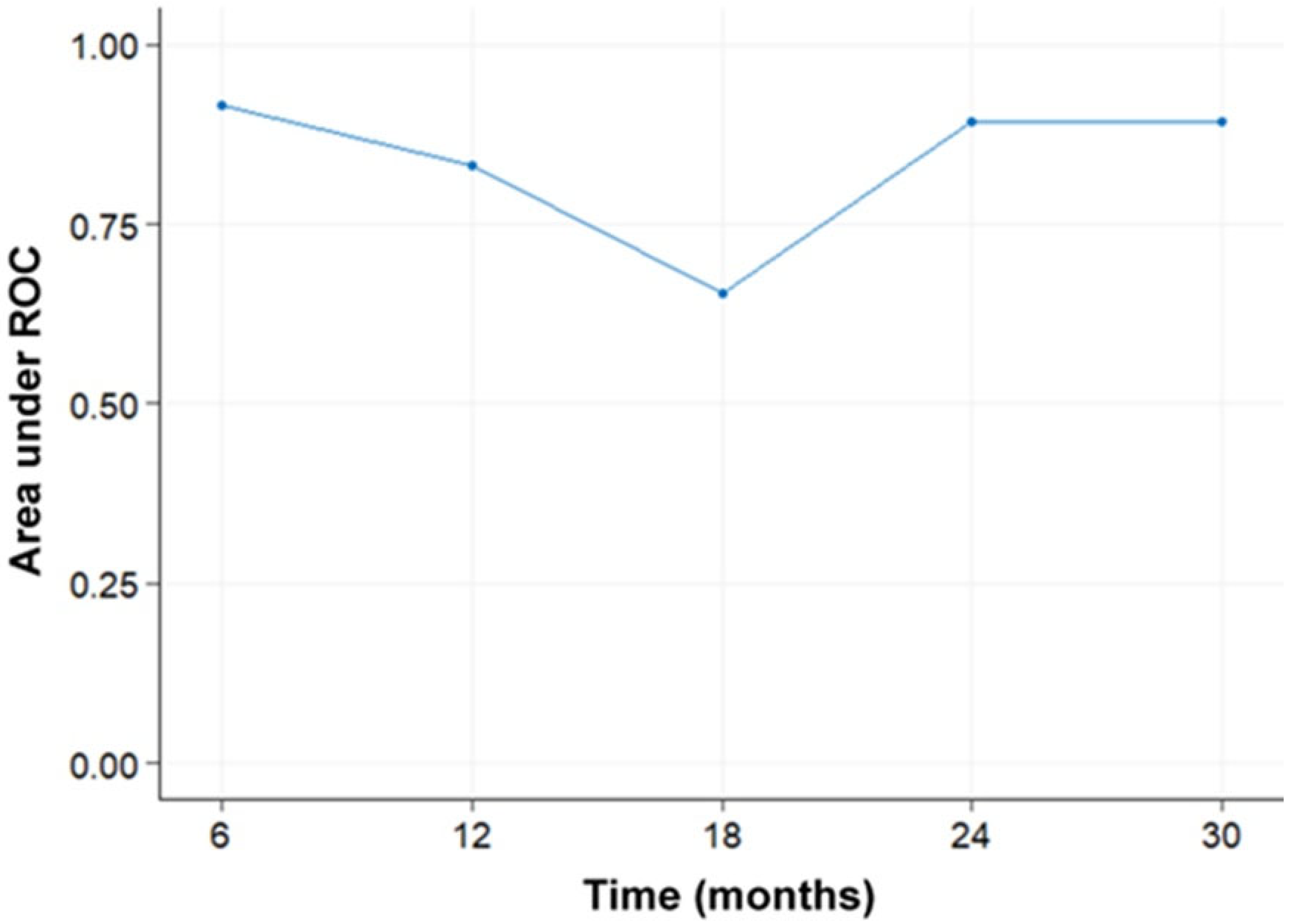
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Grochans, S.; Cybulska, A.M.; Simińska, D.; Korbecki, J.; Kojder, K.; Chlubek, D.; Baranowska-Bosiacka, I. Epidemiology of Glioblastoma Multiforme-Literature Review. Cancers 2022, 14, 2412. [Google Scholar] [CrossRef] [PubMed]
- Hanif, F.; Muzaffar, K.; Perveen, K.; Malhi, S.; Simjee, S. Glioblastoma Multiforme: A Review of Its Epidemiology and Pathogenesis through Clinical Presentation and Treatment. Asian Pac. J. Cancer Prev. 2017, 18, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Koshy, M.; Villano, J.L.; Dolecek, T.A.; Howard, A.; Mahmood, U.; Chmura, S.J.; Weichselbaum, R.R.; McCarthy, B.J. Improved Survival Time Trends for Glioblastoma Using the SEER 17 Population-Based Registries. J. Neurooncol. 2012, 107, 207–212. [Google Scholar] [CrossRef] [PubMed]
- Jovčevska, I. Sequencing the next Generation of Glioblastomas. Crit. Rev. Clin. Lab. Sci. 2018, 55, 264–282. [Google Scholar] [CrossRef] [PubMed]
- DeCordova, S.; Shastri, A.; Tsolaki, A.G.; Yasmin, H.; Klein, L.; Singh, S.K.; Kishore, U. Molecular Heterogeneity and Immunosuppressive Microenvironment in Glioblastoma. Front. Immunol. 2020, 11, 1402. [Google Scholar] [CrossRef] [PubMed]
- Komori, T. Grading of Adult Diffuse Gliomas According to the 2021 WHO Classification of Tumors of the Central Nervous System. Lab. Investig. J. Technol. Methods Pathol. 2022, 102, 126–133. [Google Scholar] [CrossRef]
- Li, W.; Tang, K.; Chen, Q.; Li, S.; Qiu, X.; Li, S.; Jiang, T. MRI Manifestions Correlate with Survival of Glioblastoma Multiforme Patients. Cancer Biol. Med. 2012, 9, 120–123. [Google Scholar] [CrossRef] [PubMed]
- Shukla, G.; Alexander, G.S.; Bakas, S.; Nikam, R.; Talekar, K.; Palmer, J.D.; Shi, W. Advanced Magnetic Resonance Imaging in Glioblastoma: A Review. Chin. Clin. Oncol. 2017, 6, 40. [Google Scholar] [CrossRef] [PubMed]
- Taha, B.; Boley, D.; Sun, J.; Chen, C.C. State of Radiomics in Glioblastoma. Neurosurgery 2021, 89, 177–184. [Google Scholar] [CrossRef]
- Mayerhoefer, M.E.; Materka, A.; Langs, G.; Häggström, I.; Szczypiński, P.; Gibbs, P.; Cook, G. Introduction to Radiomics. J. Nucl. Med. Off. Publ. Soc. Nucl. Med. 2020, 61, 488–495. [Google Scholar] [CrossRef]
- Yip, S.S.F.; Aerts, H.J.W.L. Applications and Limitations of Radiomics. Phys. Med. Biol. 2016, 61, R150–R166. [Google Scholar] [CrossRef] [PubMed]
- Chaddad, A.; Daniel, P.; Desrosiers, C.; Toews, M.; Abdulkarim, B. Novel Radiomic Features Based on Joint Intensity Matrices for Predicting Glioblastoma Patient Survival Time. IEEE J. Biomed. Health Inform. 2019, 23, 795–804. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Yoon, M.J.; Park, J.E.; Choi, E.J.; Lee, J.; Kim, H.S. Radiomics in Peritumoral Non-Enhancing Regions: Fractional Anisotropy and Cerebral Blood Volume Improve Prediction of Local Progression and Overall Survival in Patients with Glioblastoma. Neuroradiology 2019, 61, 1261–1272. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Han, Y.; Hu, X.; Wang, W.; Cui, G.; Guo, L.; Zhang, X. An Improvement of Survival Stratification in Glioblastoma Patients via Combining Subregional Radiomics Signatures. Front. Neurosci. 2021, 15, 683452. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Zhang, B.; Zhang, S.; Cheng, J.; Liu, X.; Wang, W.; Dong, Y.; Zhang, L.; Mo, X.; Chen, Q.; et al. Quantitative MRI-Based Radiomics for Noninvasively Predicting Molecular Subtypes and Survival in Glioma Patients. NPJ Precis. Oncol. 2021, 5, 72. [Google Scholar] [CrossRef] [PubMed]
- Lao, J.; Chen, Y.; Li, Z.-C.; Li, Q.; Zhang, J.; Liu, J.; Zhai, G. A Deep Learning-Based Radiomics Model for Prediction of Survival in Glioblastoma Multiforme. Sci. Rep. 2017, 7, 10353. [Google Scholar] [CrossRef] [PubMed]
- Peruzzo, D.; Bertoldo, A.; Zanderigo, F.; Cobelli, C. Automatic Selection of Arterial Input Function on Dynamic Contrast-Enhanced MR Images. Comput. Methods Programs Biomed. 2011, 104, e148–e157. [Google Scholar] [CrossRef] [PubMed]
- Boxerman, J.L.; Schmainda, K.M.; Weisskoff, R.M. Relative Cerebral Blood Volume Maps Corrected for Contrast Agent Extravasation Significantly Correlate with Glioma Tumor Grade, Whereas Uncorrected Maps Do Not. AJNR Am. J. Neuroradiol. 2006, 27, 859–867. [Google Scholar]
- van Griethuysen, J.J.M.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.H.; Fillion-Robin, J.-C.; Pieper, S.; Aerts, H.J.W.L. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef]
- Chaddad, A.; Kucharczyk, M.J.; Daniel, P.; Sabri, S.; Jean-Claude, B.J.; Niazi, T.; Abdulkarim, B. Radiomics in Glioblastoma: Current Status and Challenges Facing Clinical Implementation. Front Oncol. 2019, 9, 374. [Google Scholar] [CrossRef]
- Osman, A.F.I. A Multi-Parametric MRI-Based Radiomics Signature and a Practical ML Model for Stratifying Glioblastoma Patients Based on Survival Toward Precision Oncology. Front. Comput. Neurosci. 2019, 13, 58. [Google Scholar] [CrossRef]
- Chaddad, A.; Sabri, S.; Niazi, T.; Abdulkarim, B. Prediction of Survival with Multi-Scale Radiomic Analysis in Glioblastoma Patients. Med. Biol. Eng. Comput. 2018, 56, 2287–2300. [Google Scholar] [CrossRef] [PubMed]
- Park, J.E.; Kim, H.S.; Jo, Y.; Yoo, R.-E.; Choi, S.H.; Nam, S.J.; Kim, J.H. Radiomics Prognostication Model in Glioblastoma Using Diffusion- and Perfusion-Weighted MRI. Sci. Rep. 2020, 10, 4250. [Google Scholar] [CrossRef]
- Liu, Y.; Xu, X.; Yin, L.; Zhang, X.; Li, L.; Lu, H. Relationship between Glioblastoma Heterogeneity and Survival Time: An MR Imaging Texture Analysis. AJNR Am. J. Neuroradiol. 2017, 38, 1695–1701. [Google Scholar] [CrossRef]
- Wang, S.; Xiao, F.; Sun, W.; Yang, C.; Ma, C.; Huang, Y.; Xu, D.; Li, L.; Chen, J.; Li, H.; et al. Radiomics Analysis Based on Magnetic Resonance Imaging for Preoperative Overall Survival Prediction in Isocitrate Dehydrogenase Wild-Type Glioblastoma. Front. Neurosci. 2021, 15, 791776. [Google Scholar] [CrossRef] [PubMed]
- Chaddad, A.; Tanougast, C. Extracted Magnetic Resonance Texture Features Discriminate between Phenotypes and Are Associated with Overall Survival in Glioblastoma Multiforme Patients. Med. Biol. Eng. Comput. 2016, 54, 1707–1718. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Lu, D.; Gao, P.; Tian, Q.; Lu, H.; Xu, X.; He, X.; Liu, Y. Survival-Relevant High-Risk Subregion Identification for Glioblastoma Patients: The MRI-Based Multiple Instance Learning Approach. Eur. Radiol. 2020, 30, 5602–5610. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-X.; Lin, G.-S.; Lin, Z.-X.; Zhang, J.-D.; Liu, S.-Y.; Zhou, C.-F. Peritumoral Edema Shown by MRI Predicts Poor Clinical Outcome in Glioblastoma. World J. Surg. Oncol. 2015, 13, 97. [Google Scholar] [CrossRef]
- Kim, H.; Choi, S.H.; Kim, J.-H.; Ryoo, I.; Kim, S.C.; Yeom, J.A.; Shin, H.; Jung, S.C.; Lee, A.L.; Yun, T.J.; et al. Gliomas: Application of Cumulative Histogram Analysis of Normalized Cerebral Blood Volume on 3 T MRI to Tumor Grading. PLoS ONE 2013, 8, e63462. [Google Scholar] [CrossRef]
- Carrillo, J.A.; Lai, A.; Nghiemphu, P.L.; Kim, H.J.; Phillips, H.S.; Kharbanda, S.; Moftakhar, P.; Lalaezari, S.; Yong, W.; Ellingson, B.M.; et al. Relationship between Tumor Enhancement, Edema, IDH1 Mutational Status, MGMT Promoter Methylation, and Survival in Glioblastoma. AJNR Am. J. Neuroradiol. 2012, 33, 1349–1355. [Google Scholar] [CrossRef]
- Rathore, S.; Akbari, H.; Doshi, J.; Shukla, G.; Rozycki, M.; Bilello, M.; Lustig, R.; Davatzikos, C. Radiomic Signature of Infiltration in Peritumoral Edema Predicts Subsequent Recurrence in Glioblastoma: Implications for Personalized Radiotherapy Planning. J. Med. Imaging 2018, 5, 021219. [Google Scholar] [CrossRef] [PubMed]
- Long, H.; Zhang, P.; Bi, Y.; Yang, C.; Wu, M.; He, D.; Huang, S.; Yang, K.; Qi, S.; Wang, J. MRI Radiomic Features of Peritumoral Edema May Predict the Recurrence Sites of Glioblastoma Multiforme. Front. Oncol. 2022, 12, 1042498. [Google Scholar] [CrossRef] [PubMed]
- Eidel, O.; Burth, S.; Neumann, J.-O.; Kieslich, P.J.; Sahm, F.; Jungk, C.; Kickingereder, P.; Bickelhaupt, S.; Mundiyanapurath, S.; Bäumer, P.; et al. Tumor Infiltration in Enhancing and Non-Enhancing Parts of Glioblastoma: A Correlation with Histopathology. PLoS ONE 2017, 12, e0169292. [Google Scholar] [CrossRef] [PubMed]
- Pasquini, L.; Di Napoli, A.; Napolitano, A.; Lucignani, M.; Dellepiane, F.; Vidiri, A.; Villani, V.; Romano, A.; Bozzao, A. Glioblastoma Radiomics to Predict Survival: Diffusion Characteristics of Surrounding Nonenhancing Tissue to Select Patients for Extensive Resection. J. Neuroimaging Off. J. Am. Soc. Neuroimaging 2021, 31, 1192–1200. [Google Scholar] [CrossRef] [PubMed]
- Hammoud, M.A.; Sawaya, R.; Shi, W.; Thall, P.F.; Leeds, N.E. Prognostic Significance of Preoperative MRI Scans in Glioblastoma Multiforme. J. Neurooncol. 1996, 27, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Lacroix, M.; Abi-Said, D.; Fourney, D.R.; Gokaslan, Z.L.; Shi, W.; DeMonte, F.; Lang, F.F.; McCutcheon, I.E.; Hassenbusch, S.J.; Holland, E.; et al. A Multivariate Analysis of 416 Patients with Glioblastoma Multiforme: Prognosis, Extent of Resection, and Survival. J. Neurosurg. 2001, 95, 190–198. [Google Scholar] [CrossRef] [PubMed]
- Rathore, S.; Akbari, H.; Rozycki, M.; Abdullah, K.G.; Nasrallah, M.P.; Binder, Z.A.; Davuluri, R.V.; Lustig, R.A.; Dahmane, N.; Bilello, M.; et al. Radiomic MRI Signature Reveals Three Distinct Subtypes of Glioblastoma with Different Clinical and Molecular Characteristics, Offering Prognostic Value beyond IDH1. Sci. Rep. 2018, 8, 5087. [Google Scholar] [CrossRef] [PubMed]
- Vartanian, A.; Singh, S.K.; Agnihotri, S.; Jalali, S.; Burrell, K.; Aldape, K.D.; Zadeh, G. GBM’s Multifaceted Landscape: Highlighting Regional and Microenvironmental Heterogeneity. Neuro-Oncol. 2014, 16, 1167–1175. [Google Scholar] [CrossRef] [PubMed]
- Seker-Polat, F.; Pinarbasi Degirmenci, N.; Solaroglu, I.; Bagci-Onder, T. Tumor Cell Infiltration into the Brain in Glioblastoma: From Mechanisms to Clinical Perspectives. Cancers 2022, 14, 443. [Google Scholar] [CrossRef]
- Qin, X.; Liu, R.; Akter, F.; Qin, L.; Xie, Q.; Li, Y.; Qiao, H.; Zhao, W.; Jian, Z.; Liu, R.; et al. Peri-Tumoral Brain Edema Associated with Glioblastoma Correlates with Tumor Recurrence. J. Cancer 2021, 12, 2073–2082. [Google Scholar] [CrossRef]
- Giambra, M.; Di Cristofori, A.; Valtorta, S.; Manfrellotti, R.; Bigiogera, V.; Basso, G.; Moresco, R.M.; Giussani, C.; Bentivegna, A. The Peritumoral Brain Zone in Glioblastoma: Where We Are and Where We Are Going. J. Neurosci. Res. 2023, 101, 199–216. [Google Scholar] [CrossRef] [PubMed]
- Buonfiglioli, A.; Hambardzumyan, D. Macrophages and Microglia: The Cerberus of Glioblastoma. Acta Neuropathol. Commun. 2021, 9, 54. [Google Scholar] [CrossRef] [PubMed]
- Siker, M.L.; Wang, M.; Porter, K.; Nelson, D.F.; Curran, W.J.; Michalski, J.M.; Souhami, L.; Chakravarti, A.; Yung, W.K.A.; Delrowe, J.; et al. Age as an Independent Prognostic Factor in Patients with Glioblastoma: A Radiation Therapy Oncology Group and American College of Surgeons National Cancer Data Base Comparison. J. Neurooncol. 2011, 104, 351–356. [Google Scholar] [CrossRef]
- Jilla, S.; Prathipati, A.; Subramanian, B.V.; Das, P.; Valiyaveettil, D. Impact of Various Prognostic Factors on Survival in Glioblastoma: Tertiary Care Institutional Experience. Ecancermedicalscience 2022, 16, 1386. [Google Scholar] [CrossRef] [PubMed]


| Regressor (Significant Predictor) | Tissue | Image | Regression Coefficient |
|---|---|---|---|
| shape_Maximum2DDiameterSlice | Necrosis | T1 | −1.56 |
| glszm_ZoneVariance | Non-enhancing | Relative CBF | +2.81 |
| glcm_Idn | Edema | MTT | −7.23 |
| firstorder_Minimum | Enhancing | MD | +2.32 |
| glcm_ClusterShade | Edema | FA | +2.78 |
| glcm_Correlation | Enhancing | FA | +1.87 |
| Age | +4.62 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Styliara, E.I.; Astrakas, L.G.; Alexiou, G.; Xydis, V.G.; Zikou, A.; Kafritsas, G.; Voulgaris, S.; Argyropoulou, M.I. Survival Outcome Prediction in Glioblastoma: Insights from MRI Radiomics. Curr. Oncol. 2024, 31, 2233-2243. https://doi.org/10.3390/curroncol31040165
Styliara EI, Astrakas LG, Alexiou G, Xydis VG, Zikou A, Kafritsas G, Voulgaris S, Argyropoulou MI. Survival Outcome Prediction in Glioblastoma: Insights from MRI Radiomics. Current Oncology. 2024; 31(4):2233-2243. https://doi.org/10.3390/curroncol31040165
Chicago/Turabian StyleStyliara, Effrosyni I., Loukas G. Astrakas, George Alexiou, Vasileios G. Xydis, Anastasia Zikou, Georgios Kafritsas, Spyridon Voulgaris, and Maria I. Argyropoulou. 2024. "Survival Outcome Prediction in Glioblastoma: Insights from MRI Radiomics" Current Oncology 31, no. 4: 2233-2243. https://doi.org/10.3390/curroncol31040165
APA StyleStyliara, E. I., Astrakas, L. G., Alexiou, G., Xydis, V. G., Zikou, A., Kafritsas, G., Voulgaris, S., & Argyropoulou, M. I. (2024). Survival Outcome Prediction in Glioblastoma: Insights from MRI Radiomics. Current Oncology, 31(4), 2233-2243. https://doi.org/10.3390/curroncol31040165






