Correlation of Internal Exposure Levels of Polycyclic Aromatic Hydrocarbons to Methylation of Imprinting Genes of Sperm DNA
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Participants and Data Collection
2.2. Semen and Urine Collection
2.3. Measurement of the Concentrations of Monohydroxylated PAHs (OH-PAHs) in Urine
2.4. DNA Extraction
2.5. Bisulfite Treatment
2.6. PCR Amplification of Bisulfite-Treated Sperm DNA and Subsequent Pyrosequencing
2.7. Statistical Analysis
3. Results
3.1. Demographic Characteristics of the Study Participants
3.2. Distribution of Urine OH-PAHs among Different Demographic Groups
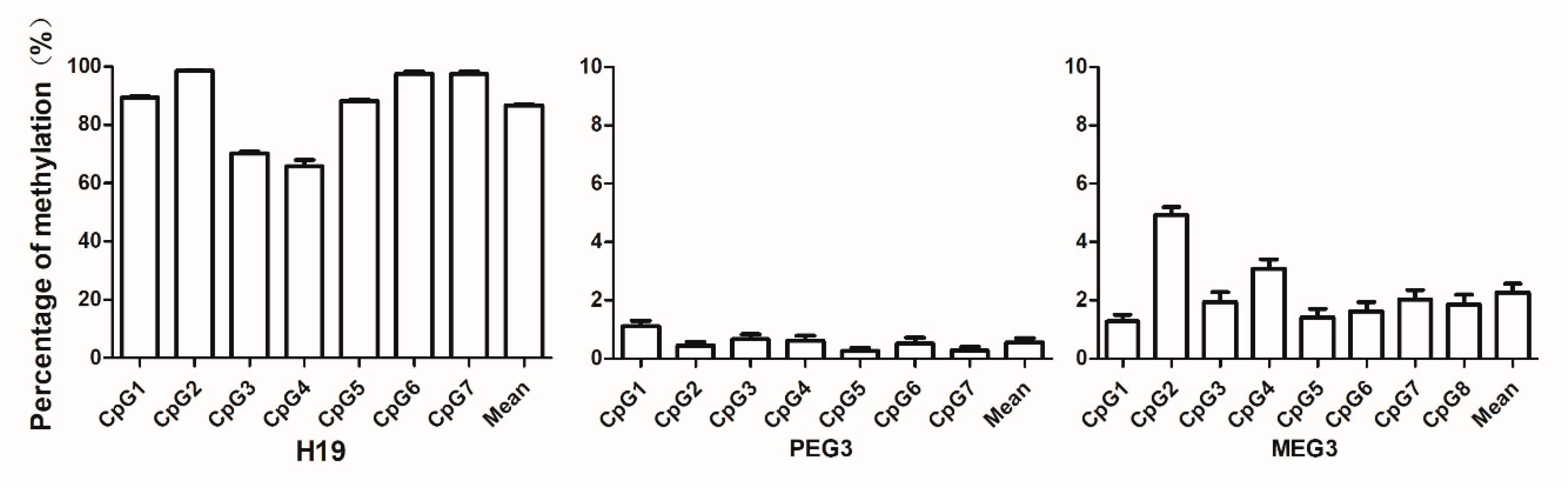
3.3. Levels of Sperm DNA Methylation of the Imprinting Genes
3.4. Correlation Analysis between Concentrations of Urinary OH-PAHs and Average Levels of Sperm DNA Imprinting Genes Methylation
4. Discussion
4.1. Sperm Chromatin is Sensitive to Environmental Pollutants
4.2. Methylation of Imprinting Genes Is Affected by Adverse Environmental Perturbations
4.3. Association between Paternal PAH Exposure and Imprinting Genes
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| DMRs | differentially methylated regions |
| MEG3 | maternally expressed gene 3 |
| PEG3 | paternally expressed gene 3 |
| PAHs | polycyclic aromatic hydrocarbons |
| OH-PAHs | hydroxylated metabolites of polycyclic aromatic hydrocarbons |
References
- Shirangi, A.; Nieuwenhuijsen, M.; Vienneau, D.; Holman, C.D.J. Living near agricultural pesticide applications and the risk of adverse reproductive outcomes: A review of the literature. Occup. Environ. Med. 2011, 68, 172–191. [Google Scholar] [CrossRef][Green Version]
- Nassar, N.; Abeywardana, P.; Barker, A.; Bower, C. Parental occupational exposure to potential endocrine disrupting chemicals and risk of hypospadias in infants. Occup. Environ. Med. 2010, 67, 585–589. [Google Scholar] [CrossRef] [PubMed]
- Hombach-Klonisch, S.; Pocar, P.; Kietz, S.; Klonisch, T. Molecular Actions of Polyhalogenated Arylhydrocarbons (PAHs) in Female Reproduction. Curr. Med. Chem. 2005, 12, 599–616. [Google Scholar] [PubMed]
- Rengarajanab, T.; Rajendranb, P.; Nandakumarc, N.; Lokeshkumard, B.; Rajendrane, P.; Nishigakib, I. Exposure to polycyclic aromatic hydrocarbons with special focus on cancer. Asian Pac. J. Trop. Biomed. 2015, 5, 182–189. [Google Scholar] [CrossRef]
- Polanska, K.; Dettbarn, G.; Jurewicz, J.; Sobala, W.; Magnus, P.; Seidel, A.; Hanke, W. Effect of Prenatal Polycyclic Aromatic Hydrocarbons Exposure on Birth Outcomes: The Polish Mother and Child Cohort Study. BioMed Res. Int. 2014, 2014, 1–10. [Google Scholar] [CrossRef]
- Choi, H.; Rauh, V.; Garfinkel, R.; Tu, Y.; Perera, F.P. Prenatal Exposure to Airborne Polycyclic Aromatic Hydrocarbons and Risk of Intrauterine Growth Restriction. Environ. Health Perspect. 2008, 116, 658–665. [Google Scholar] [CrossRef] [PubMed]
- Cordier, S.; Monfort, C.; Filippini, G.; Preston-Martin, S.; Lubin, F.; Mueller, B.A.; Holly, E.A.; Peris-Bonet, R.; McCredie, M.; Choi, W.; et al. Parental Exposure to Polycyclic Aromatic Hydrocarbons and the Risk of Childhood Brain Tumors: The SEARCH International Childhood Brain Tumor Study. Am. J. Epidemiol. 2004, 159, 1109–1116. [Google Scholar] [CrossRef] [PubMed]
- Castellanosa, P.; Olmob, E.D.; Fernández-Santosb, M.R.; Rodríguez-Estivala, J.; Gardeb, J.J.; Rafael, M. Increased chromatin fragmentation and reduced acrosome integrity in spermatozoa of red deer from lead polluted sites. Sci. Total Environ. 2015, 505, 32–38. [Google Scholar] [CrossRef]
- Mao, Z.; Xia, W.; Chang, H.; Huo, W.; Li, Y.; Xu, S. Paternal BPA exposure in early life alters Igf2 epigenetic status in sperm and induces pancreatic impairment in rat offspring. Toxicol. Lett. 2015, 238, 30–38. [Google Scholar] [CrossRef]
- Benchaib, M.; Braun, V.; Ressnikof, D.; Lornage, J.; Durand, P.; Niveleau, A.; Guérin, J. Influence of global sperm DNA methylation on IVF results. Hum. Reprod. 2005, 20, 768–773. [Google Scholar] [CrossRef]
- Bartolomei, M.S.; Ferguson-Smith, A.C. Mammalian genomic imprinting. Cold Spring Harb. Perspect Biol. 2011, 3, a002592. [Google Scholar] [CrossRef] [PubMed]
- Nicholls, R.D.; Knoll, J.H.M.; Butler, M.G.; Karam, S.; Lalande, M. Genetic imprinting suggested by maternal heterodisomy in non-deletion Prader-Willi syndrome. Nature 1989, 342, 281–285. [Google Scholar] [CrossRef] [PubMed]
- Hecht, S.S. Tobacco smoke carcinogens and breast cancer. Environ. Mol. Mutagen. 2002, 39, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.S.; Trommel, J.S.; Yan, X.J.; Ashley, D.; Watson, C.H. Determination of 14 Polycyclic Aromatic Hydrocarbons in Mainstream Smoke from Domestic Cigarettes. Environ. Sci. Technol. 2005, 39, 471–478. [Google Scholar] [CrossRef] [PubMed]
- Jongeneelen, F. Benchmark guideline for urinary 1-hydroxypyrene as biomarker of occupational exposure to polycyclic aromatic hydrocarbons. Ann. Occup. Hyg. 2001, 45, 3–13. [Google Scholar] [CrossRef]
- Suzuki, K.; Yoshinaga, J. Inhalation and dietary exposure to polycyclic aromatic hydrocarbons and urinary 1-hydroxypyrene in non-smoking university students. Int. Arch. Occup. Environ. Health 2007, 81, 115–121. [Google Scholar] [CrossRef]
- Tairova, Z.M.; Giessing, A.M.; Hansen, R.; Andersen, O. 1-Hydroxypyrene as a biomarker of PAH exposure in the marine polychaete Nereis diversicolor. Mar. Environ. Res. 2009, 67, 38–46. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Thakker, D.; Yagi, H.; Levin, W.; Wood, A.; Conney, A.; Jerina, D. Bioactivation of Foreign Compounds; Anders, M.W., Ed.; Academic Press: New York, NY, USA, 1985; pp. 177–242. [Google Scholar]
- Bauer, E.; Guo, Z.; Ueng, Y.-F.; Bell, L.C.; Zeldin, D.; Guengerich, F.P. Oxidation of Benzo[a]pyrene by Recombinant Human Cytochrome P450 Enzymes. Chem. Res. Toxicol. 1995, 8, 136–142. [Google Scholar] [CrossRef]
- Burczynski, M.E.; Harvey, R.G.; Penning, T.M. Expression and characterization of four recombinant human dihydrodiol dehydrogenase isoforms: Oxidation of trans-7, 8-dihydroxy-7,8-dihydrobenzo[a]pyrene to the activated o-quinone metabolite benzo[a]pyrene-7,8-dione. Biochemistry 1998, 37, 6781–6790. [Google Scholar] [CrossRef]
- Grimmer, G.; Jacob, J.; Dettbarn, G.; Naujack, K.-W. Determination of urinary metabolites of polycyclic aromatic hydrocarbons (PAH) for the risk assessment of PAH-exposed workers. Int. Arch. Occup. Environ. Health 1997, 69, 231–239. [Google Scholar] [CrossRef]
- Onyemauwa, F.; Rappaport, S.M.; Sobus, J.R.; Gajdošová, D.; Wu, R.; Waidyanatha, S. Using liquid chromatography–tandem mass spectrometry to quantify monohydroxylated metabolites of polycyclic aromatic hydrocarbons in urine. J. Chromatogr. B 2009, 877, 1117–1125. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Zhang, J.; Zhang, L.; Liu, W.; Weisel, C.P. Selective detection of monohydroxy metabolites of polycyclic aromatic hydrocarbons in urine using liquid chromatography/triple quadrupole tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2004, 18, 2299–2308. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.F.; Kuete, M.; Su, L.; Yang, F.; Hu, Z.Y.; Tian, B.Z.; Zhang, H.P.; Zhao, K. Comparison of three different techniques of human sperm DNA isolation for methylation assay. J. Huazhong Univ. Sci. Technol. 2015, 35, 938–942. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Ma, Y.; Gao, L.; Li, Y.; Li, Q.; Qiang, M. Urine mercury levels correlate with DNA methylation of imprinting gene H19 in the sperm of reproductive-aged men. PLoS ONE 2018, 13, e0196314. [Google Scholar] [CrossRef] [PubMed]
- Edwards, T.M.; Moore, B.C.; Guillette, L.J. Reproductive dysgenesis in wildlife: A comparative view. Int. J. Androl. 2006, 29, 109–121. [Google Scholar] [CrossRef] [PubMed]
- Somers, C.M.; Yauk, C.L.; White, P.A.; Parfett, C.L.J.; Quinn, J.S. Air pollution induces heritable DNA mutations. Proc. Natl. Acad. Sci. USA 2002, 99, 15904–15907. [Google Scholar] [CrossRef] [PubMed]
- Bishop, J.B.; Witt, K.L.; Sloane, R.A. Genetic toxicities of human teratogens. Mutat. Res. Mol. Mech. Mutagen. 1997, 396, 9–43. [Google Scholar] [CrossRef]
- Ling, X.; Zhang, G.; Sun, L.; Wang, Z.; Zou, P.; Gao, J.; Peng, K.; Chen, Q.; Yang, H.; Zhou, N.; et al. Polycyclic aromatic hydrocarbons exposure decreased sperm mitochondrial DNA copy number: A cross-sectional study (MARHCS) in Chongqing, China. Environ. Pollut. 2017, 220, 680–687. [Google Scholar] [CrossRef]
- Kotzot, D. Maternal uniparental disomy 14 dissection of the phenotype with respect to rare autosomal recessively inherited traits, trisomy mosaicism, and genomic imprinting. Annales de Génétique 2004, 47, 251–260. [Google Scholar] [CrossRef]
- Cortessis, V.K.; Thomas, D.C.; Levine, A.J.; Breton, C.V.; Mack, T.M.; Siegmund, K.D.; Haile, R.W.; Laird, P.W. Environmental epigenetics: Prospects for studying epigenetic mediation of exposure–response relationships. Qual. Life Res. 2012, 131, 1565–1589. [Google Scholar] [CrossRef]
- Zhang, W.P.; Yang, J.; Lv, Y.; Li, S.; Qiang, M. Paternal benzo[a]pyrene exposure alters the sperm DNA methylation levels of imprinting genes in F0 generation mice and their unexposed F1-2 male offspring. Chemosphere 2019, 228, 586–594. [Google Scholar] [CrossRef] [PubMed]
- Skinner, M.K. Endocrine disruptor induction of epigenetic transgenerational inheritance of disease. Mol. Cell. Endocrinol. 2014, 398, 4–12. [Google Scholar] [CrossRef] [PubMed]
| Compound | Retention Time (min) | Parent Ion (m/z) | Daughter Ion (m/z) | Collision Energy (eV) |
|---|---|---|---|---|
| 2-OHN | 11.33 | 143 | 115 | 30 |
| 1-OHN | 12.08 | 143 | 115 | 30 |
| 3-OHF | 15.08 | 181 | 180 | 30 |
| 2-OHF | 15.50 | 181 | 180 | 30 |
| 2-OHPH | 16.49 | 193 | 165 | 30 |
| 1-OHPH | 17.54 | 193 | 171 | 30 |
| 1-OHP | 20.83 | 217 | 189 | 35 |
| Groups | N | 2-OHN | 1-OHN | 3-OHF | 2-OHF | 2-OHPH | 1-OHPH | 1-OHP |
|---|---|---|---|---|---|---|---|---|
| Median (25%–75%) | Median (25%–75%) | Median (25%–75%) | Median (25%–75%) | Median (25%–75%) | Median (25%–75%) | Median (25%–75%) | ||
| Age | ||||||||
| 24–29 | 61 | 0.010 (0–0.701) | 0 (0–0.215) | 0.005 (0–0.044) | 0.023 (0–0.076) | 0.166 (0–0.351) | 0.023 (0.003–0.070) | 0.009 (0–0.071) |
| 30–32 | 61 | 0 (0–0.253) | 0 (0–0.009) | 0.001 (0–0.043) | 0.012 (0–0.047) | 0.090 (0–0.250) | 0.013 (0–0.040) | 0.009 (0–0.039) |
| 33–35 | 53 | 0 (0–0.178) | 0 (0–0.125) | 0.005 (0–0.053) | 0.014 (0–0.076) | 0.117 (0–0.340) | 0.013 (0–0.050) | 0.017 (0–0.069) |
| >35 | 44 | 0.016 (0–0.709) | 0.005 (0–0.408) | 0.006 (0–0.030) | 0.033 (0–0.094) | 0.145 (0–0.450) | 0.026 (0.011–0.075) | 0.014 (0–0.070) |
| Education | ||||||||
| Primary School or Below | 4 | 0.196 (0.034–4.908) | 0 (0–0.148) | 0 (0–0.005) | 0.024 (0–0.121) | 0.023 (0–1.083) | 0.036 (0.005–0.172) | 0.006 (0.001–0.046) |
| Junior high | 57 | 0.011 (0–0.658) | 0 (0–0.024) | 0.006 (0–0.049) | 0.027 (0–0.087) | 0.138 (0–0.332) | 0.010 (0.001–0.047) | 0.019 (0–0.075) |
| High school | 56 | 0.003 (0–0.739) | 0.006 (0–0.042) | 0.035 (0–0.054) | 0.028 (0–0.078) | 0.095 (0–0.275) | 0.015 (0–0.065) | 0.010 (0–0.054) |
| College and above | 102 | 0 (0–0.084) | 0 (0–0.009) | 0.035 (0–0.028) | 0.015 (0–0.061) | 0.146 (0–0.332) | 0021 (0.003–0.048) | 0.008 (0–0.052) |
| Smoking (years) | ||||||||
| 0 | 80 | 0 (0–0.160) | 0 (0–0.009) | 0.005 (0–0.030) | 0.014 (0–0.050) | 0.134 (0.007–0.367) | 0.018 (0.006–0.050) | 0.009 (0–0.057) |
| 1–3 | 9 | 0 (0–0.128) | 0 (0–0.004) | 0.009 (0.001–0.104) | 0.025 (0.001–0.080) | 0.211 (0.094–0.352) | 0.010 (0.003–0.060) | 0.008 (0–0.038) |
| 3–5 | 17 | 0.010 (0–0.389) | 0 (0–0.006) | 0 (0–0.028) | 0.011 (0–0.059) | 0.091 (0–0.201) | 0.010 (0–0.060) | 0 (0–0.011) |
| 6–10 | 43 | 0 (0–1.234) | 0 (0–0.040) | 0 (0–0.042) | 0.014 (0–0.083) | 0.129 (0–0.249) | 0.014 (0–0.051) | 0.011 (0–0.052) |
| >10 | 70 | 0.039 (0–0.504) | 0.006 (0–0.036) | 0.004 (0–0.040) | 0.035 (0–0.100) | 0.075 (0–0.317) | 0.018 (0–0.061) | 0.019 (0–0.074) |
| Drinking (years) | ||||||||
| 0 | 134 | 0 (0–0.427) | 0 (0–0.020) | 0.002 (0–0.038) | 0.025 (0–0.073) | 0.162 (0–0.366) | 0.028 (0.002–0.058) | 0.013 (0–0.060) |
| 0–3 | 7 | 0 (0–0.573) | 0 (0–0.005) | 0.018 (0–0.213) | 0 (0–0.050) | 0 (0–0.508) | 0.014 (0–0.080) | 0 (0–0.060) |
| 3–5 | 9 | 0.067 (0.003–4.017) | 0 (0–0.184) | 0.007 (0–0.092) | 0 (0–0.131) | 0.036 (0–0.191) | 0.025 (0–0.064) | 0.001 (0–0.092) |
| 6–10 | 28 | 0 (0–0.776) | 0 (0–0.026) | 0.006 (0–0.079) | 0.003 (0–0.068) | 0.071 (0–0.294) | 0.009 (0–0.038) | 0.011 (0–0.065) |
| >10 | 41 | 0 (0–0.095) | 0 (0–0.016) | 0.003 (0–0.026) | 0.014 (0–0.049) | 0.100 (0–0.243) | 0.011 (0.003–0.032) | 0.009 (0–0.042) |
| Bacon * | ||||||||
| 0 | 180 | 0 (0–0.536) | 0 (0–0.020) | 0.002 (0–0.031) | 0.024 (0–0.062) | 0.141 (0–0.329) | 0.016 (0–0.054) | 0.009 (0–0.056) |
| 1–2 | 32 | 0 (0–0.191) | 0 (0–0.007) | 0.007 (0–0.089) | 0.018 (0–0.079) | 0.078 (0–0.233) | 0.011 (0–0.037) | 0.018 (0–0.074) |
| 3–4 | 6 | 0 (0–0.1152) | 0 (0–0.014) | 0.024 (0–0.295) | 0.005 (0–0.066) | 0 (0–0.748) | 0.027 (0.003–0.065) | 0.013 (0.003–0.063) |
| ≧5 | 1 | - | - | - | - | - | - | - |
| OH-PAH | N | Min. | Max. | 25% | Median | 75% |
|---|---|---|---|---|---|---|
| 2-OHN | 219 | 0.000 | 98.346 | 0.000 | 0.000 | 0.428 |
| 1-OHN | 219 | 0.000 | 1.996 | 0.000 | 0.000 | 0.016 |
| 3-OHF | 219 | 0.000 | 0.944 | 0.000 | 0.004 | 0.039 |
| 2-OHF | 219 | 0.000 | 0.881 | 0.000 | 0.023 | 0.069 |
| 2-OHPH | 219 | 0.000 | 4.224 | 0.000 | 0.129 | 0.319 |
| 1-OHPH | 219 | 0.000 | 0.418 | 0.000 | 0.015 | 0.052 |
| 1-OHP | 219 | 0.000 | 1.837 | 0.000 | 0.010 | 0.058 |
| OH-PAH | H19 | PEG3 | MEG3 |
|---|---|---|---|
| 2-OHN | −0.027 | −0.041 | −0.028 |
| 1-OHN | 0.029 | 0.012 | 0.021 |
| 3-OHF | 0.097 | 0.012 | 0.061 |
| 2-OHF | 0.125 | −0.006 | −0.067 |
| 2-OHPH | 0.102 | 0.034 | 0.026 |
| 1-OHPH | 0.189 * | 0.190 * | 0.130 |
| 1-OHP | −0.127 | 0.147 | 0.054 |
| Gene Sites | 1-OHPH | 1-OHP | ||
|---|---|---|---|---|
| r | p | r | p | |
| H19-CpG4 | 0.170 | 0.024 | - | - |
| H19-CpG6 | 0.158 | 0.037 | - | - |
| PEG3-CpG1 | 0.207 | 0.006 | 0.156 | 0.034 |
| MEG3-CpG2 | 0.201 | 0.008 | - | - |
| B | SE | t | q (FDR) | |
|---|---|---|---|---|
| 1-OHP | −3.62 | 1.05 | −3.43 | <0.01 |
| 1-OHPH | 17.72 | 6.63 | 2.67 | 0.01 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, Y.; Lu, Z.; Wang, L.; Qiang, M. Correlation of Internal Exposure Levels of Polycyclic Aromatic Hydrocarbons to Methylation of Imprinting Genes of Sperm DNA. Int. J. Environ. Res. Public Health 2019, 16, 2606. https://doi.org/10.3390/ijerph16142606
Ma Y, Lu Z, Wang L, Qiang M. Correlation of Internal Exposure Levels of Polycyclic Aromatic Hydrocarbons to Methylation of Imprinting Genes of Sperm DNA. International Journal of Environmental Research and Public Health. 2019; 16(14):2606. https://doi.org/10.3390/ijerph16142606
Chicago/Turabian StyleMa, Yufeng, Zhaoxu Lu, Li Wang, and Mei Qiang. 2019. "Correlation of Internal Exposure Levels of Polycyclic Aromatic Hydrocarbons to Methylation of Imprinting Genes of Sperm DNA" International Journal of Environmental Research and Public Health 16, no. 14: 2606. https://doi.org/10.3390/ijerph16142606
APA StyleMa, Y., Lu, Z., Wang, L., & Qiang, M. (2019). Correlation of Internal Exposure Levels of Polycyclic Aromatic Hydrocarbons to Methylation of Imprinting Genes of Sperm DNA. International Journal of Environmental Research and Public Health, 16(14), 2606. https://doi.org/10.3390/ijerph16142606





