Age-Related Differences in miRNA Expression in Mexican-American Newborns and Children
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Subjects
2.2. Blood Collection and Processing
2.3. RNA Isolation
2.4. nCounter miRNA Expression Assay
2.5. Cell Composition
2.6. Statistical Analysis
3. Results
3.1. Characteristics of Study Participants
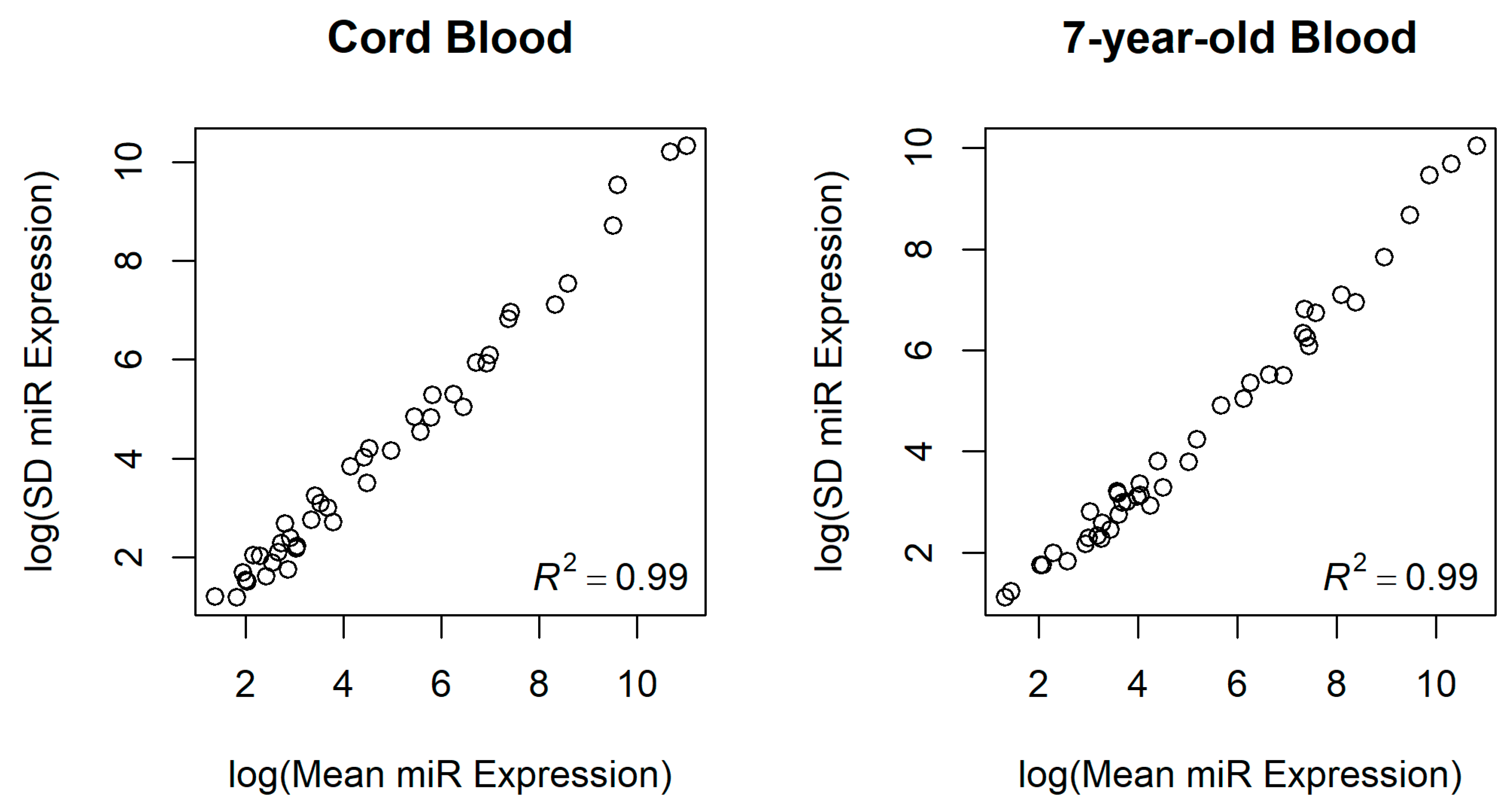
3.2. miRNA Expression in Newborns and 7-Year-Old Children
3.3. miRNA Expression and Blood Cell Count Proportions
3.4. Differential miRNA Expression by Sex in Newborns and 7-Year-Old Children
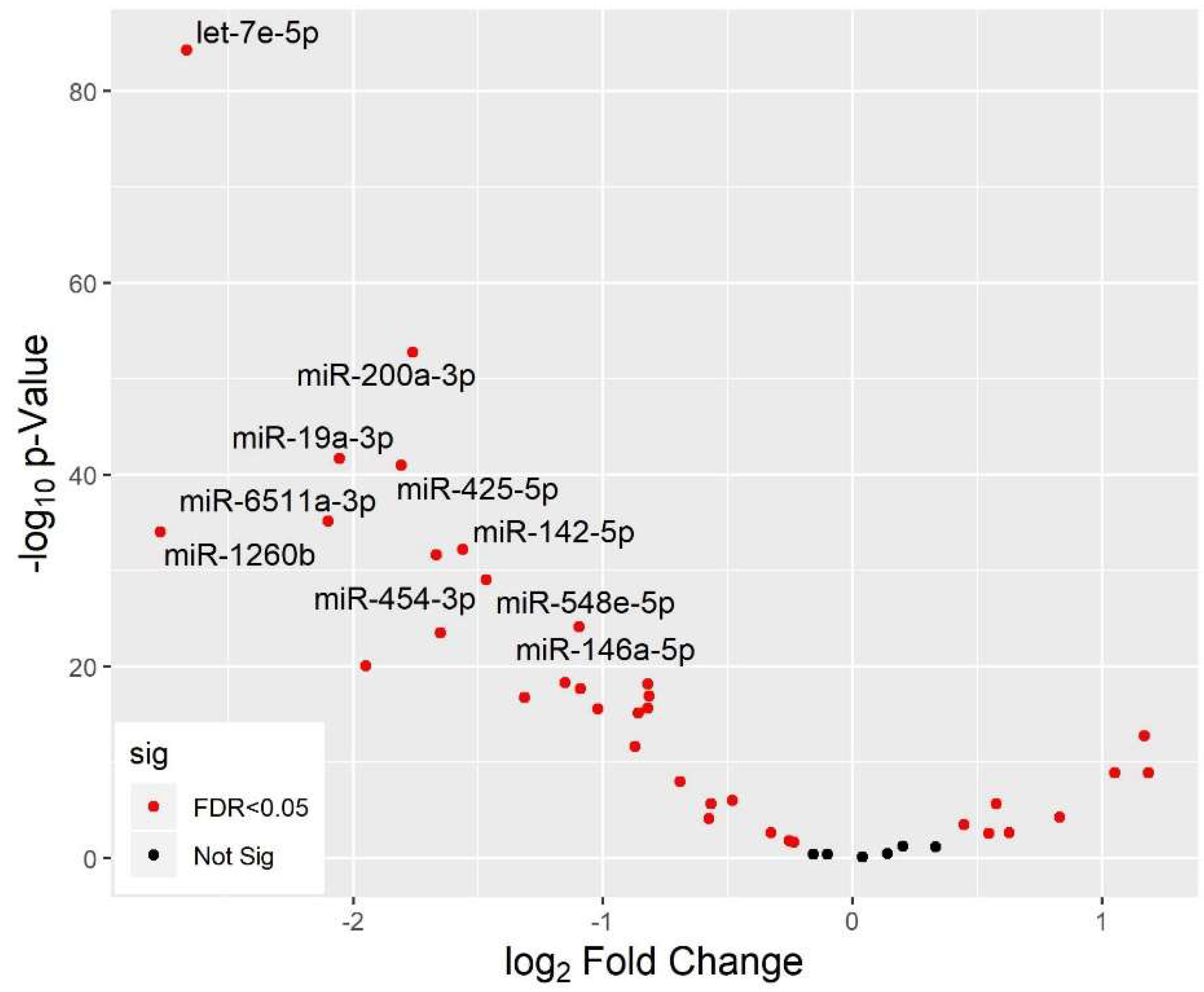
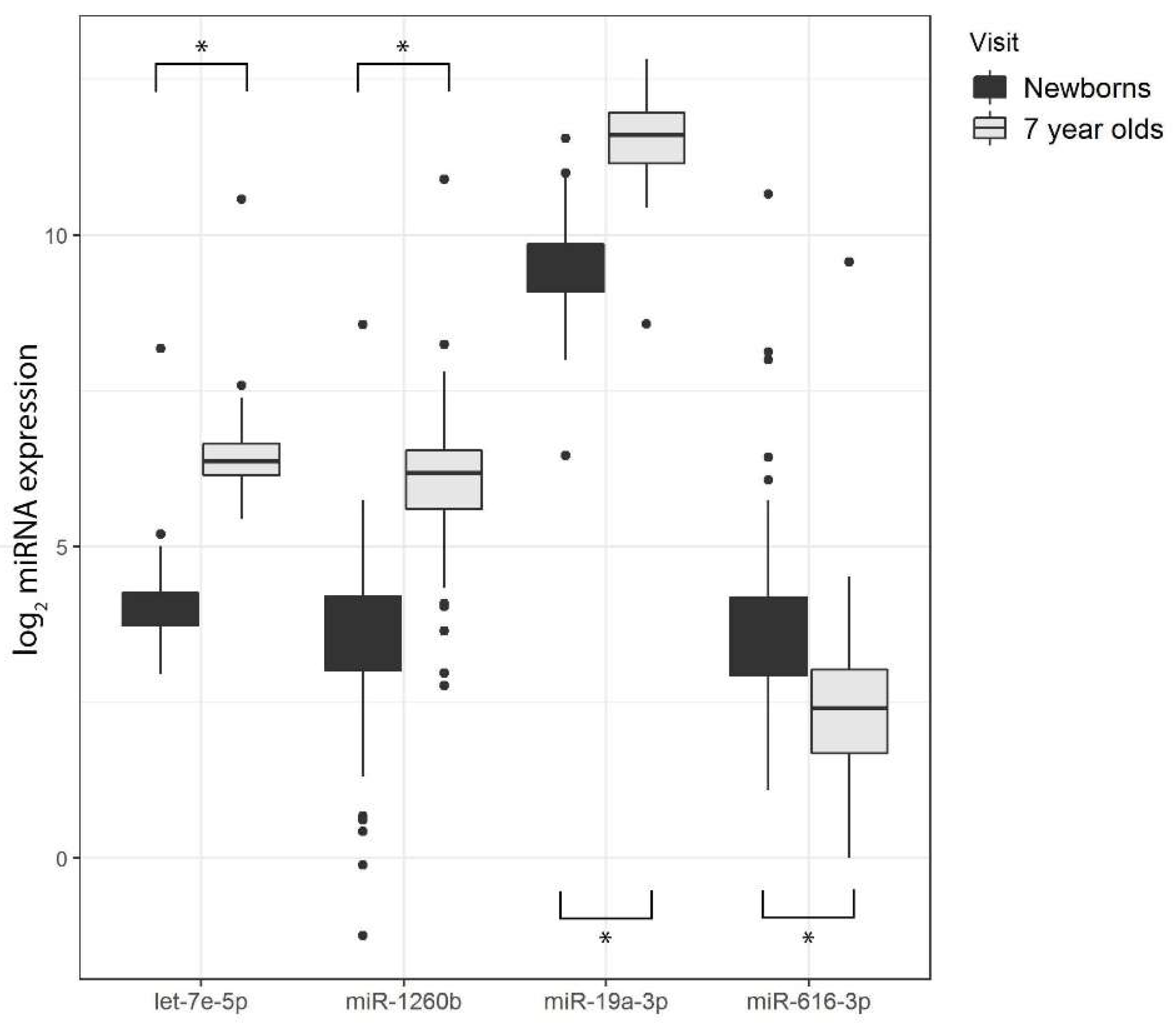
3.5. Differential miRNA Expression by Age
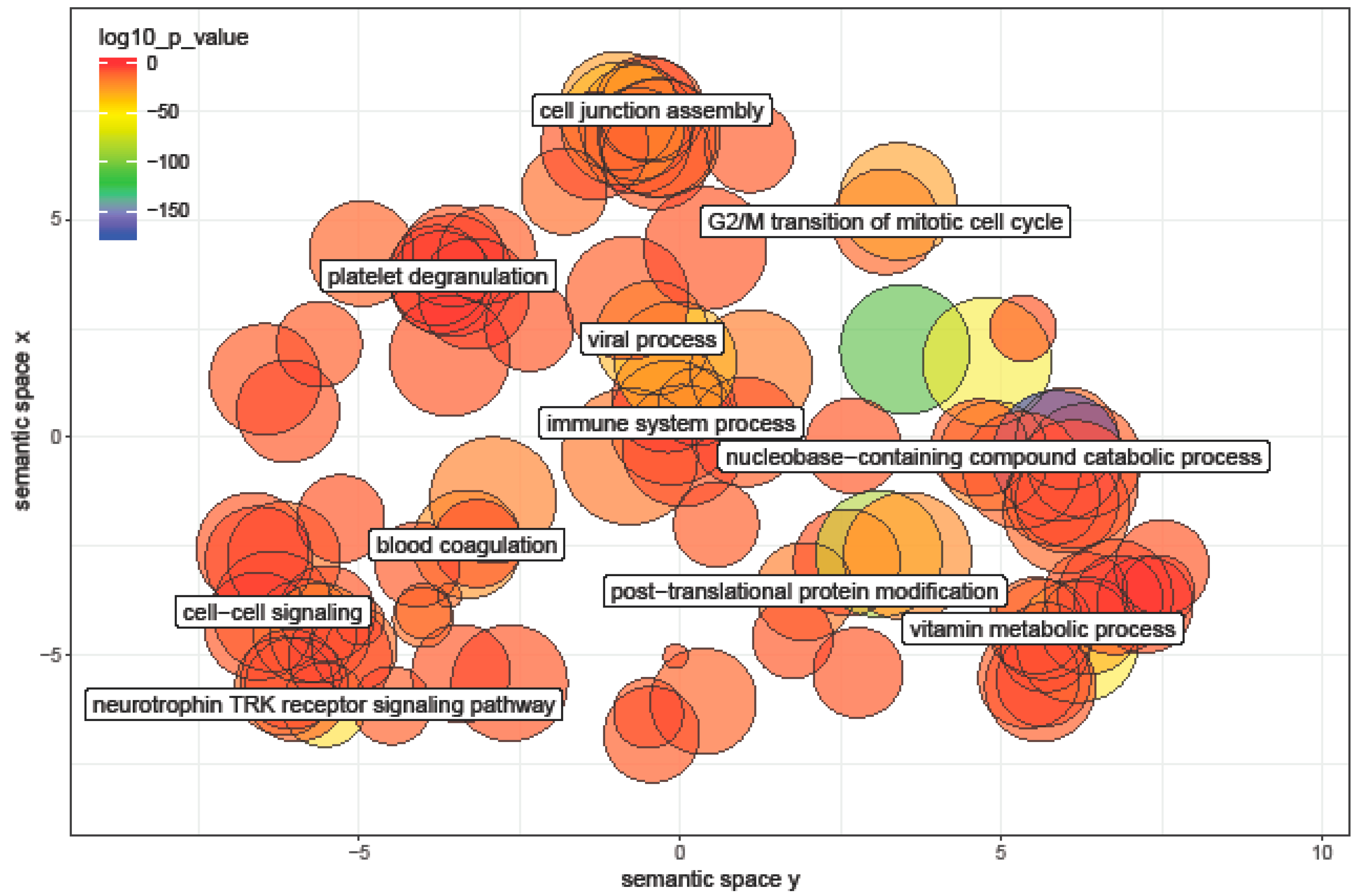
3.6. Pathway Analysis of miRNA Differentially Expressed between Ages
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pennisi, E. Behind the Scenes of Gene Expression. Science 2001, 293, 1064–1067. [Google Scholar] [CrossRef] [PubMed]
- Bowers, E.C.; McCullough, S.D. Linking the Epigenome with Exposure Effects and Susceptibility: The Epigenetic Seed and Soil Model. Toxicol. Sci. 2017, 155, 302–314. [Google Scholar] [CrossRef] [PubMed]
- Foley, D.L.; Craig, J.M.; Morley, R.; Olsson, C.A.; Dwyer, T.; Smith, K.; Saffery, R. Prospects for Epigenetic Epidemiology. Am. J. Epidemiol. 2009, 169, 389–400. [Google Scholar] [CrossRef] [PubMed]
- Solomon, O.; Yousefi, P.; Huen, K.; Gunier, R.B.; Escudero-Fung, M.; Barcellos, L.F.; Eskenazi, B.; Holland, N. Prenatal Phthalate Exposure and Altered Patterns of DNA Methylation in Cord Blood. Environ. Mol. Mutagen. 2017, 58, 398–410. [Google Scholar] [CrossRef] [PubMed]
- Cardenas, A.; Rifas-Shiman, S.L.; Agha, G.; Hivert, M.; Litonjua, A.A.; DeMeo, D.L.; Lin, X.; Amarasiriwardena, C.J.; Oken, E.; Gillman, M.W.; et al. Persistent DNA Methylation Changes Associated with Prenatal Mercury Exposure and Cognitive Performance during Childhood. Sci. Rep. 2017, 7, 288. [Google Scholar] [CrossRef] [PubMed]
- Simpkin, A.J.; Suderman, M.; Gaunt, T.R.; Lyttleton, O.; McArdle, W.L.; Ring, S.M.; Tilling, K.; Davey Smith, G.; Relton, C.L. Longitudinal Analysis of DNA Methylation Associated with Birth Weight and Gestational Age. Hum. Mol. Genet. 2015, 24, 3752–3763. [Google Scholar] [PubMed]
- Poirier, J.T.; Gardner, E.E.; Connis, N.; Moreira, A.L.; De Stanchina, E.; Hann, C.L.; Rudin, C.M. DNA Methylation in Small Cell Lung Cancer Defines Distinct Disease Subtypes and Correlates with High Expression of EZH2. Oncogene 2015, 34, 5869–5878. [Google Scholar] [CrossRef]
- Wang, J.; Chen, J.; Sen, S. MicroRNA as Biomarkers and Diagnostics. J. Cell. Physiol. 2016, 231, 25–30. [Google Scholar] [CrossRef]
- LaRocca, J.; Binder, A.M.; McElrath, T.F.; Michels, K.B. First-Trimester Urine Concentrations of Phthalate Metabolites and Phenols and Placenta miRNA Expression in a Cohort of U.S. Women. Environ. Health Perspect. 2016, 124, 380–387. [Google Scholar] [CrossRef]
- Vrijens, K.; Bollati, V.; Nawrot, T.S. MicroRNAs as Potential Signatures of Environmental Exposure or Effect: A Systematic Review. Environ. Health Perspect. 2015, 123, 399–411. [Google Scholar] [CrossRef] [PubMed]
- Rodosthenous, R.S.; Burris, H.H.; Sanders, A.P.; Just, A.C.; Dereix, A.E.; Svensson, K.; Solano, M.; Téllez-Rojo, M.M.; Wright, R.O.; Baccarelli, A.A. Second Trimester Extracellular microRNAs in Maternal Blood and Fetal Growth: An Exploratory Study. Epigenetics 2017, 12, 804–810. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Lin, J.; Kong, D.; Huang, M.; Xu, C.; Kim, T.; Etheridge, A.; Luo, Y.; Ding, Y.; Wang, K. Current State of Circulating microRNAs as Cancer Biomarkers. Clin. Chem. 2015, 61, 1138–1155. [Google Scholar] [CrossRef] [PubMed]
- Small, E.M.; Olson, E.N. Pervasive Roles of microRNAs in Cardiovascular Biology. Nature 2011, 469, 336–342. [Google Scholar] [CrossRef] [PubMed]
- Ameling, S.; Kacprowski, T.; Chilukoti, R.K.; Malsch, C.; Liebscher, V.; Suhre, K.; Pietzner, M.; Friedrich, N.; Homuth, G.; Hammer, E.; et al. Associations of Circulating Plasma microRNAs with Age, Body Mass Index and Sex in a Population-Based Study. BMC Med. Genomics 2015, 8, 61. [Google Scholar] [CrossRef] [PubMed]
- Lizarraga, D.; Huen, K.; Combs, M.; Escudero-Fung, M.; Eskenazi, B.; Holland, N. miRNAs Differentially Expressed by Next-Generation Sequencing in Cord Blood Buffy Coat Samples of Boys and Girls. Epigenomics 2016, 8, 1619–1635. [Google Scholar] [CrossRef] [PubMed]
- Burgess, K.S.; Philips, S.; Benson, E.A.; Desta, Z.; Gaedigk, A.; Gaedigk, R.; Segar, M.W.; Liu, Y.; Skaar, T.C. Age-Related Changes in MicroRNA Expression and Pharmacogenes in Human Liver. Clin. Pharmacol. Ther. 2015, 98, 205–215. [Google Scholar] [CrossRef] [PubMed]
- Noren Hooten, N.; Abdelmohsen, K.; Gorospe, M.; Ejiogu, N.; Zonderman, A.B.; Evans, M.K. microRNA Expression Patterns Reveal Differential Expression of Target Genes with Age. PLoS ONE 2010, 5, e10724. [Google Scholar] [CrossRef]
- Eskenazi, B.; Bradman, A.; Gladstone, E.A.; Jaramillo, S.; Birch, K.; Holland, N. CHAMACOS, a Longitudinal Birth Cohort Study: Lessons from the Fields. J. Child. Health 2003, 1, 3–27. [Google Scholar] [CrossRef]
- Bollati, V.; Marinelli, B.; Apostoli, P.; Bonzini, M.; Nordio, F.; Hoxha, M.; Pegoraro, V.; Motta, V.; Tarantini, L.; Cantone, L.; et al. Exposure to Metal-Rich Particulate Matter Modifies the Expression of Candidate microRNAs in Peripheral Blood Leukocytes. Environ. Health Perspect. 2010, 118, 763–768. [Google Scholar] [CrossRef]
- Donzelli, S.; Blandino, G.; Muti, P. Use of Buffy Coat miRNA Profiling for Breast Cancer Prediction in Healthy Women. Methods Mol. Biol. 2016, 1379, 13–19. [Google Scholar]
- Georgiadis, P.; Hebels, D.G.; Valavanis, I.; Liampa, I.; Bergdahl, I.A.; Johansson, A.; Palli, D.; Chadeau-Hyam, M.; Chatziioannou, A.; Jennen, D.G.; et al. Omics for Prediction of Environmental Health Effects: Blood Leukocyte-Based Cross-Omic Profiling Reliably Predicts Diseases Associated with Tobacco Smoking. Sci. Rep. 2016, 6, 20544. [Google Scholar] [CrossRef] [PubMed]
- Hebels, D.G.; Georgiadis, P.; Keun, H.C.; Athersuch, T.J.; Vineis, P.; Vermeulen, R.; Portengen, L.; Bergdahl, I.A.; Hallmans, G.; Palli, D.; et al. Performance in Omics Analyses of Blood Samples in Long-Term Storage: Opportunities for the Exploitation of Existing Biobanks in Environmental Health Research. Environ. Health Perspect. 2013, 121, 480–487. [Google Scholar] [CrossRef] [PubMed]
- Voellenkle, C.; van Rooij, J.; Cappuzzello, C.; Greco, S.; Arcelli, D.; Di Vito, L.; Melillo, G.; Rigolini, R.; Costa, E.; Crea, F.; et al. MicroRNA Signatures in Peripheral Blood Mononuclear Cells of Chronic Heart Failure Patients. Physiol. Genomics 2010, 42, 420–426. [Google Scholar] [CrossRef] [PubMed]
- Aryee, M.J.; Jaffe, A.E.; Corrada-Bravo, H.; Ladd-Acosta, C.; Feinberg, A.P.; Hansen, K.D.; Irizarry, R.A. Minfi: A Flexible and Comprehensive Bioconductor Package for the Analysis of Infinium DNA Methylation Microarrays. Bioinformatics 2014, 30, 1363–1369. [Google Scholar] [CrossRef] [PubMed]
- Yousefi, P.; Huen, K.; Dave, V.; Barcellos, L.; Eskenazi, B.; Holland, N. Sex Differences in DNA Methylation Assessed by 450 K BeadChip in Newborns. BMC Genomics 2015, 16, 911. [Google Scholar] [CrossRef] [PubMed]
- Huen, K.; Solomon, O.; Kogut, K.; Eskenazi, B.; Holland, N. PON1 DNA Methylation and Neurobehavior in Mexican-American Children with Prenatal Organophosphate Exposure. Environ. Int. 2018, 121, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Bakulski, K.M.; Feinberg, J.I.; Andrews, S.V.; Yang, J.; Brown, S.; McKenney, S.L.; Witter, F.; Walston, J.; Feinberg, A.P.; Fallin, M.D. DNA Methylation of Cord Blood Cell Types: Applications for Mixed Cell Birth Studies. Epigenetics 2016, 11, 354–362. [Google Scholar] [CrossRef]
- Houseman, E.A.; Accomando, W.P.; Koestler, D.C.; Christensen, B.C.; Marsit, C.J.; Nelson, H.H.; Wiencke, J.K.; Kelsey, K.T. DNA Methylation Arrays as Surrogate Measures of Cell Mixture Distribution. BMC Bioinformatics 2012, 13, 86. [Google Scholar] [CrossRef]
- Vlachos, I.S.; Zagganas, K.; Paraskevopoulou, M.D.; Georgakilas, G.; Karagkouni, D.; Vergoulis, T.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA-miRPath V3.0: Deciphering microRNA Function with Experimental Support. Nucleic Acids Res. 2015, 43, W460–W466. [Google Scholar] [CrossRef]
- Supek, F.; Bošnjak, M.; Škunca, N.; Šmuc, T. REVIGO Summarizes and Visualizes Long Lists of Gene Ontology Terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Noren Hooten, N.; Fitzpatrick, M.; Wood, W.H., 3rd; De, S.; Ejiogu, N.; Zhang, Y.; Mattison, J.A.; Becker, K.G.; Zonderman, A.B.; Evans, M.K. Age-Related Changes in microRNA Levels in Serum. Aging (Albany NY) 2013, 5, 725–740. [Google Scholar] [CrossRef] [PubMed]
- Huan, T.; Chen, G.; Liu, C.; Bhattacharya, A.; Rong, J.; Chen, B.H.; Seshadri, S.; Tanriverdi, K.; Freedman, J.E.; Larson, M.G.; et al. Age-Associated microRNA Expression in Human Peripheral Blood is Associated with all-Cause Mortality and Age-Related Traits. Aging Cell 2018, 17. [Google Scholar] [CrossRef] [PubMed]
- Adalsteinsson, B.T.; Gudnason, H.; Aspelund, T.; Harris, T.B.; Launer, L.J.; Eiriksdottir, G.; Smith, A.V.; Gudnason, V. Heterogeneity in White Blood Cells has Potential to Confound DNA Methylation Measurements. PLoS ONE 2012, 7, e46705. [Google Scholar] [CrossRef] [PubMed]
- Breton, C.V.; Marsit, C.J.; Faustman, E.; Nadeau, K.; Goodrich, J.M.; Dolinoy, D.C.; Herbstman, J.; Holland, N.; LaSalle, J.M.; Schmidt, R.; et al. Small-Magnitude Effect Sizes in Epigenetic End Points are Important in Children’s Environmental Health Studies: The Children’s Environmental Health and Disease Prevention Research Center’s Epigenetics Working Group. Environ. Health Perspect. 2017, 125, 511–526. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.J.; Islam, S.A.; Edgar, R.D.; Kobor, M.S. Adjusting for Cell Type Composition in DNA Methylation Data using a Regression-Based Approach. Methods Mol. Biol. 2017, 1589, 99–106. [Google Scholar] [PubMed]
- Juzenas, S.; Venkatesh, G.; Hubenthal, M.; Hoeppner, M.P.; Du, Z.G.; Paulsen, M.; Rosenstiel, P.; Senger, P.; Hofmann-Apitius, M.; Keller, A.; et al. A Comprehensive, Cell Specific microRNA Catalogue of Human Peripheral Blood. Nucleic Acids Res. 2017, 45, 9290–9301. [Google Scholar] [CrossRef]
- Sharma, S.; Eghbali, M. Influence of Sex Differences on microRNA Gene Regulation in Disease. Biol. Sex Differ. 2014, 5, 3. [Google Scholar] [CrossRef]
- Cui, C.; Yang, W.; Shi, J.; Zhou, Y.; Yang, J.; Cui, Q.; Zhou, Y. Identification and Analysis of Human Sex-Biased MicroRNAs. Genom. Proteom. Bioinf. 2018, 16, 200–211. [Google Scholar] [CrossRef]
- Teruel-Montoya, R.; Kong, X.; Abraham, S.; Ma, L.; Kunapuli, S.P.; Holinstat, M.; Shaw, C.A.; McKenzie, S.E.; Edelstein, L.C.; Bray, P.F. MicroRNA Expression Differences in Human Hematopoietic Cell Lineages Enable Regulated Transgene Expression. PLoS ONE 2014, 9, e102259. [Google Scholar] [CrossRef]
- Lussier, Y.A.; Stadler, W.M.; Chen, J.L. Advantages of Genomic Complexity: Bioinformatics Opportunities in microRNA Cancer Signatures. J. Am. Med. Inform. Assoc. 2012, 19, 156–160. [Google Scholar] [CrossRef] [PubMed]

| Child Characteristics | n (%) | Mean (SD) |
|---|---|---|
| Newborns | ||
| Sex | ||
| Boy | 51 (42) | |
| Girl | 70 (58) | |
| Gestational Age (weeks) | 38.7 (1.5) | |
| Birthweight | ||
| Low (≤2500 g) | 3 (2) | |
| Normal (>2500 g) | 118 (98) | |
| 7-year-olds | ||
| Sex | ||
| Boy | 64 (45) | |
| Girl | 78 (55) | |
| Age at 7-year assessment (years) | 7.1 (0.3) | |
| Obesity Status at age 7 years | ||
| Normal (≤85th percentile) | 67 (47) | |
| Overweight (>85th, <95th percentile) | 27 (19) | |
| Obese (≥95th percentile) | 48 (34) |
| miRNA Name | MIMAT ID | Newborns | 7-Year-Olds | Level of Expression | ||||
|---|---|---|---|---|---|---|---|---|
| Mean | SD | IQR | Mean | SD | IQR | |||
| hsa-miR-223-3p | MIMAT0000280 | 60218.9 | 31065.5 | 34513.5 | 49912.1 | 23174.2 | 33122.0 | High Expression (mean > 500) * |
| hsa-miR-150-5p | MIMAT0000451 | 42279.6 | 27052.2 | 28918.5 | 29763.7 | 16302.7 | 17273.0 | |
| hsa-miR-25-3p | MIMAT0000081 | 14566.5 | 13796.9 | 8073.7 | 19238.3 | 13050.8 | 12311.9 | |
| hsa-let-7b-5p | MIMAT0000063 | 13089.6 | 6215.4 | 6115.7 | 12935.8 | 5898.9 | 6897.7 | |
| hsa-miR-26b-5p | MIMAT0000083 | 5192.5 | 1967.3 | 2439.9 | 7633.9 | 2618.9 | 3315.9 | |
| hsa-let-7d-5p | MIMAT0000065 | 4024.1 | 1316.9 | 1234.8 | 4313.4 | 1095.8 | 1406.0 | |
| hsa-miR-185-5p | MIMAT0000455 | 1640.3 | 1000.4 | 688.2 | 1531.7 | 609.6 | 633.6 | |
| hsa-miR-92a-3p | MIMAT0000092 | 1623.7 | 1045.0 | 1031.2 | 1534.0 | 913.7 | 970.0 | |
| hsa-miR-26a-5p | MIMAT0000082 | 1108.7 | 513.9 | 412.6 | 1977.9 | 931.9 | 694.0 | |
| hsa-miR-130a-3p | MIMAT0000425 | 1057.6 | 628.1 | 278.4 | 1695.1 | 441.6 | 448.8 | |
| hsa-miR-19a-3p | MIMAT0000073 | 808.2 | 383.2 | 378.4 | 3263.7 | 1234.5 | 1729.3 | |
| hsa-miR-107 | MIMAT0000104 | 628.2 | 159.8 | 175.3 | 1018.3 | 249.5 | 276.9 | |
| hsa-miR-425-5p | MIMAT0003393 | 509.1 | 202.4 | 235.4 | 1609.7 | 527.8 | 645.9 | |
| hsa-miR-199a-5p | MIMAT0000231 | 336.1 | 197.1 | 189.1 | 286.1 | 135.9 | 164.9 | Medium Expression (50 < mean < 500) * |
| hsa-let-7c-5p | MIMAT0000064 | 335.3 | 179.6 | 142.9 | 536.8 | 277.9 | 236.1 | |
| hsa-miR-146a-5p | MIMAT0000449 | 259.5 | 250.7 | 100.6 | 474.6 | 292.9 | 188.4 | |
| hsa-miR-454-3p | MIMAT0003885 | 254.3 | 95.9 | 115.4 | 754.8 | 250.3 | 293.7 | |
| hsa-miR-301a-3p | MIMAT0000688 | 145.0 | 64.6 | 59.0 | 176.8 | 71.0 | 65.3 | |
| hsa-miR-495-3p | MIMAT0002817 | 131.9 | 376.5 | 64.4 | 45.6 | 126.9 | 28.0 | |
| hsa-miR-136-5p | MIMAT0000448 | 114.9 | 389.5 | 49.5 | 45.1 | 127.1 | 27.7 | |
| hsa-miR-199b-5p | MIMAT0000263 | 95.1 | 73.8 | 69.8 | 57.5 | 39.0 | 35.1 | |
| hsa-miR-664a-3p | MIMAT0005949 | 89.2 | 35.4 | 47.0 | 151.1 | 47.5 | 63.4 | |
| hsa-miR-130b-3p | MIMAT0000691 | 54.5 | 104.2 | 15.2 | 81.4 | 155.0 | 21.9 | |
| hsa-miR-1289 | MIMAT0005879 | 52.2 | 128.7 | 21.0 | 53.0 | 125.7 | 20.9 | |
| hsa-miR-141-3p | MIMAT0000432 | 52.0 | 333.6 | 10.4 | 94.6 | 699.8 | 20.4 | |
| hsa-miR-505-3p | MIMAT0002876 | 46.2 | 94.8 | 17.1 | 28.3 | 62.6 | 10.4 | Low Expression (mean < 50) * |
| hsa-miR-4461 | MIMAT0018983 | 41.2 | 217.2 | 9.9 | 46.1 | 253.6 | 11.9 | |
| hsa-miR-377-3p | MIMAT0000730 | 37.5 | 61.7 | 26.1 | 22.2 | 34.1 | 18.2 | |
| hsa-miR-616-3p | MIMAT0004805 | 32.9 | 149.5 | 10.5 | 11.9 | 63.5 | 4.9 | |
| hsa-miR-335-5p | MIMAT0000765 | 29.5 | 22.9 | 14.8 | 54.9 | 35.0 | 25.0 | |
| hsa-miR-142-5p | MIMAT0000433 | 29.3 | 85.9 | 9.6 | 74.2 | 220.0 | 22.5 | |
| hsa-miR-139-5p | MIMAT0000250 | 26.9 | 174.8 | 6.1 | 17.6 | 127.3 | 5.6 | |
| hsa-let-7e-5p | MIMAT0000066 | 18.7 | 25.3 | 5.8 | 98.1 | 123.1 | 30.4 | |
| hsa-miR-766-3p | MIMAT0003888 | 18.5 | 49.2 | 9.5 | 49.0 | 126.0 | 23.8 | |
| hsa-miR-371b-5p | MIMAT0019892 | 17.7 | 94.7 | 5.5 | 5.3 | 31.8 | 1.8 | |
| hsa-miR-1260b | MIMAT0015041 | 17.7 | 34.3 | 10.4 | 92.1 | 159.3 | 46.6 | |
| hsa-miR-1275 | MIMAT0005929 | 17.4 | 47.9 | 7.4 | 23.2 | 62.9 | 11.1 | |
| hsa-miR-6511a-3p | MIMAT0025479 | 16.3 | 94.7 | 3.9 | 54.6 | 349.4 | 14.9 | |
| hsa-miR-92b-3p | MIMAT0003218 | 12.8 | 67.4 | 5.7 | 5.8 | 31.8 | 3.1 | |
| hsa-miR-1254 | MIMAT0005905 | 11.5 | 84.0 | 2.3 | 30.2 | 254.9 | 7.0 | |
| hsa-miR-200a-3p | MIMAT0000682 | 11.1 | 10.4 | 3.6 | 32.6 | 31.3 | 11.5 | |
| hsa-miR-548e-5p | MIMAT0026736 | 10.5 | 34.4 | 4.2 | 26.8 | 94.7 | 10.8 | |
| hsa-miR-103a-3p | MIMAT0000101 | 6.7 | 14.8 | 2.8 | 14.7 | 31.4 | 6.7 | |
| miRNA Name | cd8+T | cd4+T | NK Cells | B Cells | Monocytes | Granulocytes | nRBCs | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β (95% CI) | p-Value | β (95% CI) | p-Value | β (95% CI) | p-Value | β (95% CI) | p-Value | β (95% CI) | p-Value | β (95% CI) | p-Value | β (95% CI) | p-Value | |
| Newborns | ||||||||||||||
| miR-495-3p | 12.38 (−4.25, 29.01) | 0.143 | 16.39 (1.33, 31.46) | 0.033 | 14.13 (−3.48, 31.75) | 0.115 | 13.10 (−2.47, 28.67) | 0.098 | 19.66 (0.28, 39.04) | 0.047 | 13.21 (−2.29, 28.71) | 0.094 | 14.62 (−1.07, 30.31) | 0.068 |
| miR-377-3p | 16.37 (−0.78, 33.52) | 0.061 | 20.27 (4.74, 35.80) | 0.011 | 12.37 (−5.80, 30.53) | 0.180 | 19.31 (3.25, 35.36) | 0.019 | 22.68 (2.69, 42.66) | 0.027 | 17.09 (1.10, 33.07) | 0.036 | 19.19 (3.01, 35.37) | 0.021 |
| miR-301a-3p | 6.22 (−1.73, 14.17) | 0.124 | 9.01 (1.81, 16.21) | 0.015 | 8.77 (0.35, 17.19) | 0.041 | 6.21 (−1.23, 13.65) | 0.101 | 8.8 (−0.46, 18.07) | 0.062 | 8.27 (0.86, 15.68) | 0.029 | 9.07 (1.57, 16.57) | 0.018 |
| miR-26b-5p | −6.64 (−15.92, 2.63) | 0.159 | −8.92 (−17.32, −0.51) | 0.038 | −5.3 (−15.12, 4.53) | 0.288 | −9.01 (−17.69, −0.32) | 0.042 | −10.66 (−21.46, 0.15) | 0.053 | −8.04 (−16.69, 0.60) | 0.068 | −7.92 (−16.68, 0.83) | 0.075 |
| miR-199b-5p | 10.68 (−1.81, 23.17) | 0.093 | 12.48 (1.17, 23.80) | 0.031 | 7.31 (−5.93, 20.54) | 0.276 | 11.45 (−0.24, 23.15) | 0.055 | 14.98 (0.42, 29.54) | 0.044 | 9.31 (−2.34, 20.95) | 0.116 | 9.96 (−1.83, 21.74) | 0.097 |
| miR-136-5p | 18.89 (0.51, 37.28) | 0.044 | 20.04 (3.39, 36.70) | 0.019 | 19.54 (0.06, 39.03) | 0.049 | 19.37 (2.15, 36.58) | 0.028 | 24.31 (2.88, 45.73) | 0.027 | 18.40 (1.26, 35.54) | 0.036 | 19.55 (2.20, 36.90) | 0.028 |
| let-7e-5p | 5.29 (−2.21, 12.80) | 0.165 | 6.87 (0.07, 13.67) | 0.048 | 3.06 (−4.89, 11.01) | 0.447 | 7.17 (0.14, 14.20) | 0.046 | 9.36 (0.61, 18.11) | 0.036 | 7.10 (0.10, 14.10) | 0.047 | 6.57 (−0.51, 13.65) | 0.069 |
| let-7b-5p | −7.17 (−15.64, 1.29) | 0.096 | −8.35 (−16.02, −0.69) | 0.033 | −2.94 (−11.91, 6.02) | 0.517 | −8.72 (−16.64, −0.80) | 0.031 | −8.16 (−18.02, 1.70) | 0.104 | −6.52 (−14.40, 1.37) | 0.104 | −6.86 (−14.84, 1.13) | 0.092 |
| 7-year-olds | ||||||||||||||
| miR-495-3p | 20.71 (0.67, 40.75) | 0.043 | 25.52 (3.32, 47.73) | 0.025 | 23.61 (3.33, 43.89) | 0.023 | 16.21 (−2.84, 35.25) | 0.095 | 20.36 (−0.35, 41.07) | 0.054 | 23.59 (3.10, 44.09) | 0.024 | ||
| miRNA Name | logFC * | FC | log2 Mean Expression | FDR-Adjusted p-Value |
|---|---|---|---|---|
| miR-1260b | −2.77 | −6.84 | 7.66 | 7.29 × 10−34 |
| let-7e-5p | −2.67 | −6.36 | 8.08 | 2.47 × 10−83 |
| miR-6511a-3p | −2.10 | −4.29 | 6.34 | 6.14 × 10−35 |
| miR-19a-3p | −2.06 | −4.16 | 13.43 | 2.95 × 10−41 |
| miR-1254 | −1.95 | −3.86 | 4.80 | 2.94 × 10−20 |
| miR-425-5p | −1.81 | −3.50 | 12.61 | 1.24 × 10−40 |
| miR-200a-3p | −1.76 | −3.39 | 6.88 | 3.86 × 10−52 |
| miR-454-3p | −1.67 | −3.18 | 11.57 | 1.16 × 10−31 |
| miR-766-3p | −1.65 | −3.14 | 7.14 | 1.28 × 10−23 |
| miR-142-5p | −1.56 | −2.95 | 7.82 | 3.74 × 10−32 |
| miR-548e-5p | −1.47 | −2.76 | 6.17 | 4.32 × 10−29 |
| miR-103a-3p | −1.31 | −2.49 | 5.67 | 4.52 × 10−17 |
| miR-141-3p | −1.15 | −2.22 | 7.32 | 1.53 × 10−18 |
| miR-146a-5p | −1.10 | −2.14 | 11.07 | 2.99 × 10−24 |
| miR-335-5p | −1.09 | −2.13 | 7.94 | 6.37 × 10−18 |
| miR-26a-5p | −1.02 | −2.03 | 13.25 | 6.30 × 10−16 |
| let-7c-5p | −0.87 | −1.83 | 11.43 | 4.65 × 10−12 |
| miR-664a-3p | −0.86 | −1.81 | 9.59 | 1.62 × 10−15 |
| miR-130a-3p | −0.82 | −1.77 | 13.12 | 2.02 × 10−18 |
| miR-107 | −0.82 | −1.77 | 12.45 | 5.57 × 10−16 |
| miR-130b-3p | −0.82 | −1.76 | 8.53 | 3.18 × 10−17 |
| miR-1275 | −0.69 | −1.61 | 6.54 | 1.82 × 10−8 |
| miR-25-3p | −0.57 | −1.49 | 16.60 | 1.06 × 10−4 |
| miR-26b-5p | −0.57 | −1.48 | 15.40 | 3.12 × 10−6 |
| miR-4461 | −0.48 | −1.39 | 7.20 | 1.53 × 10−6 |
| miR-301a-3p | −0.33 | −1.25 | 10.03 | 2.77 × 10−3 |
| let-7d-5p | −0.25 | −1.19 | 14.81 | 1.83 × 10−2 |
| miR-1289 | −0.24 | −1.18 | 8.01 | 2.30 × 10−2 |
| miR-92a-3p | −0.16 | −1.11 | 13.24 | 3.95 × 10−1 |
| let-7b-5p | −0.10 | −1.07 | 16.38 | 4.18 × 10−1 |
| miR-185-5p | −0.10 | −1.07 | 13.29 | 4.18 × 10−1 |
| miR-223-3p | 0.04 | 1.03 | 18.40 | 6.96 × 10−1 |
| miR-199a-5p | 0.14 | 1.10 | 10.85 | 3.47 × 10−1 |
| miR-505-3p | 0.20 | 1.15 | 7.43 | 5.84 × 10−2 |
| miR-139-5p | 0.34 | 1.26 | 5.44 | 6.93 × 10−2 |
| miR-199b-5p | 0.45 | 1.36 | 8.72 | 4.00 × 10−4 |
| miR-92b-3p | 0.55 | 1.46 | 4.59 | 3.10 × 10−3 |
| miR-150-5p | 0.58 | 1.49 | 17.72 | 3.26 × 10−6 |
| miR-377-3p | 0.63 | 1.55 | 6.94 | 2.77 × 10−3 |
| miR-136-5p | 0.83 | 1.78 | 7.92 | 8.28 × 10−5 |
| miR-495-3p | 1.05 | 2.07 | 8.22 | 2.37 × 10−9 |
| miR-371b-5p | 1.17 | 2.25 | 4.67 | 3.27 × 10−13 |
| miR-616-3p | 1.19 | 2.28 | 5.71 | 2.37 × 10−9 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huen, K.; Lizarraga, D.; Kogut, K.; Eskenazi, B.; Holland, N. Age-Related Differences in miRNA Expression in Mexican-American Newborns and Children. Int. J. Environ. Res. Public Health 2019, 16, 524. https://doi.org/10.3390/ijerph16040524
Huen K, Lizarraga D, Kogut K, Eskenazi B, Holland N. Age-Related Differences in miRNA Expression in Mexican-American Newborns and Children. International Journal of Environmental Research and Public Health. 2019; 16(4):524. https://doi.org/10.3390/ijerph16040524
Chicago/Turabian StyleHuen, Karen, Daneida Lizarraga, Katherine Kogut, Brenda Eskenazi, and Nina Holland. 2019. "Age-Related Differences in miRNA Expression in Mexican-American Newborns and Children" International Journal of Environmental Research and Public Health 16, no. 4: 524. https://doi.org/10.3390/ijerph16040524
APA StyleHuen, K., Lizarraga, D., Kogut, K., Eskenazi, B., & Holland, N. (2019). Age-Related Differences in miRNA Expression in Mexican-American Newborns and Children. International Journal of Environmental Research and Public Health, 16(4), 524. https://doi.org/10.3390/ijerph16040524




