Abstract
ROS1 rearrangements are identified in 1–2% of lung adenocarcinoma cases, and reflex testing is guideline-recommended. We developed a decision model for population-based ROS1 testing from a Canadian public healthcare perspective to determine the strategy that optimized detection of true-positive (TP) cases while minimizing costs and turnaround time (TAT). Eight diagnostic strategies were compared, including reflex single gene testing via immunohistochemistry (IHC) screening, fluorescence in-situ hybridization (FISH), next-generation sequencing (NGS), and biomarker-informed (EGFR/ALK/KRAS wildtype) testing initiated by pathologists and clinician-initiated strategies. Reflex IHC screening with FISH confirmation of positive cases yielded the best results for TAT, TP detection rate, and cost. IHC screening saved CAD 1,000,000 versus reflex FISH testing. NGS was the costliest reflex strategy. Biomarker-informed testing was cost-efficient but delayed TAT. Clinician-initiated testing was the least costly but resulted in long TAT and missed TP cases, highlighting the importance of reflex testing. Thus, reflex IHC screening for ROS1 with FISH confirmation provides a cost-efficient strategy with short TAT and maximizes the number of TP cases detected.
1. Introduction
Advances in targeted therapies in lung cancer have changed the management and subsequent disease trajectory of lung cancer patients. There are now several genomic aberrations contributing to tumorigenesis that have shown improved patient outcomes with targeted therapy; these include mutations in EGFR, BRAF, and MET and rearrangements in ALK, NTRK, RET, and ROS1, among others [1]. ROS1 gene rearrangements are present in 1–2% of all NSCLC [2,3] and are more commonly found in adenocarcinoma, younger patients, and never-smokers [3]. Crizotinib and entrectinib have been approved for the treatment of patients with ROS1-rearranged advanced non-squamous NSCLC [4,5] and have markedly improved outcomes in this patient population [6].
Methods for detecting ROS1 rearrangements include immunohistochemistry (IHC) [7], fluorescence and other in-situ hybridization techniques (FISH), and next-generation sequencing (NGS). FISH is considered the validated standard, although NGS has emerged as an important alternative [8]. Guidelines from the International Association for the Study of Lung Cancer/College of American Pathologists, American Society of Clinical Oncology (ASCO), European Society for Medical Oncology (ESMO), and the Pan-Asian adapted ESMO Clinical Practice Guidelines all recommend that patients with advanced non-squamous NSCLC have reflex testing for ROS1, irrespective of clinical characteristics [1,9,10,11]. ASCO/Ontario Health [12], ESMO [10], NCCN [13], and other groups recommend targeted treatment for this subgroup of patients.
However, not all jurisdictions have the same access to testing and targeted therapy. For example, in Ontario, Canada, public funding for ROS1 testing and targeted treatment has only recently been approved [14]. While some jurisdictions can afford routine reflex NGS testing for multiple actionable markers in lung cancer samples, many cannot. The challenges of implementing ROS1 testing as a population-based routine reflex diagnostic test include the low frequency of ROS1 rearrangements, tissue requirements, and costs associated with testing. In publicly funded health care systems and those with lesser resources, identifying the most cost-efficient diagnostic testing strategy for single-gene tests like ROS1 continues to be clinically relevant. Not all patients have funding or sufficient tissue for NGS, and not all patients are able to have repeat biopsies.
We developed a population-based ROS1 testing model for advanced non-squamous NSCLC to determine the most cost-efficient ROS1 testing strategy from a Canadian public perspective that maximizes the detection of true positive cases (TP) while minimizing costs and turnaround time (TAT). We also included biomarker-informed testing after exclusion of EGFR-, ALK-, and KRAS-positive cases in order to explore potential cost savings and clinician-initiated testing.
2. Methods
2.1. Model Structure and Diagnostic Testing Strategies
A test-based decision modeling approach was developed using ROS1 IHC, FISH, and NGS sensitivity and specificity in addition to the prevalence of ROS1 fusions. For IHC screening, positive predictive values (PPV) and negative predictive values (NPV) were calculated for each arm of the model to determine the proportion of patients who would first test positive after IHC screening and the subsequent proportion of patients who would undergo FISH confirmation (Figure 1, Tables S1–S9) [15]. Two models were developed with the calculated parameters to compare cost to true positive rates and cost to turnaround time of eight diagnostic testing strategies in patients with advanced non-squamous NSCLC (Figure S1). The target population was treatment-naïve patients with advanced non-squamous NSCLC in Ontario, Canada, estimated at 4000 new cases per year [16]. A total of 1000 patients were used for baseline value calculations. This analysis was conducted from the perspective of the Ontario health care system (University Health Network, Cancer Care Ontario, Toronto, ON, Canada), a publicly funded health care system [17].
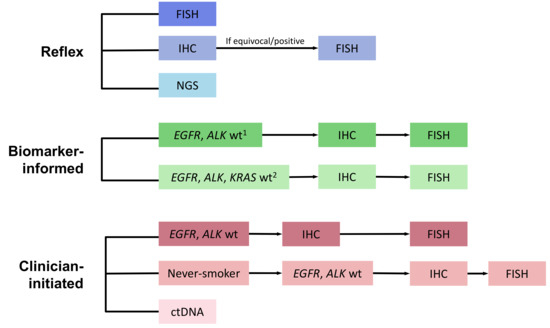
Figure 1.
ROS-1 Testing Model Schema. ctDNA—circulating tumor DNA; FISH—Fluorescence in-situ hybridization; IHC—Immunohistochemistry; NGS—Next-generation sequencing; wt—wildtype. Footnotes: 1 If EGFR, ALK, or KRAS positive, then no further testing performed; wt—wildtype; 2 PCR-based sequencing.
Eight diagnostic strategies were compared, including reflex testing, serial testing informed by biomarker results, and clinician-initiated testing (Figure 1). Methods for reflex testing at the time of diagnosis in all non-squamous NSCLC cases included: (1) FISH; (2) IHC screening followed by FISH confirmation for positive cases [18,19,20], and (3) comprehensive NGS (DNA, RNA panel) [21]. The model assumed that actionable genomic alterations were mutually exclusive. Methods for serial testing informed by biomarker results included pathologist-initiated reflex testing of EGFR, ALK, and/or KRAS wild-type cases with IHC screening and FISH confirmation (hereafter described as “biomarker-informed”). Clinician-initiated testing strategies incorporated a selection of cases with EGFR and ALK wildtype, either limited or not limited to never-smokers. Finally, circulating tumor DNA (ctDNA) testing using a commercial assay in all patients was also included as a clinician-initiated strategy [22].
2.2. Model Assumptions
All patients were assumed to have accepted testing [23]. The model did not incorporate test failure rates or the option of repeat biopsies. The model accounted for the sensitivity and specificity of the ROS1 assay using IHC (D4D6 clone (Cell Signaling) [17], FISH, NGS, and ctDNA. The sensitivities and specificities of the EGFR, ALK, and KRAS assays were not included in the model.
2.3. Costs
All costs for diagnostic assays were estimated based on current University Health Network (UHN) Laboratory Medicine Program costs and provided in Canadian dollars (Table 1). The base case values included an estimate of reagent costs, sample processing, and handling costs and were based on expert opinion. Strategies involving serial testing included costs for EGFR and KRAS testing (as appropriate), based on validated PCR assays, and ALK testing using IHC [24,25]. Costs of ctDNA were based on estimated costs of a marketed commercial assay (e.g., Guardant360™, Guardant Health, Inc., Redwood City, CA, USA). Professional fees and administrative costs were not included. Costs were varied by 20% for sensitivity analysis unless otherwise indicated. Discounting was not applied, given the fixed time horizon included. Pricing, including for commercial liquid biopsy assays, is based on testing available to Ontario clinicians at the time the model was developed.

Table 1.
Diagnostic test costs and turnaround time (TAT).
2.4. Parameter Inputs
Parameter inputs used for the model are outlined in Table S1. The prevalence of ROS1 gene rearrangements in patients with advanced NSCLC was not varied and was estimated at 1.5%. The frequencies of EGFR, ALK, and KRAS mutations in patients presenting with advanced non-squamous NSCLC were estimated as follows: EGFR mutation frequency was estimated at 17% [27], ALK at 3% [28], and KRAS at 25% [29]. We estimated the proportion of never-smokers in patients with advanced non-squamous NSCLC to be 20% (this was based on the mean of four studies that examined the proportion of never-smokers among patients presenting with NSCLC) [30,31,32,33]. The proportion of EGFR- and ALK-positive cases among never-smokers was estimated at 60% [34]. Lastly, the probability of ROS1 fusions among EGFR and ALK wild-type never-smokers was estimated at 12% [35]. We assumed that ROS1 fusions were exclusive of EGFR- and ALK-detected aberrations, such that all ROS1-positive cases would be detected among EGFR and ALK wild-type cases.
2.5. Outcomes
The primary outcomes were result turnaround time (TAT) and the proportion of true-positive (TP) cases detected. The TAT (in days) for each diagnostic testing strategy included laboratory and sample processing time and was based on current UHN laboratory practices. The time to see an oncologist was based on the current Canadian Cancer Care Ontario recommendation of a 14-day target [36]. The effectiveness score for true-positive outcomes was set at 1 at terminal nodes. All other nodes were given a score of zero (this included true negatives, false positives, and false negatives). The model did not include final patient outcomes, i.e., quality-adjusted life years or treatment outcomes.
2.6. Base Case, Cost-Effectiveness, and Sensitivity Analysis
TreeAge Pro 2020 (TreeAge Software, Inc., Williamstown, MA, USA) was used to construct the decision trees and to complete base-case and cost- analyses (Figure S1). Base-case estimates were obtained, and variations around each were used for sensitivity analysis to assess the impact of variation around model inputs. Probabilities and assay sensitivities and specificities were varied as indicated in Table S1. Univariate sensitivity analysis was conducted on the parameter of cost and reported as a Tornado plot. The statistical software Matlab was used to generate the three-dimensional graph of TAT, TP detection rate, and cost for each testing strategy.
3. Results
The rate of TP cases detected, TAT, and mean cost for each strategy are listed in Table 2. Although FISH was assumed to be the gold standard for this analysis, upfront NGS and IHC screening with FISH confirmation also yielded a high rate of TP cases. The strategy with the lowest yield of TP cases was clinician-initiated testing of patient samples that were EGFR/ALK wild-type in never smokers, missing 41% of TP cases (Figure 2). This strategy was also the least costly due to only a small proportion of patient samples undergoing testing, with a mean TAT of 25 days, longer than with pathologist-initiated reflex testing strategies. The other biomarker-informed testing strategies, i.e., testing of only EGFR/ALK wild-type samples, were also less costly due to a smaller number of cases undergoing testing. However, these all added significant length to resulting TAT. Incorporating KRAS testing into the biomarker-informed approach added cost and yielded the longest TAT (32 days, see Table 2).

Table 2.
Base case analysis of cost, turnaround time (TAT), and true-positive (TP) cases.
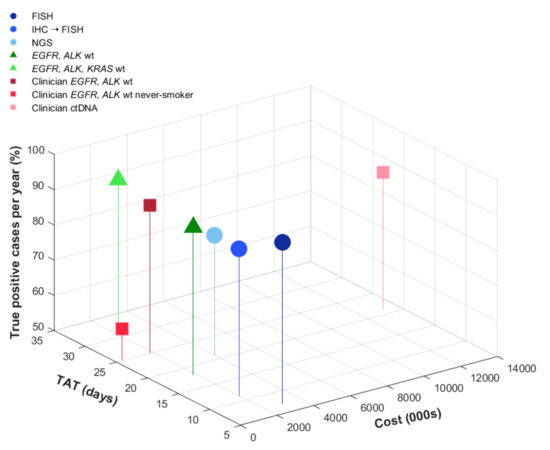
Figure 2.
Cost, true-positive cases, and turnaround time. The graph shows base case analysis results of true-positive cases detected, turnaround time (in days), and cost (CAD). Reflex strategies initiated at diagnosis are shown in blue, biomarker-informed pathologist-initiated testing in green, and clinician-initiated strategies in red.
Balancing shorter TAT and higher TP rates, reflex IHC screening for ROS1 followed by FISH confirmation was the most cost-efficient strategy. This yielded an additional CAD 28 per case or CAD 112,000 for 4000 cases compared with molecular selection for EGFR and ALK wild-type cases (Table 2). Assuming 4000 cases tested per year, this approach would cost CAD 1,940,000, which would save CAD 1,020,000 compared to initial reflex testing with FISH alone. In addition, given the high specificity of ROS1 IHC, negative cases could be rapidly signed out with the shortest TAT, approximately 3 days, allowing results to be available at the time of clinician visit (Figure 3). The proportion of TP detected by this strategy would be similar to the biomarker informed testing strategies, assuming that ROS1 fusions are mutually exclusive from EGFR mutations and ALK fusions (92%).
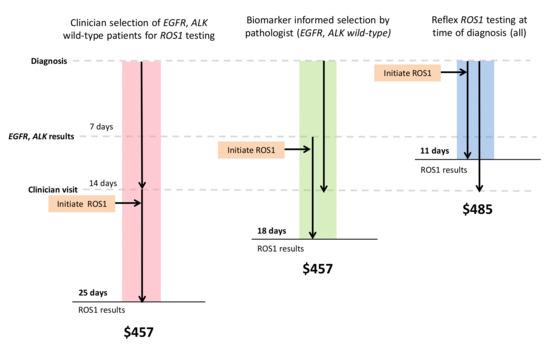
Figure 3.
IHC screening with FISH confirmation most cost-efficient. Color bars represent the three strategies that are most cost-efficient in each arm of the model (clinician-initiated, biomarker-informed, and reflex testing). Black arrows demonstrate the time at which ROS1 testing is initiated. Result turnaround time and estimated costs are labeled.
More advanced sequencing technologies required longer TAT and had a higher cost. Among reflex testing strategies, NGS had the longest TAT (21 days) and was estimated to cost approximately CAD 4 million for 4000 cases. This approach would capture 84% (78–99%) of TP cases based on a sensitivity of 0.84 (base-value) and specificity of 0.99. The strategy of clinician-initiated ctDNA testing using commercial assays was most costly, estimated at CAD 13 million for 4000 cases (testing costs only).
Sensitivity Analysis
One-way sensitivity analysis was performed to test the influence of various parameters on cost. Reflex strategies were included in the analysis. The proportion of cases testing positive by IHC, which includes the sum of true positives and false-positive cases, had the greatest impact on cost (Figure 4). False-positive cases were also an important driver of cost in the case of the lower specificity of the ROS1 IHC assay. With a higher false-positive rate by IHC, more cases require FISH confirmation, increasing overall cost. The cost of the IHC and FISH assay also influenced overall costs.
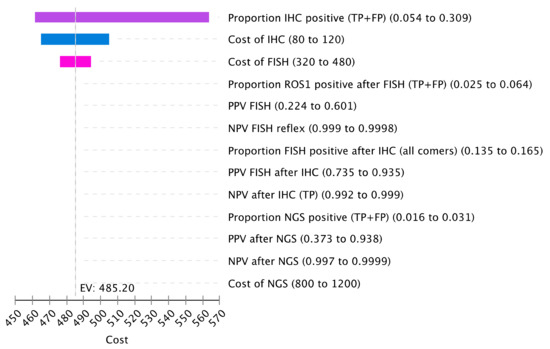
Figure 4.
Sensitivity Analysis of Cost Drivers. The tornado diagram represents the one-way sensitivity analysis for cost. Select variables for cost were included for this plot. Input parameters are on the right with the listed variation. Variable definitions are listed in Table S9.
4. Discussion
Current biomarker testing strategies for patients with advanced NSCLC include testing for mutations and gene fusions that have approved targeted therapies with evidence of improved outcomes [1]. This includes aberrations in EGFR, ALK, expression of PD-L1, among several others [37]. Although more recent, the recommendation for ROS1 testing is well-established, but the gold standard method to detect these gene rearrangements has been FISH testing, which is costly to apply broadly at a population level [19,20,38]. In this study, we used a decision-tree-based analytic model to compare the cost, result TAT, and TP detection rate between different diagnostic testing strategies to detect ROS1 gene rearrangements. We show that reflex testing for ROS1 fusions using IHC screening followed by FISH confirmation is the most cost-efficient approach. As part of reflex or routine testing, this strategy provides short TAT and a high proportion of TP detected as compared with other testing strategies. This is in large part due to the high sensitivity of the IHC assay, and concerns regarding lower specificity can be overcome with confirmation of positive cases by FISH testing [39].
Clinician-initiated testing is suboptimal for several reasons. Clinician-initiated strategies added 14 days to result TAT, based on current wait times to see a clinician from the diagnostic biopsy. We have previously shown that delays in molecular result TAT lead to delays in initiation of patient treatment and missed opportunities for targeted therapy [40]. The case for reflex testing is even more compelling when we consider that clinician-initiated testing is not systematic. We believe our strategy of clinicians requesting ROS-1 testing preferentially in patients that are never-smokers with EGFR/ALK wild-type tumors is common practice in systems without reflex testing. In our model, this approach missed up to 41% of TP cases and had prolonged TAT. Our model further highlights the importance of reflex testing, as lack of systematic testing will lead to less access to targeted therapy and inferior outcomes for patients.
The limitations of our study include the use of three single-gene assays for EGFR, ALK, and ROS-1, a current reality in our public system, in comparison to the growing practice of broader NGS testing for multiple targets now relevant in the care of advanced NSCLC patients (Figure 5, [41]). The costs of additional genomic targets in BRAF, NTRK, RET, MET, and emerging targets in KRAS, ERBB2, and NRG-1 were not included in our model as these are not yet funded as a standard of care in our jurisdiction. NGS testing platforms will become more widespread and will hopefully be incorporated as reflex testing around the world, in keeping with international guidelines [1], as they offer more complete genotyping, including the detection of uncommon genomic aberrations [42] and multiplexed target analysis, which reduces tissue consumption, all of which can impact clinical outcomes [43,44].
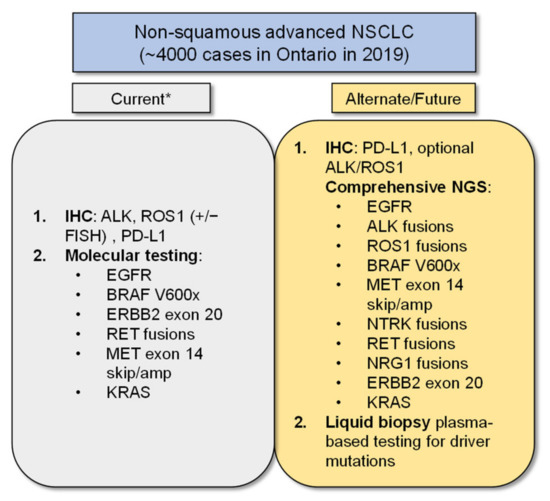
Figure 5.
Ideal Biomarker Testing. The schematic shows current and future biomarker testing options for patients. Footnote: * At our institution, comprehensive NGS testing in addition to reflex PD-L1 IHC testing is now standard practice, but this has not been implemented across the province or country at the time this manuscript was written. This figure was adapted with modifications from [41].
In our model, NGS was associated with the longest TAT and significant cost, although these costs may fall over time. In an EGFR mutant predominant population, upfront tissue NGS did not lead to significant increases in cost compared with standard of care molecular testing, and detected additional patients eligible for targeted therapy with short TAT [45]. In the United States, NGS has been shown to be cost-effective based on testing for seven or more genomic targets, although these results are not necessarily generalizable to other jurisdictions [46]. Similarly, our costs, based on the Ontario (Canada) public health system, may also have limited generalizability. For patients without comprehensive genomic tumor testing, those with small diagnostic samples, and those in jurisdictions with restricted health care funding, we believe that single-gene tests will remain relevant. However, it is hoped that NGS will become more accessible worldwide to allow more complete genotyping with a growing number of genomic targets needed for treatment decision-making for patients with advanced lung cancer.
Liquid biopsy is an emerging and important technology to ensure that patients can have complete genotyping and has increasingly demonstrated clinical validity and utility in several prospective studies, especially in cases of insufficient tissue [26,47,48]. In our analysis, we used commercial list prices for this technology, resulting in routine ctDNA testing in all patients plus EGFR/ALK testing in tissue as the most expensive population-based testing strategy. Recent recommendations for plasma ctDNA testing include NGS-based platforms in treatment-naïve patients and in those who have developed drug resistance, now allowing a growing list of guideline-recommended targets to be assessed [49]. Although such platforms offer broad-based testing of known and unknown variants, non-NGS PCR-based ctDNA assays (either as a single gene or multiplexed), known to be highly sensitive for detecting a select few targetable alterations, may still offer a lower-cost alternative in resource-limited settings. However, liquid biopsy has been shown to help to avoid repeat tissue biopsy in patients with small samples, increase the identification of patients with actionable alterations, which may offset costs and offer short TAT [26,50]. Furthermore, it is expected that the costs of liquid biopsy technology will decrease over time, and the use of non-commercial assays or price negotiation may make this strategy more affordable in future.
Another limitation of our analysis is that our clinical estimates of probabilities are based on published literature and expert opinion. A key concern with the use of ROS1 IHC screening relates to the variable specificity of current assays. With two antibody clones available and multiple ways of scoring the IHC assay (based on staining patterns, staining strengths, and H-score cutoffs) [1], confirmation by FISH (or NGS) remains necessary as part of the current standard. Validating the assay in accredited laboratories to ensure best practice continues to be of great importance in ensuring optimal patient outcomes. Lastly, the model does not take into account treatment outcomes, treatment costs, and the potential impact of false-negative results.
5. Conclusions
In conclusion, we found that reflex ROS1 testing with IHC followed by FISH confirmation in positive cases is a cost-efficient approach with a high yield of true-positive cases and favorable result TAT (3 days or less for negative cases). Our study, which is conducted from the perspective of a publicly funded health care system (Ontario, Canada), underscores the importance of system-wide reflex testing for key biomarkers in lung cancer to optimize TAT and TP rates. Clinician-led strategies resulted in much longer TAT and the potential for missed cases. Biomarker-informed selection led to smaller testing volumes and therefore decreased costs, but significantly prolonged TAT. As the affordability of broad-spectrum NGS testing improves over time, we expect to see a shift from single-gene testing approaches to multiplexed approaches, even in jurisdictions with restricted healthcare funding. Until then, we believe our model demonstrates a feasible and affordable way to incorporate routine ROS-1 testing in those jurisdictions that continue to struggle with the rising costs of cancer care, including genomic testing.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/curroncol28050284/s1, Figure S1: Decision tree model of testing strategies, Table S1: Model Parameter inputs, Table S2: FISH test characteristics, Table S3: IHC screening (followed by FISH confirmation) test characteristics, Table S4: NGS test characteristics, Table S5: Biomarker informed selection of EGFR, ALK wildtype samples for ROS1 IHC screening and FISH confirmation, Table S6: Biomarker informed selection of EGFR, ALK, KRAS wildtype samples for ROS1 IHC screening and FISH confirmation, Table S7: Clinical selection of EGFR, ALK wildtype samples in never-smokers for ROS1 IHC screening and FISH confirmation, Table S8: ctDNA test characteristics, Table S9: Variable definitions.
Author Contributions
M.M.: Conception and Design, Collection and Assembly of Data, Analysis and Interpretation, Manuscript Writing (first draft, editing), and Final Approval; D.A.E.: Conception and Design, Collection and Assembly of Data, Analysis and Interpretation, Manuscript Writing, and Final Approval; A.C.S.: Conception and Design, Collection and Assembly of Data, Analysis and Interpretation, Supervision, Manuscript Writing, and Final Approval; J.J.N.L.: Collection and Assembly of Data, Manuscript Writing, and Final Approval; J.H.L.: Collection and Assembly of Data, Manuscript Writing, and Final Approval; M.-S.T.: Conception and Design, Collection and Assembly of Data, Analysis and Interpretation, Supervision, Manuscript Writing, and Final Approval; N.B.L.: Conception and Design, Collection and Assembly of Data, Analysis and Interpretation, Supervision, Manuscript Writing, and Final Approval. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Princess Margaret Cancer Foundation (OSI Pharmaceuticals Foundation Chair (N.B.L.) and Qasim Choksi Chair (M.-S.T.)).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data generated or analyzed during this study are included in this article (and its supplementary information files).
Conflicts of Interest
M.M., D.A.E., A.C.S., J.J.N.L., and J.H.L. have no conflict to declare; N.B.L. has received unrelated institutional research funding from Guardant Health, Pfizer, and Roche and is supported through the Princess Margaret Cancer Foundation (OSI Pharmaceuticals Foundation Chair). M.-S.T. received research grants (Institution) and honorarium (personal) from Pfizer.
References
- Lindeman, N.I.; Cagle, P.T.; Aisner, D.L.; Arcila, M.E.; Beasley, M.B.; Bernicker, E.H.; Colasacco, C.; Dacic, S.; Hirsch, F.R.; Kerr, K.; et al. Updated Molecular Testing Guideline for the Selection of Lung Cancer Patients for Treatment with Targeted Tyrosine Kinase Inhibitors: Guideline from the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. Arch. Pathol. Lab. Med. 2018, 142, 321–346. [Google Scholar] [CrossRef]
- Gainor, J.F.; Shaw, A.T. Novel Targets in Non-Small Cell Lung Cancer: ROS1 and RET Fusions. Oncologist 2013, 18, 865–875. [Google Scholar] [CrossRef]
- Bergethon, K.; Shaw, A.T.; Ou, S.-H.I.; Katayama, R.; Lovly, C.M.; McDonald, N.T.; Massion, P.P.; Siwak-Tapp, C.; Gonzalez, A.; Fang, R.; et al. ROS1 rearrangements define a unique molecular class of lung cancers. J. Clin. Oncol. 2012, 30, 863–870. [Google Scholar] [CrossRef]
- Regulatory Decision Summary–Xalkori–Health Canada. Available online: https://hpr-rps.hres.ca/reg-content/regulatory-decision-summary-detail.php?lang=en&linkID=RDS00285 (accessed on 24 January 2020).
- ROZLYTREK (Entrectinib) Product Monograph. Available online: https://www.rochecanada.com/content/dam/rochexx/roche-ca/products/ConsumerInformation/MonographsandPublicAdvisories/Rozlytrek/Rozlytrek_PM_E.pdf (accessed on 16 May 2021).
- Shaw, A.; Riely, G.; Bang, Y.-J.; Kim, D.-W.; Camidge, D.; Solomon, B.; Varella-Garcia, M.; Iafrate, A.; Shapiro, G.; Usari, T.; et al. Crizotinib in ROS1-rearranged advanced non-small-cell lung cancer (NSCLC): Updated results, including overall survival, from PROFILE 1001. Ann. Oncol. 2019, 30, 1121–1126. [Google Scholar] [CrossRef]
- Sholl, L.M.; Sun, H.; Butaney, M.; Zhang, C.; Lee, C.; Jänne, P.A.; Rodig, S.J. ROS1 Immunohistochemistry for Detection of ROS1-Rearranged Lung Adenocarcinomas. Am. J. Surg. Pathol. 2013, 37, 1441–1449. [Google Scholar] [CrossRef] [PubMed]
- Rossi, G.; Jocollé, G.; Conti, A.; Tiseo, M.; Marino, F.Z.; Donati, G.; Franco, R.; Bono, F.; Barbisan, F.; Facchinetti, F. Detection of ROS1 rearrangement in non-small cell lung cancer: Current and future perspectives. Lung Cancer Targets Ther. 2017, 8, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Kalemkerian, G.P.; Narula, N.; Kennedy, E.B. Molecular Testing Guideline for the Selection of Lung Cancer Patients for Treatment with Targeted Tyrosine Kinase Inhibitors: American Society of Clinical Oncology Endorsement Summary of the College of American Pathologists/International Association for the Study of Lung Cancer/Association for Molecular Pathology Clinical Practice Guideline Update. J. Oncol. Pract. 2018, 14, 323–327. [Google Scholar] [PubMed]
- Planchard, D.; Popat, S.; Kerr, K.; Novello, S.; Smit, E.F.; Faivre-Finn, C.; Mok, T.S.; Reck, M.; Van Schil, P.E.; Hellmann, M.D.; et al. Metastatic non-small cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2018, 29, iv192–iv237. [Google Scholar] [CrossRef]
- Wu, Y.-L.; Planchard, D.; Lu, S.; Sun, H.; Yamamoto, N.; Kim, D.-W.; Tan, D.; Yang, J.C.-H.; Azrif, M.; Mitsudomi, T.; et al. Pan-Asian adapted Clinical Practice Guidelines for the management of patients with metastatic non-small-cell lung cancer: A CSCO–ESMO initiative endorsed by JSMO, KSMO, MOS, SSO and TOS. Ann. Oncol. 2019, 30, 171–210. [Google Scholar] [CrossRef] [PubMed]
- Hanna, N.H.; Robinson, A.G.; Temin, S.; Baker, S.; Brahmer, J.R.; Ellis, P.M.; Gaspar, L.E.; Haddad, R.Y.; Hesketh, P.J.; Jain, D.; et al. Therapy for Stage IV Non–Small-Cell Lung Cancer with Driver Alterations: ASCO and OH (CCO) Joint Guideline Update. J. Clin. Oncol. 2021, 39, 1040–1091. [Google Scholar] [CrossRef]
- Ettinger, D.S.; Wood, D.E.; Aisner, D.L.; Akerley, W.; Bauman, J.R.; Bharat, A.; Bruno, D.S.; Chang, J.Y.; Chirieac, L.R.; D’Amico, T.A.; et al. NCCN Guidelines Insights: Non–Small Cell Lung Cancer, Version 2.2021. J. Natl. Compr. Cancer Netw. 2021, 19, 254–266. [Google Scholar] [CrossRef]
- Rozlytrek—Notice of Compliance with Conditions—Qualifying Notice. Available online: https://www.canada.ca/en/health-canada/services/drugs-health-products/drug-products/notice-compliance/conditions/rozlytrek-qualifying-notice.html (accessed on 22 August 2021).
- Rautenberg, T.; Gerritsen, A.; Downes, M. Health Economic Decision Tree Models of Diagnostics for Dummies: A Pictorial Primer. Diagnostics 2020, 10, 158. [Google Scholar] [CrossRef] [PubMed]
- Canadian Cancer Society’s Advisory Committee on Cancer Statistics. Canadian Cancer Statistics, Special Topic: Pancreatic Cancer; Canadian Cancer Society: Toronto, ON, Canada, 2017. [Google Scholar]
- Cheung, C.C.; Smith, A.C.; Albadine, R.; Bigras, G.; Bojarski, A.; Couture, C.; Cutz, J.-C.; Huang, W.-Y.; Ionescu, D.; Itani, D.; et al. Canadian ROS proto-oncogene 1 Study (CROS) for multi-institutional implementation of ROS1 testing in non-small cell lung cancer. Lung Cancer 2021, in press. [Google Scholar] [CrossRef]
- Lin, J.J.; Shaw, A.T. Recent Advances in Targeting ROS1 in Lung Cancer. J. Thorac. Oncol. 2017, 12, 1611–1625. [Google Scholar] [CrossRef]
- Shan, L.; Lian, F.; Guo, L.; Qiu, T.; Ling, Y.; Ying, J.; Lin, D. Detection of ROS1 Gene Rearrangement in Lung Adenocarcinoma: Comparison of IHC, FISH and Real-Time RT-PCR. PLoS ONE 2015, 10, e0120422. [Google Scholar] [CrossRef] [PubMed]
- Viola, P.; Maurya, M.; Croud, J.; Gazdova, J.; Suleman, N.; Lim, E.; Newsom-Davis, T.; Plowman, N.; Rice, A.; Montero, M.A.; et al. A Validation Study for the Use of ROS1 Immunohistochemical Staining in Screening for ROS1 Translocations in Lung Cancer. J. Thorac. Oncol. 2016, 11, 1029–1039. [Google Scholar] [CrossRef]
- Velizheva, N.P.; Rechsteiner, M.P.; Valtcheva, N.; Freiberger, S.N.; Wong, C.E.; Vrugt, E.; Zhong, Q.; Wagner, U.; Moch, H.; Hillinger, S.; et al. Targeted next-generation-sequencing for reliable detection of targetable rearrangements in lung adenocarcinoma-a single center retrospective study. Pathol. Res. Pract. 2018, 214, 572–578. [Google Scholar] [CrossRef]
- Lanman, R.B.; Mortimer, S.A.; Zill, O.A.; Sebisanovic, D.; Lopez, R.; Blau, S.; Collisson, E.A.; Divers, S.G.; Hoon, D.; Kopetz, S.; et al. Analytical and Clinical Validation of a Digital Sequencing Panel for Quantitative, Highly Accurate Evaluation of Cell-Free Circulating Tumor DNA. PLoS ONE 2015, 10, e0140712. [Google Scholar] [CrossRef]
- Cuffe, S.; Hon, H.; Qiu, X.; Tobros, K.; Wong, C.-K.A.; De Souza, B.; McFarlane, G.; Masroor, S.; Azad, A.K.; Hasani, E.; et al. Cancer patients’ acceptance, understanding, and willingness-to-pay for pharmacogenomic testing. Pharm. Genom. 2014, 24, 348–355. [Google Scholar] [CrossRef]
- Fiset, P.O.; Labbé, C.; Young, K.; Craddock, K.J.; Smith, A.C.; Tanguay, J.; Pintilie, M.; Wang, R.; Torlakovic, E.; Cheung, C.; et al. Anaplastic lymphoma kinase 5A4 immunohistochemistry as a diagnostic assay in lung cancer: A Canadian reference testing center’s results in population-based reflex testing. Cancer 2019, 125, 4043–4051. [Google Scholar] [CrossRef]
- Martin, P.; Shiau, C.J.; Pasic, M.; Tsao, M.; Kamel-Reid, S.; Lin, S.; Tudor, R.; Cheng, S.; Higgins, B.; Burkes, R.; et al. Clinical impact of mutation fraction in epidermal growth factor receptor mutation positive NSCLC patients. Br. J. Cancer 2016, 114, 616–622. [Google Scholar] [CrossRef][Green Version]
- Leighl, N.B.; Page, R.D.; Raymond, V.M.; Daniel, D.B.; Divers, S.G.; Reckamp, K.L.; Villalona-Calero, M.A.; Dix, D.; Odegaard, J.I.; Lanman, R.B.; et al. Clinical Utility of Comprehensive Cell-free DNA Analysis to Identify Genomic Biomarkers in Patients with Newly Diagnosed Metastatic Non–small Cell Lung Cancer. Clin. Cancer Res. 2019, 25, 4691–4700. [Google Scholar] [CrossRef]
- Ellis, P.M.; Verma, S.; Sehdev, S.; Younus, J.; Leighl, N.B. Challenges to implementation of an epidermal growth factor receptor testing strategy for non-small-cell lung cancer in a publicly funded health care system. J. Thorac. Oncol. 2013, 8, 1136–1141. [Google Scholar] [CrossRef] [PubMed]
- Hallberg, B.; Palmer, R.H. Mechanistic insight into ALK receptor tyrosine kinase in human cancer biology. Nat. Rev. Cancer 2013, 13, 685–700. [Google Scholar] [CrossRef] [PubMed]
- Kris, M.G.; Johnson, B.E.; Kwiatkowski, D.J.; Iafrate, A.J.; Wistuba, I.I.; AronsonJ, S.L.; Engelman, A.; Shyr, Y.; Khuri, F.R.; Rudin, C.M.; et al. Identification of driver mutations in tumor specimens from 1000 patients with lung adenocarcinoma: The NCI’s Lung Cancer Mutation Consortium (LCMC). J. Clin. Oncol. 2011, 29 (Suppl. S15), CRA7506. [Google Scholar] [CrossRef]
- Cho, J.; Choi, S.M.; Lee, J.; Lee, C.-H.; Lee, S.-M.; Kim, D.-W.; Yim, J.-J.; Kim, Y.T.; Yoo, C.-G.; Kim, Y.W.; et al. Proportion and clinical features of never-smokers with non-small cell lung cancer. Chin. J. Cancer 2017, 36, 20. [Google Scholar] [CrossRef]
- Pelosof, L.; Ahn, C.; Gao, A.; Horn, L.; Madrigales, A.; Cox, J.; McGavic, D.; Minna, J.D.; Gazdar, A.F.; Schiller, J. Proportion of Never-Smoker Non–Small Cell Lung Cancer Patients at Three Diverse Institutions. J. Natl. Cancer Inst. 2017, 109, 295. [Google Scholar] [CrossRef] [PubMed]
- Santoro, I.L.; Ramos, R.P.; Franceschini, J.; Jamnik, S.; Fernandes, A.L.G. Non-small cell lung cancer in never smokers: A clinical entity to be identified. Clinics 2011, 66, 1873–1877. [Google Scholar] [CrossRef] [PubMed]
- Dias, M.; Linhas, R.; Campainha, S.; Conde, S.; Barroso, A. Lung cancer in never-smokers–what are the differences? Acta Oncol. 2017, 56, 931–935. [Google Scholar] [CrossRef]
- Korpanty, G.J.; Kamel-Reid, S.; Pintilie, M.; Hwang, D.M.; Zer, A.; Liu, G.; Leighl, N.B.; Feld, R.; Siu, L.L.; Bedard, P.L.; et al. Lung cancer in never smokers from the Princess Margaret Cancer Centre. Oncotarget 2018, 9, 22559–22570. [Google Scholar] [CrossRef]
- Marchetti, A.; Barberis, M.; Di Lorito, A.; Pace, M.V.; Di Lisio, C.; Felicioni, L.; Guerini-Rocco, E.; Vingiani, A.; D’Antuono, T.; Liberatore, M.; et al. ROS1 Gene Fusion in Advanced Lung Cancer in Women: A Systematic Analysis, Review of the Literature, and Diagnostic Algorithm. JCO Precis. Oncol. 2017, 2017, 1–9. [Google Scholar] [CrossRef]
- Lim, C.; Sekhon, H.S.; Cutz, J.C.; Hwang, D.M.; Kamel-Reid, S.; Carter, R.F.; da Cunha Santos, G.; Waddell, T.; Binnie, M.; Patel, M.; et al. Improving molecular testing and personalized medicine in non-small-cell lung cancer in Ontario. Curr. Oncol. 2017, 24, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Kerr, K.M.; Bubendorf, L.; Edelman, M.; Marchetti, A.; Mok, T.; Novello, S.; O’Byrne, K.; Stahel, R.; Peters, S.; Felip, E.; et al. Second ESMO consensus conference on lung cancer: Pathology and molecular biomarkers for non-small-cell lung cancer. Ann. Oncol. 2014, 25, 1681–1690. [Google Scholar] [CrossRef]
- Yoshida, A.; Tsuta, K.; Wakai, S.; Arai, Y.; Asamura, H.; Shibata, T.; Furuta, K.; Kohno, T.; Kushima, R. Immunohistochemical detection of ROS1 is useful for identifying ROS1 rearrangements in lung cancers. Mod. Pathol. 2014, 27, 711–720. [Google Scholar] [CrossRef]
- Selinger, C.I.; Lisa, H.; Pavlakis, N.; Links, M.; Gill, A.J.; Lee, A.; Clarke, S.; Tran, T.N.; Lum, T.; Yip, P.Y.; et al. Screening forROS1gene rearrangements in non-small-cell lung cancers using immunohistochemistry with FISH confirmation is an effective method to identify this rare target. Histopathology 2017, 70, 402–411. [Google Scholar] [CrossRef]
- Lim, C.; Tsao, M.; Le, L.W.; Shepherd, F.A.; Feld, R.; Burkes, R.L.; Liu, G.; Kamel-Reid, S.; Hwang, D.; Tanguay, J.; et al. Biomarker testing and time to treatment decision in patients with advanced nonsmall-cell lung cancer. Ann. Oncol. 2015, 26, 1415–1421. [Google Scholar] [CrossRef]
- Bebb, D.G.; Banerji, S.; Blais, N.; Desmeules, P.; Gill, S.; Grin, A.; Feilotter, H.; Hansen, A.R.; Hyrcza, M.; Krzyzanowska, M.; et al. Canadian Consensus for Biomarker Testing and Treatment of TRK Fusion Cancer in Adults. Curr. Oncol. 2021, 28, 523–548. [Google Scholar] [CrossRef] [PubMed]
- Russo, A.; Franchina, T.; Ricciardi, G.; Battaglia, A.; Picciotto, M.; Adamo, V. Heterogeneous Responses to Epidermal Growth Factor Receptor (EGFR) Tyrosine Kinase Inhibitors (TKIs) in Patients with Uncommon EGFR Mutations: New Insights and Future Perspectives in this Complex Clinical Scenario. Int. J. Mol. Sci. 2019, 20, 1431. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.M.; Kim, E.Y.; Kim, H.R.; Ali, S.M.; Greenbowe, J.R.; Shim, H.S.; Chang, H.; Lim, S.; Paik, S.; Cho, B.C.; et al. Genomic profiling of lung adenocarcinoma patients reveals therapeutic targets and confers clinical benefit when standard molecular testing is negative. Oncotarget 2016, 7, 24172–24178. [Google Scholar] [CrossRef] [PubMed]
- Drilon, A.; Wang, L.; Arcila, M.E.; Balasubramanian, S.; Greenbowe, J.R.; Ross, J.S.; Stephens, P.; Lipson, D.; Miller, V.A.; Kris, G.M.; et al. Broad, Hybrid Capture-Based Next-Generation Sequencing Identifies Actionable Genomic Alterations in Lung Adenocarcinomas Otherwise Negative for Such Alterations by Other Genomic Testing Approaches. Clin. Cancer Res. 2015, 21, 3631–3639. [Google Scholar] [CrossRef]
- Tan, A.C.; Lai, G.G.; Tan, G.S.; Poon, S.Y.; Doble, B.; Lim, T.H.; Aung, Z.W.; Takano, A.; Tan, W.L.; Ang, M.-K.; et al. Utility of incorporating next-generation sequencing (NGS) in an Asian non-small cell lung cancer (NSCLC) population: Incremental yield of actionable alterations and cost-effectiveness analysis. Lung Cancer 2020, 139, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Pennell, N.; Mutebi, A.; Zhou, Z.-Y.; Ricculli, M.L.; Tang, W.; Wang, H.; Guerin, A.; Arnhart, T.; Dalal, A.; Sasane, M.; et al. Economic Impact of Next-Generation Sequencing Versus Single-Gene Testing to Detect Genomic Alterations in Metastatic Non–Small-Cell Lung Cancer Using a Decision Analytic Model. JCO Precis. Oncol. 2019, 2019, 1–9. [Google Scholar] [CrossRef]
- Remon, J.; Lacroix, L.; Jovelet, C.; Caramella, C.; Howarth, K.; Plagnol, V.; Rosenfeld, N.; Morris, C.; Mezquita, L.; Pannet, C.; et al. Real-World Utility of an Amplicon-Based Next-Generation Sequencing Liquid Biopsy for Broad Molecular Profiling in Patients with Advanced Non–Small-Cell Lung Cancer. JCO Precis. Oncol. 2019, 3, 1–14. [Google Scholar] [CrossRef]
- Zugazagoitia, J.; Ramos, I.; Trigo, J.M.; Palka, M.; Gómez-Rueda, A.; Jantus-Lewintre, E.; Camps, C.; Isla, D.; Iranzo, P.; Ponce-Aix, S.; et al. Clinical utility of plasma-based digital next-generation sequencing in patients with advance-stage lung adenocarcinomas with insufficient tumor samples for tissue genotyping. Ann. Oncol. 2019, 30, 290–296. [Google Scholar] [CrossRef]
- Rolfo, C.; Mack, P.; Scagliotti, G.V.; Aggarwal, C.; Arcila, M.E.; Barlesi, F.; Bivona, T.; Diehn, M.; Dive, C.; Dziadziuszko, R.; et al. Liquid Biopsy for Advanced Non-Small Cell Lung Cancer: A Consensus Statement from The International Association for the Study of Lung Cancer (IASLC). J. Thorac. Oncol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Leighl, N.B.; Kamel-Reid, S.; Cheema, P.K.; Laskin, J.; Karsan, A.; Zhang, T.; Stockley, T.; Barnes, T.A.; Tudor, R.A.; Liu, G.; et al. Multicenter Validation Study to Implement Plasma Epidermal Growth Factor Receptor T790M Testing in Clinical Laboratories. JCO Precis. Oncol. 2020, 2020, 520–533. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).