Classification of Brain Tumor from Magnetic Resonance Imaging Using Vision Transformers Ensembling
Abstract
1. Introduction
2. Experimental Methods
2.1. Dataset
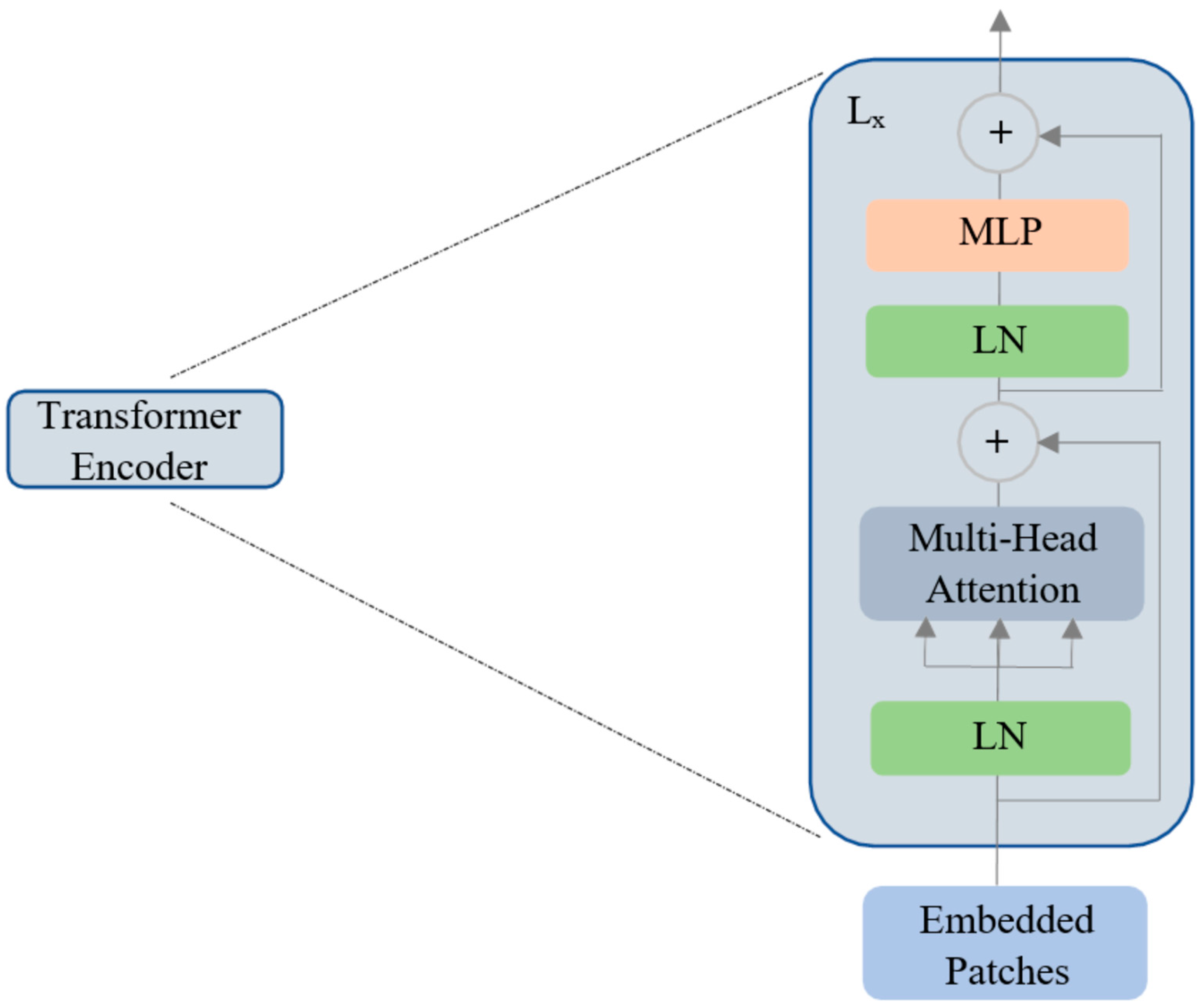
2.2. Vision Transformer
2.3. Computational Infrastructure
2.4. Model Ensembling
2.5. Performance Metrics
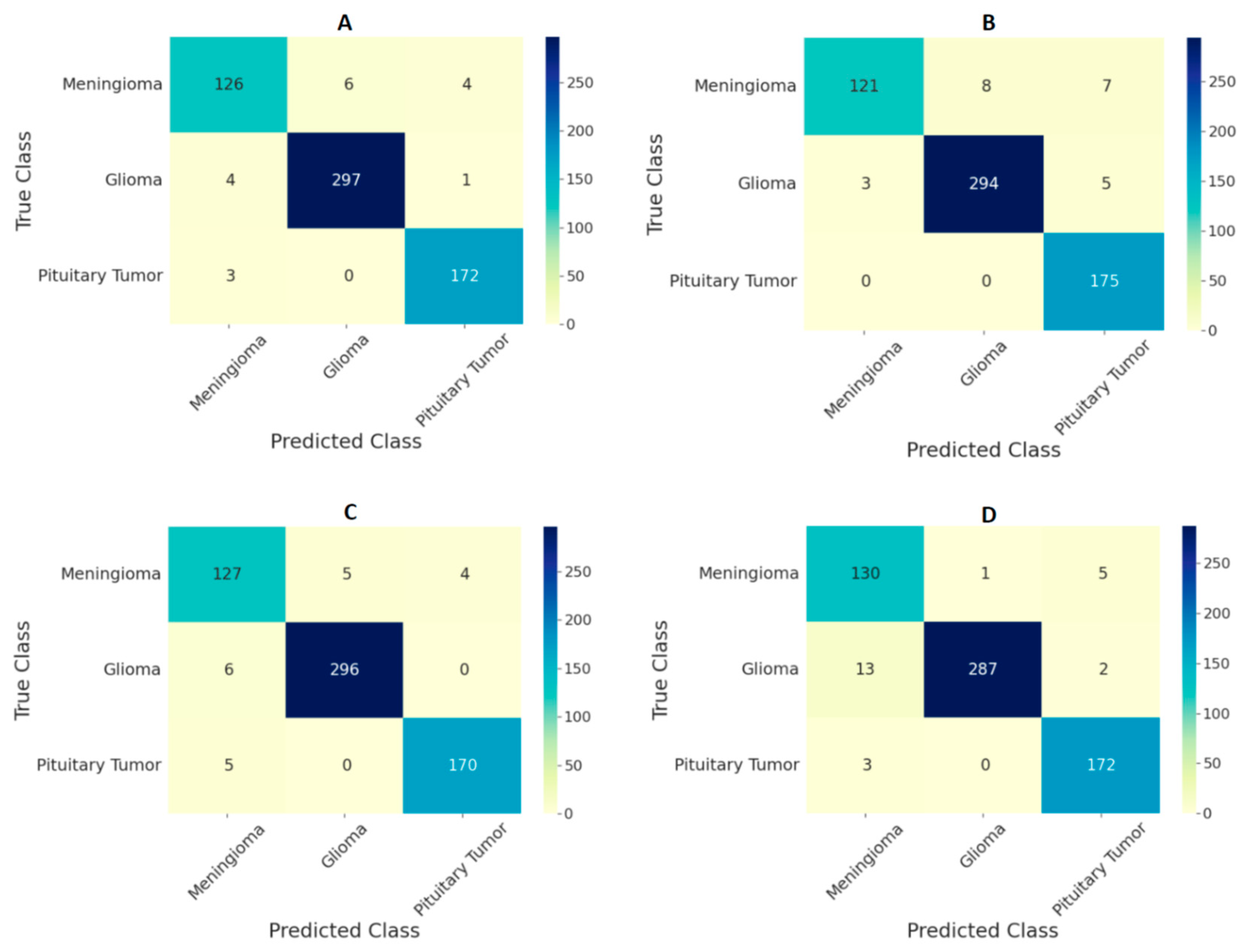
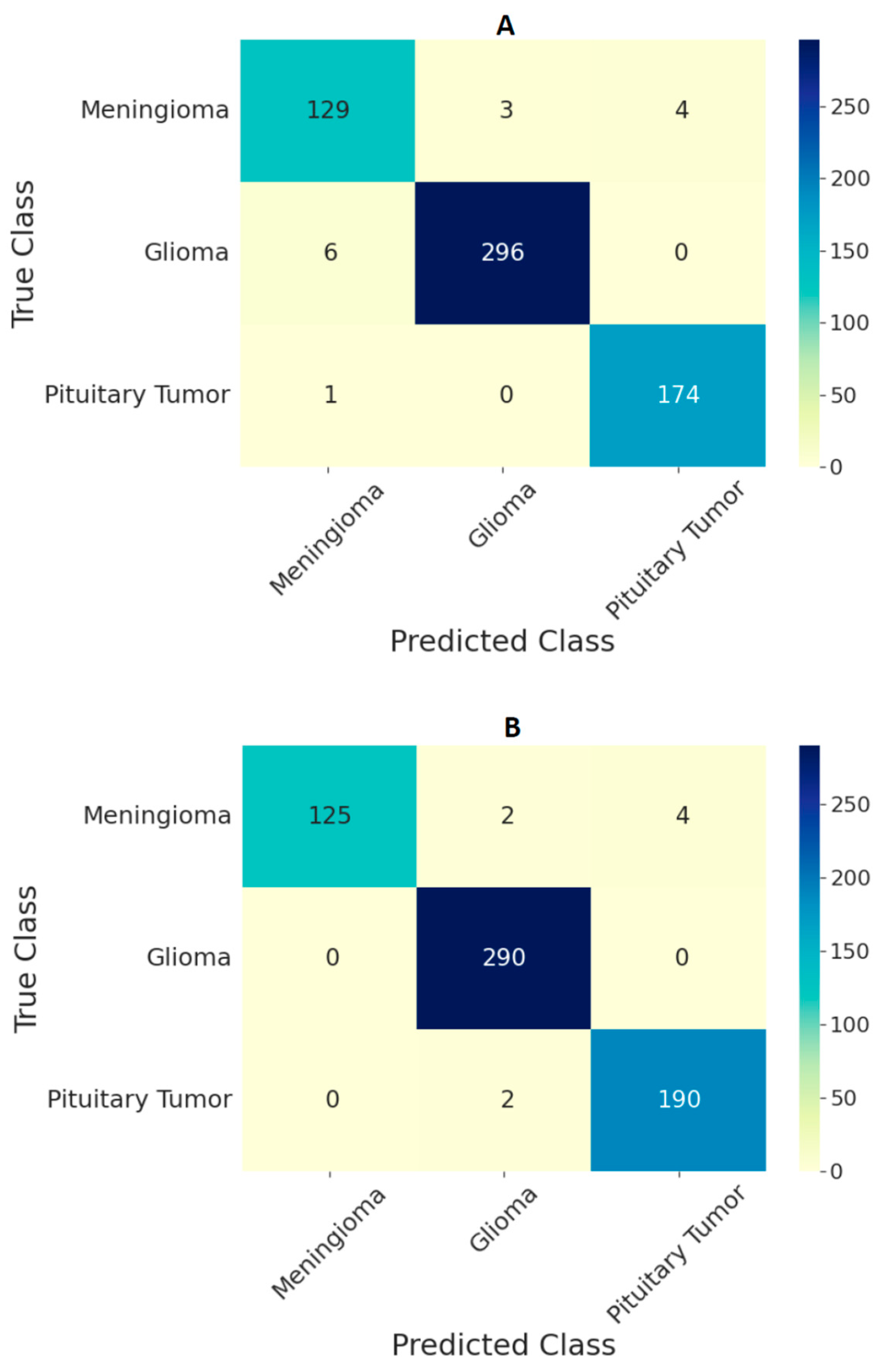
3. Results
4. Discussion
| Work | Method | Image Resolution | Training Data | Accuracy |
|---|---|---|---|---|
| J. Cheng [40] | GLCM-BoW | 512 × 512 | 80% | 91.28% |
| M.R. Ismael [48] | DWT-2D Gabor | 512 × 512 | 70% | 91.90% |
| A. Pashaei [49] | CNN-ELM | 512 × 512 | 70% | 93.68% |
| P. Afshar [50] | CapsuleNet | 128 × 128 | - | 90.89% |
| S. Deepak [22] | CNN-SVM-kNN | 224 × 224 | 80% | 97.80% |
| O. Polat [19] | Transfer Learning | 224 × 224 | 70% | 99.02% |
| B. Ahmad [18] | GAN-VAEs | 512 × 512 | 60% | 96.25% |
| N.S. Shaik [25] | MANet | 224 × 224 | - | 96.51% |
| Present study | Ensemble of ViTs | 224 × 224 | 70% | 97.71% |
| 384 × 384 | 70% | 98.70% |
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rasheed, S.; Rehman, K.; Akash, M.S.H. An insight into the risk factors of brain tumors and their therapeutic interventions. Biomed. Pharmacother. 2021, 143, 112119. [Google Scholar] [CrossRef] [PubMed]
- Sánchez Fernández, I.; Loddenkemper, T. Seizures caused by brain tumors in children. Seizure 2017, 44, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Chintagumpala, M.; Gajjar, A. Brain tumors. Pediatr. Clin. N. Am. 2015, 62, 167–178. [Google Scholar] [CrossRef]
- Herholz, K.; Langen, K.J.; Schiepers, C.; Mountz, J.M. Brain tumors. Semin. Nucl. Med. 2012, 42, 356–370. [Google Scholar] [CrossRef] [PubMed]
- Boire, A.; Brastianos, P.K.; Garzia, L.; Valiente, M. Brain metastasis. Nat. Rev. Cancer 2020, 20, 4–11. [Google Scholar] [CrossRef]
- Kontogeorgos, G. Classification and pathology of pituitary tumors. Endocrine 2005, 28, 27–35. [Google Scholar] [CrossRef]
- Viallon, M.; Cuvinciuc, V.; Delattre, B.; Merlini, L.; Barnaure-Nachbar, I.; Toso-Patel, S.; Becker, M.; Lovblad, K.O.; Haller, S. State-of-the-art MRI techniques in neuroradiology: Principles, pitfalls, and clinical applications. Neuroradiology 2015, 57, 441–467. [Google Scholar] [CrossRef] [PubMed]
- Villanueva-Meyer, J.E.; Mabray, M.C.; Cha, S. Current Clinical Brain Tumor Imaging. Neurosurgery 2017, 81, 397–415. [Google Scholar] [CrossRef] [PubMed]
- Kavin Kumar, K.; Meera Devi, T.; Maheswaran, S. An Efficient Method for Brain Tumor Detection Using Texture Features and SVM Classifier in MR Images. Asian Pac. Cancer Prev. 2018, 19, 2789–2794. [Google Scholar] [CrossRef]
- Kang, J.; Ullah, Z.; Gwak, J. MRI-Based Brain Tumor Classification Using Ensemble of Deep Features and Machine Learning Classifiers. Sensors 2021, 21, 2222. [Google Scholar] [CrossRef] [PubMed]
- Zacharaki, E.I.; Wang, S.; Chawla, S.; Yoo, D.S.; Wolf, R.; Melhem, E.R.; Davatzikos, C. Classification of brain tumor type and grade using MRI texture and shape in a machine learning scheme. Magn. Reson. Med. 2009, 62, 1609. [Google Scholar] [CrossRef]
- Shrot, S.; Salhov, M.; Dvorski, N.; Konen, E.; Averbuch, A.; Hoffmann, C. Application of MR morphologic, diffusion tensor, and perfusion imaging in the classification of brain tumors using machine learning scheme. Neuroradiology 2019, 61, 757–765. [Google Scholar] [CrossRef] [PubMed]
- Deepak, S.; Ameer, P.M. Retrieval of brain MRI with tumor using contrastive loss based similarity on GoogLeNet encodings. Comput. Biol. Med. 2020, 125, 103993. [Google Scholar] [CrossRef] [PubMed]
- Swati, Z.N.K.; Zhao, Q.; Kabir, M.; Ali, F.; Ali, Z.; Ahmed, S.; Lu, J. Brain tumor classification for MR images using transfer learning and fine-tuning. Comput. Med. Imaging Graph. 2019, 75, 34–46. [Google Scholar] [CrossRef] [PubMed]
- Zhuge, Y.; Ning, H.; Mathen, P.; Cheng, J.Y.; Krauze, A.V.; Camphausen, K.; Miller, R.W. Automated glioma grading on conventional MRI images using deep convolutional neural networks. Med. Phys. 2020, 47, 3044–3053. [Google Scholar] [CrossRef]
- Pomponio, R.; Erus, G.; Habes, M.; Doshi, J.; Srinivasan, D.; Mamourian, E.; Bashyam, V.; Nasrallah, I.M.; Satterthwaite, T.D.; Fan, Y.; et al. Harmonization of large MRI datasets for the analysis of brain imaging patterns throughout the lifespan. NeuroImage 2019, 208, 116450. [Google Scholar] [CrossRef]
- Naser, M.A.; Deen, M.J. Brain tumor segmentation and grading of lower-grade glioma using deep learning in MRI images. Comput. Biol. Med. 2020, 121, 103758. [Google Scholar] [CrossRef]
- Ahmad, B.; Sun, J.; You, Q.; Palade, V.; Mao, Z. Brain Tumor Classification Using a Combination of Variational Autoencoders and Generative Adversarial Networks. Biomedicines 2022, 10, 223. [Google Scholar] [CrossRef]
- Polat, Ö.; Güngen, C. Classification of brain tumors from MR images using deep transfer learning. Supercomputing 2021, 77, 7236–7252. [Google Scholar] [CrossRef]
- Khan, H.A.; Jue, W.; Mushtaq, M.; Mushtaq, M.U.; Khan, H.A.; Jue, W.; Mushtaq, M.; Mushtaq, M.U. Brain tumor classification in MRI image using convolutional neural network. Math. Biosci. Eng. 2020, 17, 6203–6216. [Google Scholar] [CrossRef]
- Badža, M.M.; Barjaktarović, M.C. Classification of Brain Tumors from MRI Images Using a Convolutional Neural Network. Appl. Sci. 2020, 10, 1999. [Google Scholar] [CrossRef]
- Deepak, S.; Ameer, P.M. Brain tumor classification using deep CNN features via transfer learning. Comput. Biol. Med. 2019, 111, 103345. [Google Scholar] [CrossRef] [PubMed]
- Haq, E.U.; Jianjun, H.; Li, K.; Haq, H.U.; Zhang, T. An MRI-based deep learning approach for efficient classification of brain tumors. Ambient Intell. Humaniz. Comput. 2021, 2021, 1–22. [Google Scholar] [CrossRef]
- Sekhar, A.; Biswas, S.; Hazra, R.; Sunaniya, A.K.; Mukherjee, A.; Yang, L. Brain tumor classification using fine-tuned GoogLeNet features and machine learning algorithms: IoMT enabled CAD system. IEEE Biomed. Health Inform. 2021, 26, 983–991. [Google Scholar] [CrossRef] [PubMed]
- Shaik, N.S.; Cherukuri, T.K. Multi-level attention network: Application to brain tumor classification. Signal Image Video Process. 2021, 16, 817–824. [Google Scholar] [CrossRef]
- Alanazi, M.F.; Ali, M.U.; Hussain, S.J.; Zafar, A.; Mohatram, M.; Irfan, M.; Alruwaili, R.; Alruwaili, M.; Ali, N.H.; Albarrak, A.M. Brain Tumor/Mass Classification Framework Using Magnetic-Resonance-Imaging-Based Isolated and Developed Transfer Deep-Learning Model. Sensors 2022, 22, 372. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention Is All You Need. Adv. Neural Inf. Process. Syst. 2017, 30, 5999–6009. [Google Scholar] [CrossRef]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image is Worth 16 × 16 Words: Transformers for Image Recognition at Scale. arXiv 2020, arXiv:2010.11929. [Google Scholar] [CrossRef]
- Steiner, A.; Kolesnikov, A.; Zhai, X.; Wightman, R.; Uszkoreit, J.; Beyer, L. How to Train Your ViT? Data, Augmentation, and Regularization in Vision Transformers, (n.d.). Available online: https://github.com/rwightman/pytorch-image-models. (accessed on 10 March 2022).
- Wu, Y.; Qi, S.; Sun, Y.; Xia, S.; Yao, Y.; Qian, W. A vision transformer for emphysema classification using CT images. Phys. Med. Biol. 2021, 66, 245016. [Google Scholar] [CrossRef]
- Gheflati, B.; Rivaz, H. Vision Transformer for Classification of Breast Ultrasound Images. In Proceedings of the 2022 44th Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Glasgow, UK, 11–15 July 2022. [Google Scholar] [CrossRef]
- Shamshad, F.; Khan, S.; Zamir, S.W.; Khan, M.H.; Hayat, M.; Khan, F.S.; Fu, H. Transformers in Medical Imaging: A Survey. arXiv 2022, arXiv:2201.09873. [Google Scholar] [CrossRef]
- Wang, J.; Fang, Z.; Lang, N.; Yuan, H.; Su, M.Y.; Baldi, P. A multi-resolution approach for spinal metastasis detection using deep Siamese neural networks. Comput. Biol. Med. 2017, 84, 137–146. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Gao, Y.; Liu, F. TransMed: Transformers Advance Multi-modal Medical Image Classification. Diagnostics 2021, 11, 1384. [Google Scholar] [CrossRef] [PubMed]
- Gheflati, B.; Rivaz, H. Vision transformers for classification of breast ultrasound images. arXiv 2021. [Google Scholar] [CrossRef]
- Mondal, A.K.; Bhattacharjee, A.; Singla, P.; Prathosh, A.P. xViTCOS: Explainable Vision Transformer Based COVID-19 Screening Using Radiography. IEEE Transl. Eng. Health Med. 2022, 10, 1100110. [Google Scholar] [CrossRef]
- Ayan, E.; Karabulut, B.; Ünver, H.M. Diagnosis of Pediatric Pneumonia with Ensemble of Deep Convolutional Neural Networks in Chest X-Ray Images. Arab. Sci. Eng. 2022, 47, 2123–2139. [Google Scholar] [CrossRef]
- Ko, H.; Ha, H.; Cho, H.; Seo, K.; Lee, J. Pneumonia Detection with Weighted Voting Ensemble of CNN Models. In Proceedings of the 2019 2nd International Conference on Artificial Intelligence and Big Data (ICAIBD), Chengdu, China, 25–28 May 2019; pp. 306–310. [Google Scholar] [CrossRef]
- Afifi, A.; Hafsa, N.E.; Ali, M.A.S.; Alhumam, A.; Alsalman, S. An Ensemble of Global and Local-Attention Based Convolutional Neural Networks for COVID-19 Diagnosis on Chest X-ray Images. Symmetry 2021, 13, 113. [Google Scholar] [CrossRef]
- Cheng, J.; Huang, W.; Cao, S.; Yang, R.; Yang, W.; Yun, Z.; Wang, Z.; Feng, Q. Enhanced Performance of Brain Tumor Classification via Tumor Region Augmentation and Partition. PLoS ONE 2015, 10, e0140381. [Google Scholar] [CrossRef]
- Cheng, J.; Yang, W.; Huang, M.; Huang, W.; Jiang, J.; Zhou, Y.; Yang, R.; Zhao, J.; Feng, Y.; Feng, Q.; et al. Retrieval of Brain Tumors by Adaptive Spatial Pooling and Fisher Vector Representation. PLoS ONE 2016, 11, e0157112. [Google Scholar] [CrossRef]
- Marosi, C.; Hassler, M.; Roessler, K.; Reni, M.; Sant, M.; Mazza, E.; Vecht, C. Meningioma. Crit. Rev. Oncol. Hematol. 2008, 67, 153–171. [Google Scholar] [CrossRef]
- Ostrom, Q.T.; Gittleman, H.; Stetson, L.; Virk, S.M.; Barnholtz-Sloan, J.S. Epidemiology of gliomas. Cancer Treat. Res. 2015, 163, 1–14. [Google Scholar] [CrossRef]
- Devlin, J.; Chang, M.W.; Lee, K.; Toutanova, K. BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. arXiv 2018, arXiv:1810.04805. [Google Scholar] [CrossRef]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin Transformer: Hierarchical Vision Transformer using Shifted Windows. IEEE/CVF Int. Conf. Comput. Vis. 2021, 2021, 10012–10022. [Google Scholar] [CrossRef]
- Touvron, H.; Cord, M.; Douze, M.; Massa, F.; Sablayrolles, A.; Jégou, H.; Ai, F. Training data-efficient image transformers & distillation through attention. In Proceedings of the International Conference on Machine Learning, PMLR, Virtual Conference. 13–18 July 2020. [Google Scholar] [CrossRef]
- Han, K.; Xiao, A.; Wu, E.; Guo, J.; Xu, C.; Wang, Y. Transformer in Transformer. Adv. Neural Inf. Process. Syst. 2021, 34, 15908–15919. [Google Scholar] [CrossRef]
- Ismael, M.R.; Abdel-Qader, I. Brain Tumor Classification via Statistical Features and Back-Propagation Neural Network. In Proceedings of the 2018 IEEE International Conference on Electro/Information Technology (EIT), Rochester, MI, USA, 3–5 May 2018; pp. 252–257. [Google Scholar] [CrossRef]
- Pashaei, A.; Sajedi, H.; Jazayeri, N. Brain tumor classification via convolutional neural network and extreme learning machines. In Proceedings of the 2018 8th International Conference on Computer and Knowledge Engineering (ICCKE), Mashhad, Iran, 25–26 October 2018; pp. 314–319. [Google Scholar] [CrossRef]
- Afshar, P.; Plataniotis, K.N.; Mohammadi, A. Capsule Networks for Brain Tumor Classification Based on MRI Images and Coarse Tumor Boundaries. In Proceedings of the ICASSP 2019-2019 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Brighton, UK, 12–17 May 2019; pp. 1368–1372. [Google Scholar] [CrossRef]
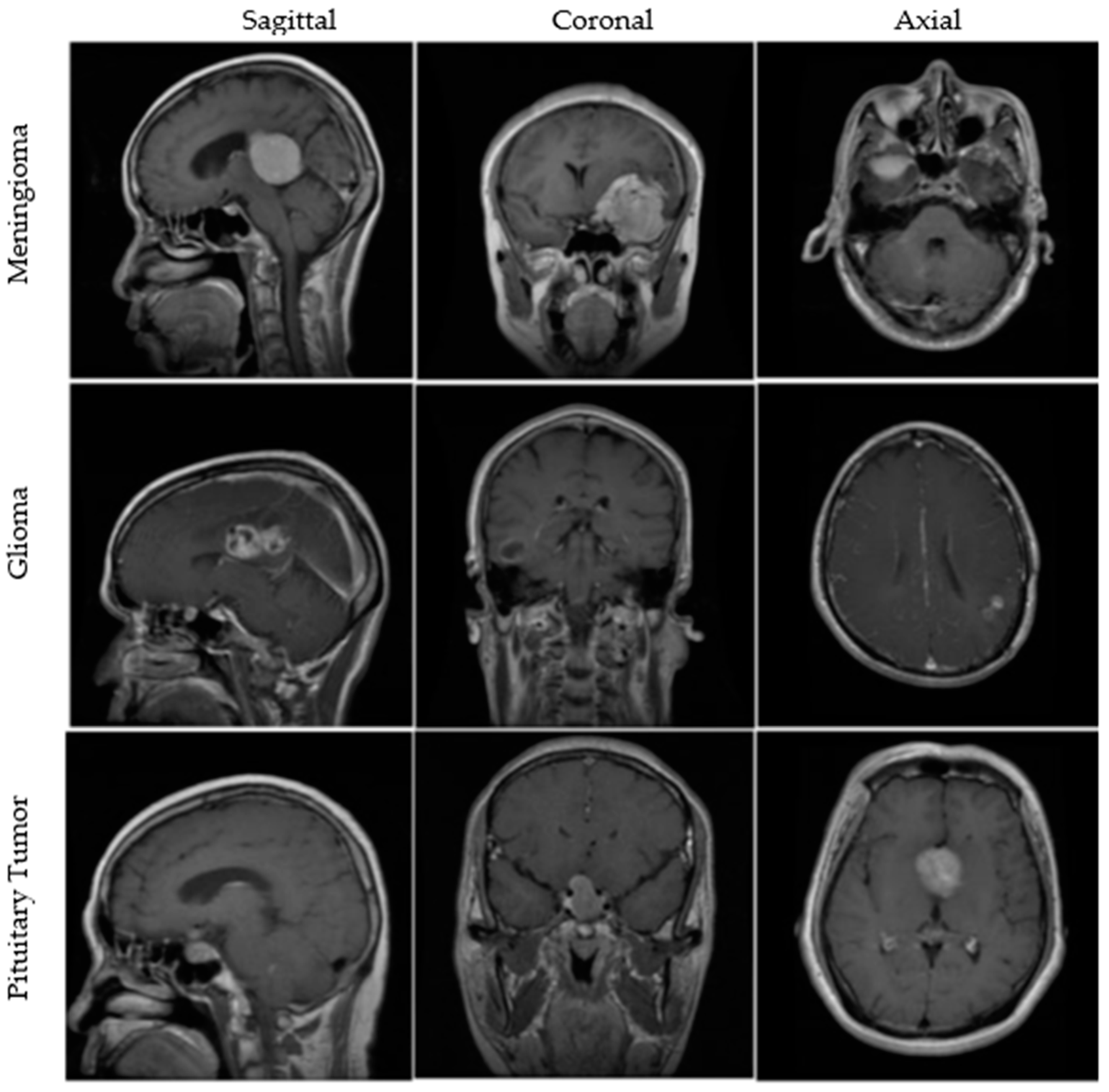
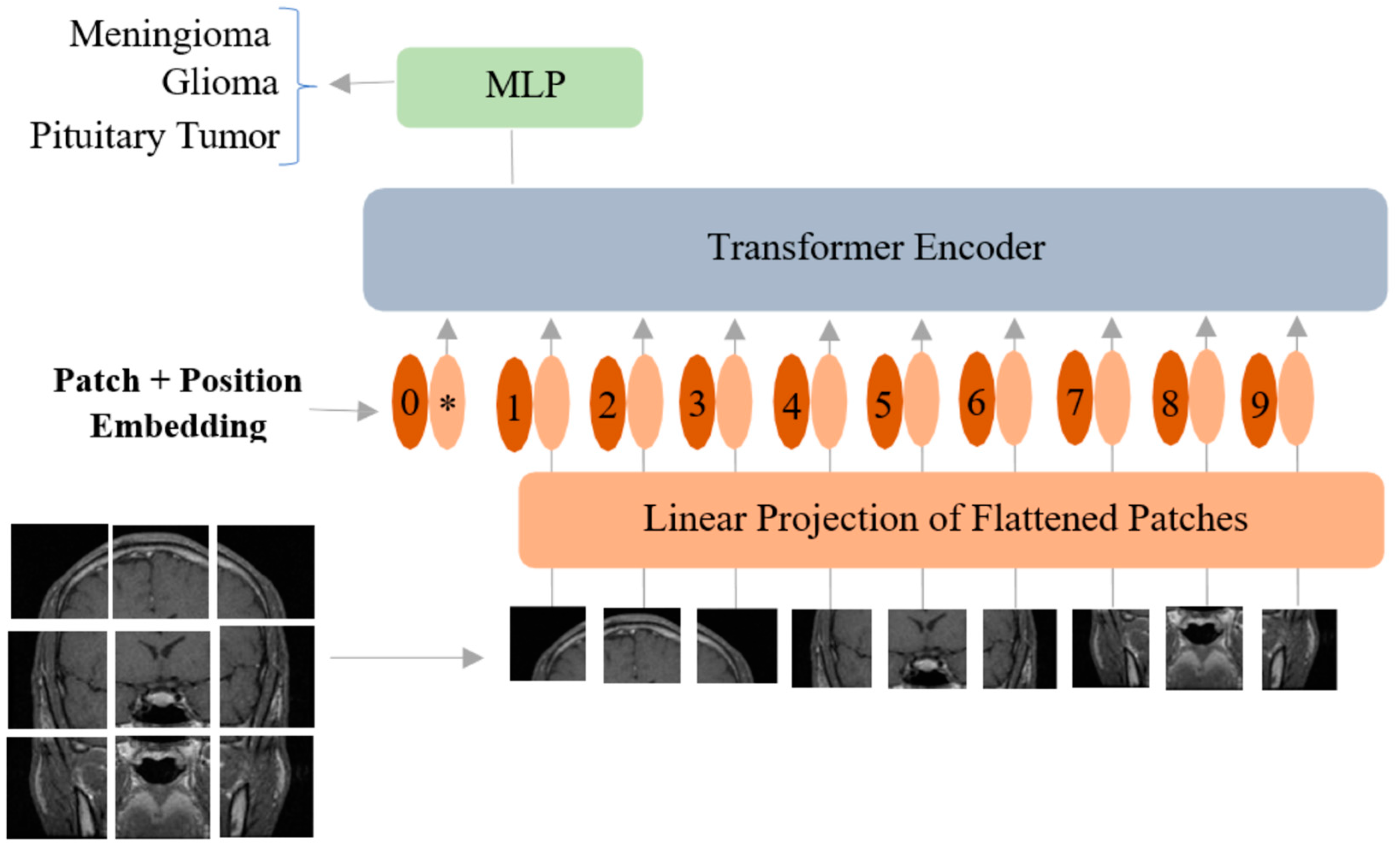

| BT Type | Total Images | Training | Validation | Testing |
|---|---|---|---|---|
| Meningioma | 708 | 502 | 75 | 131 |
| Glioma | 1426 | 988 | 148 | 290 |
| Pituitary Tumor | 930 | 647 | 91 | 192 |
| Total (N) | 3064 | 2137 | 314 | 613 |
| Resolution | Optimizers and Hyperparameters | Validation Accuracy in Percentage | |||
|---|---|---|---|---|---|
| ViT-B/16 | ViT-B/32 | ViT-L/16 | ViT-L/32 | ||
| 224 × 224 | |||||
| 384 × 384 | |||||
| Resolution | Accuracy | |||
|---|---|---|---|---|
| ViT-B/16 | ViT-B/32 | ViT-L/16 | ViT-L/32 | |
| 224 × 224 | 97.06 | 96.25 | 96.74 | 96.01 |
| 384 × 384 | 97.72 | 97.87 | 97.55 | 98.21 |
| Ensemble Model | Accuracy | Sensitivity | Specificity |
|---|---|---|---|
| All ViT models at 224 × 224 resolution | 97.71 | 96.87 | 99.10 |
| All ViT models at 384 × 384 resolution | 98.70 | 97.78 | 99.42 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tummala, S.; Kadry, S.; Bukhari, S.A.C.; Rauf, H.T. Classification of Brain Tumor from Magnetic Resonance Imaging Using Vision Transformers Ensembling. Curr. Oncol. 2022, 29, 7498-7511. https://doi.org/10.3390/curroncol29100590
Tummala S, Kadry S, Bukhari SAC, Rauf HT. Classification of Brain Tumor from Magnetic Resonance Imaging Using Vision Transformers Ensembling. Current Oncology. 2022; 29(10):7498-7511. https://doi.org/10.3390/curroncol29100590
Chicago/Turabian StyleTummala, Sudhakar, Seifedine Kadry, Syed Ahmad Chan Bukhari, and Hafiz Tayyab Rauf. 2022. "Classification of Brain Tumor from Magnetic Resonance Imaging Using Vision Transformers Ensembling" Current Oncology 29, no. 10: 7498-7511. https://doi.org/10.3390/curroncol29100590
APA StyleTummala, S., Kadry, S., Bukhari, S. A. C., & Rauf, H. T. (2022). Classification of Brain Tumor from Magnetic Resonance Imaging Using Vision Transformers Ensembling. Current Oncology, 29(10), 7498-7511. https://doi.org/10.3390/curroncol29100590








