Selective Utilization of Polyguluronate by the Human Gut Bacteroides Species
Abstract
1. Introduction
2. Results
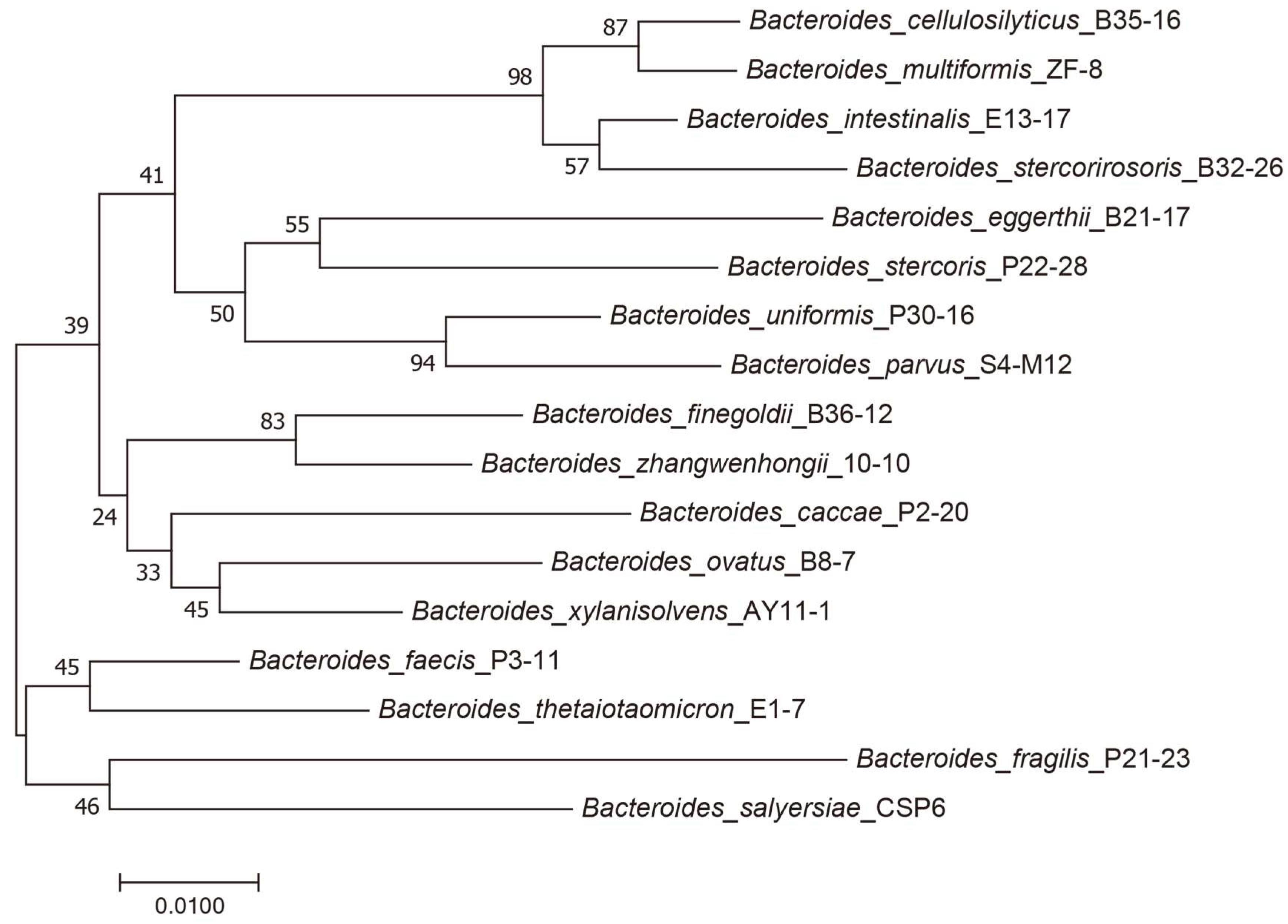
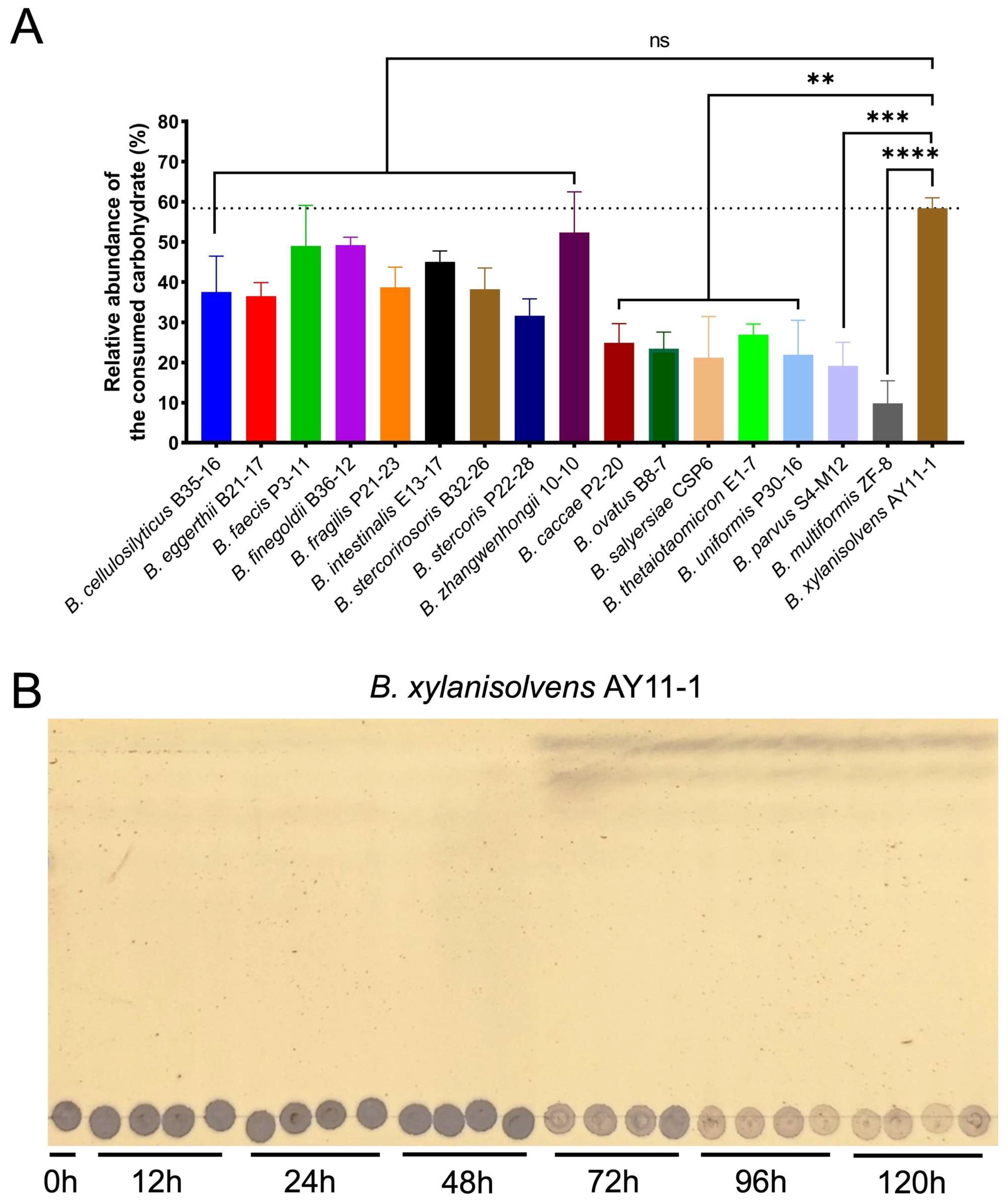
2.1. Each Bacteroides Species Was Characterized with a Unique Capability for PG Utilization
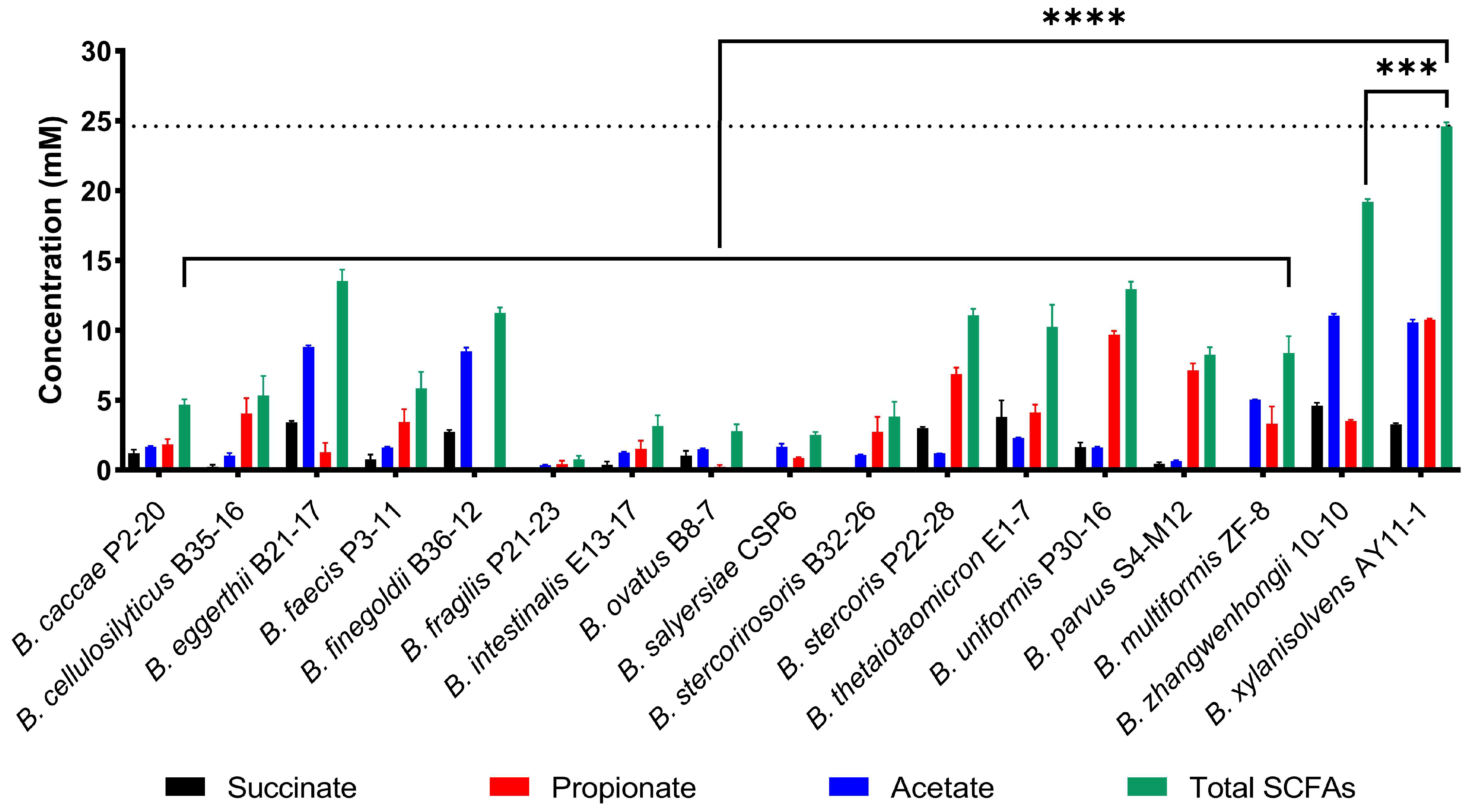
2.2. B. xylanisolvens Was Potentially a Keystone Bacterium Responsible for PG Utilization
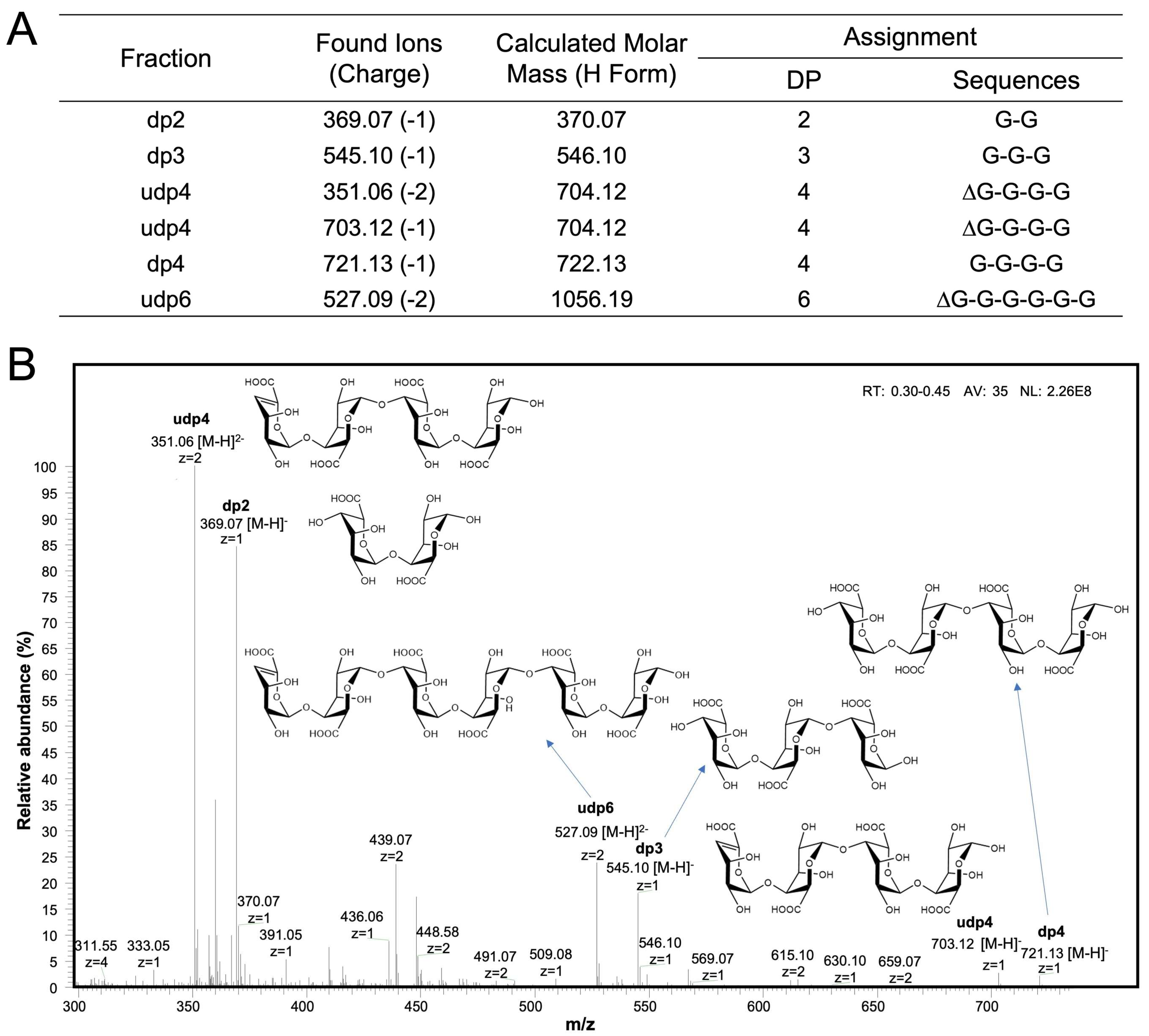
2.3. PG Was Degraded into a Series of Oligosaccharides by B. xylanisolvens
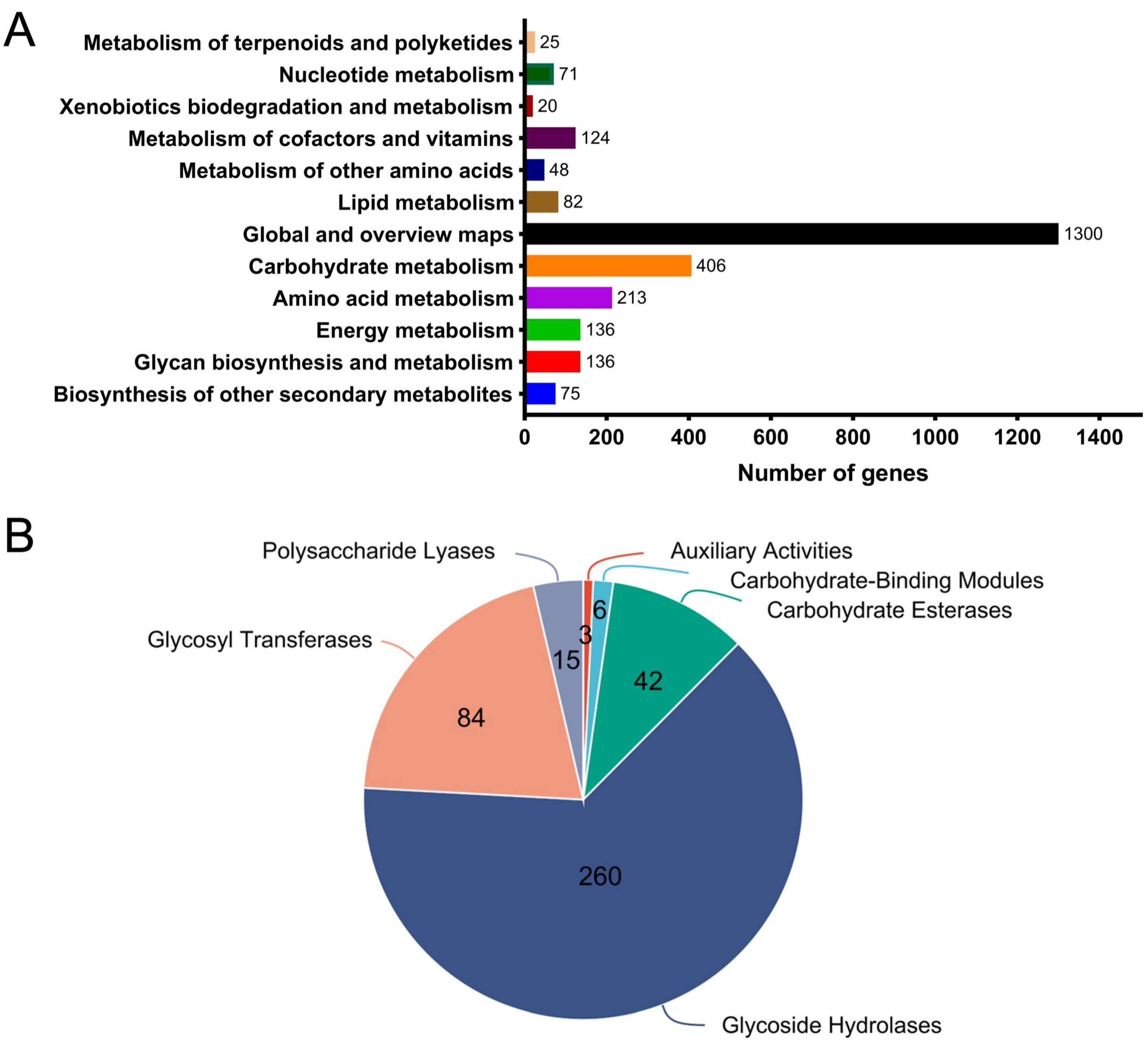
2.4. Genomic Analysis Linked PL17_2 and PL6_1 to PG Degradation in B. xylanisolvens
3. Discussion
3.1. Strengths and Key Findings of the Study
3.2. Outlooks and Future Directions in the Field
3.3. Study Limitations
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Bacterial Strains
4.3. In Vitro Fermentation
4.4. Carbohydrate Utilization Analysis
4.5. SCFAs Analysis
4.6. Degradation Products Analysis
4.7. Genome Sequencing and Bioinformatics Analysis
4.8. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| PG | Polyguluronate |
| OD | Optical density |
| TLC | Thin-layer chromatography |
| SCFAs | Short-chain fatty acids |
| MS | Mass spectrometry |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| CAZymes | Carbohydrate active enzymes |
| PLs | Polysaccharide lyases |
| GHs | Glycoside hydrolases |
| AAs | Auxiliary activities |
| GTs | Glycosyltransferases |
| CBMs | Carbohydrate-binding modules |
| CEs | Carbohydrate esterases |
| Sus | Starch utilization system |
| PM | Polymannuronate |
| HPLC | High-performance liquid chromatography |
| PULs | Polysaccharide utilization loci |
References
- Bi, D.; Yang, X.; Yao, L.; Hu, Z.; Li, H.; Xu, X.; Lu, J. Potential food and nutraceutical applications of alginate: A review. Mar. Drugs 2022, 20, 564. [Google Scholar] [CrossRef]
- Guo, X.; Wang, Y.; Qin, Y.M.; Shen, P.L.; Peng, Q. Structures, properties and application of alginic acid: A review. Int. J. Biol. Macromol. 2020, 162, 618–628. [Google Scholar] [CrossRef]
- Łętocha, A.; Miastkowska, M.; Sikora, E. Preparation and characteristics of alginate microparticles for food, pharmaceutical and cosmetic applications. Polymers 2022, 14, 3834. [Google Scholar] [CrossRef]
- Gao, Y.Y.; Liu, W.; Wang, W.; Zhao, X.; Wang, F.H. Polyguluronate sulfate (PGS) attenuates immunological liver injury in vitro and in vivo. Int. J. Biol. Macromol. 2018, 114, 592–598. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Bi, D.; Yi, J.; Yao, L.; Cao, J.; Yang, P.; Li, M.; Wu, Y.; Xu, H.; Hu, Z.; et al. Soy protein isolate-polyguluronate nanoparticles loaded with resveratrol for effective treatment of colitis. Food Chem. 2023, 410, 135418. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Ma, M.; Wang, Y.; Dai, W.; Fu, T.; Wang, L.; Shang, Q.; Yu, G. Polyguluronate alleviates ulcerative colitis by targeting the gut commensal Lactobacillus murinus and its anti-inflammatory metabolites. Int. J. Biol. Macromol. 2024, 257, 128592. [Google Scholar] [CrossRef]
- Zheng, L.X.; Chen, X.Q.; Cheong, K.L. Current trends in marine algae polysaccharides: The digestive tract, microbial catabolism, and prebiotic potential. Int. J. Biol. Macromol. 2020, 151, 344–354. [Google Scholar] [CrossRef]
- Shang, Q.; Jiang, H.; Cai, C.; Hao, J.; Li, G.; Yu, G. Gut microbiota fermentation of marine polysaccharides and its effects on intestinal ecology: An overview. Carbohydr. Polym. 2018, 179, 173–185. [Google Scholar] [CrossRef] [PubMed]
- Cheong, K.-L.; Yu, B.; Chen, J.; Zhong, S. A comprehensive review of the cardioprotective effect of marine algae polysaccharide on the gut microbiota. Foods 2022, 11, 3550. [Google Scholar] [CrossRef]
- Ramnani, P.; Chitarrari, R.; Tuohy, K.; Grant, J.; Hotchkiss, S.; Philp, K.; Campbell, R.; Gill, C.; Rowland, I. In vitro fermentation and prebiotic potential of novel low molecular weight polysaccharides derived from agar and alginate seaweeds. Anaerobe 2012, 18, 1–6. [Google Scholar] [CrossRef]
- Shannon, E.; Conlon, M.; Hayes, M. Seaweed components as potential modulators of the gut microbiota. Mar. Drugs 2021, 19, 358. [Google Scholar] [CrossRef] [PubMed]
- Bai, S.; Chen, H.; Zhu, L.; Liu, W.; Yu, H.D.; Wang, X.; Yin, Y. Comparative study on the in vitro effects of Pseudomonas aeruginosa and seaweed alginates on human gut microbiota. PLoS ONE 2017, 12, e0171576. [Google Scholar] [CrossRef]
- Fu, T.; Pan, L.; Shang, Q.; Yu, G. Fermentation of alginate and its derivatives by different enterotypes of human gut microbiota: Towards personalized nutrition using enterotype-specific dietary fibers. Int. J. Biol. Macromol. 2021, 183, 1649–1659. [Google Scholar] [CrossRef]
- Lozupone, C.A.; Stombaugh, J.I.; Gordon, J.I.; Jansson, J.K.; Knight, R. Diversity, stability and resilience of the human gut microbiota. Nature 2012, 489, 220–230. [Google Scholar] [CrossRef]
- McCallum, G.; Tropini, C. The gut microbiota and its biogeography. Nat. Rev. Microbiol. 2024, 22, 105–118. [Google Scholar] [CrossRef] [PubMed]
- Tanca, A.; Abbondio, M.; Palomba, A.; Fraumene, C.; Manghina, V.; Cucca, F.; Fiorillo, E.; Uzzau, S. Potential and active functions in the gut microbiota of a healthy human cohort. Microbiome 2017, 5, 79. [Google Scholar] [CrossRef]
- Xiong, W.; Abraham, P.E.; Li, Z.; Pan, C.; Hettich, R.L. Microbial metaproteomics for characterizing the range of metabolic functions and activities of human gut microbiota. Proteomics 2015, 15, 3424–3438. [Google Scholar] [CrossRef]
- Franzosa, E.A.; Sirota-Madi, A.; Avila-Pacheco, J.; Fornelos, N.; Haiser, H.J.; Reinker, S.; Vatanen, T.; Hall, A.B.; Mallick, H.; McIver, L.J.; et al. Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat. Microbiol. 2019, 4, 293–305. [Google Scholar] [CrossRef]
- Zafar, H.; Saier, M.H., Jr. Gut Bacteroides species in health and disease. Gut Microbes 2021, 13, 1848158. [Google Scholar] [CrossRef]
- Shin, J.H.; Tillotson, G.; MacKenzie, T.N.; Warren, C.A.; Wexler, H.M.; Goldstein, E.J.C. Bacteroides and related species: The keystone taxa of the human gut microbiota. Anaerobe 2024, 85, 102819. [Google Scholar] [CrossRef] [PubMed]
- Sonnenburg, E.D.; Zheng, H.; Joglekar, P.; Higginbottom, S.K.; Firbank, S.J.; Bolam, D.N.; Sonnenburg, J.L. Specificity of polysaccharide use in intestinal Bacteroides species determines diet-induced microbiota alterations. Cell 2010, 141, 1241–1252. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Hu, J.; Geng, F.; Nie, S. Bacteroides utilization for dietary polysaccharides and their beneficial effects on gut health. Food Sci. Hum. Wellness 2022, 11, 1101–1110. [Google Scholar] [CrossRef]
- Qu, Z.; Liu, H.; Yang, J.; Zheng, L.; Huang, J.; Wang, Z.; Xie, C.; Zuo, W.; Xia, X.; Sun, L.; et al. Selective utilization of medicinal polysaccharides by human gut Bacteroides and Parabacteroides species. Nat. Commun. 2025, 16, 638. [Google Scholar] [CrossRef] [PubMed]
- Fu, T.; Wang, Y.; Ma, M.; Dai, W.; Pan, L.; Shang, Q.; Yu, G. Isolation of alginate-degrading bacteria from the human gut microbiota and discovery of Bacteroides xylanisolvens AY11-1 as a novel anti-colitis probiotic bacterium. Nutrients 2023, 15, 1352. [Google Scholar] [CrossRef]
- Wang, Y.; Ma, M.; Dai, W.; Shang, Q.; Yu, G. Bacteroides salyersiae is a potent chondroitin sulfate-degrading species in the human gut microbiota. Microbiome 2024, 12, 41. [Google Scholar] [CrossRef]
- Koh, A.; De Vadder, F.; Kovatcheva-Datchary, P.; Bäckhed, F. From dietary fiber to host physiology: Short-chain fatty acids as key bacterial metabolites. Cell 2016, 165, 1332–1345. [Google Scholar] [CrossRef]
- Morrison, D.J.; Preston, T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes 2016, 7, 189–200. [Google Scholar] [CrossRef]
- Fusco, W.; Lorenzo, M.B.; Cintoni, M.; Porcari, S.; Rinninella, E.; Kaitsas, F.; Lener, E.; Mele, M.C.; Gasbarrini, A.; Collado, M.C.; et al. Short-chain fatty-acid-producing bacteria: Key components of the human gut microbiota. Nutrients 2023, 15, 2211. [Google Scholar] [CrossRef]
- El Kaoutari, A.; Armougom, F.; Gordon, J.I.; Raoult, D.; Henrissat, B. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat. Rev. Genet. 2013, 11, 497–504. [Google Scholar] [CrossRef]
- Flint, H.J.; Scott, K.P.; Duncan, S.H.; Louis, P.; Forano, E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes 2012, 3, 289–306. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, T.; Ghosh, T.S.; Mande, S.S. Global profiling of carbohydrate active enzymes in human gut microbiome. PLoS ONE 2015, 10, e0142038. [Google Scholar] [CrossRef] [PubMed]
- Wardman, J.F.; Bains, R.K.; Rahfeld, P.; Withers, S.G. Carbohydrate-active enzymes (CAZymes) in the gut microbiome. Nat. Rev. Microbiol. 2022, 20, 542–556. [Google Scholar] [CrossRef]
- Despres, J.; Forano, E.; Lepercq, P.; Comtet-Marre, S.; Jubelin, G.; Chambon, C.; Yeoman, C.J.; Miller, M.E.B.; Fields, C.J.; Martens, E.; et al. Xylan degradation by the human gut Bacteroides xylanisolvens XB1AT involves two distinct gene clusters that are linked at the transcriptional level. BMC Genom. 2016, 17, 326. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Zheng, L.; Guo, Z.; Tang, T.C.; Zhu, B.W. Alginate degrading enzymes: An updated comprehensive review of the structure, catalytic mechanism, modification method and applications of alginate lyases. Crit. Rev. Biotechnol. 2021, 41, 953–968. [Google Scholar] [CrossRef]
- Lozada, M.; Dionisi, H.M. Insights into putative alginate lyases from epipelagic and mesopelagic communities of the global ocean. Sci. Rep. 2025, 15, 8111. [Google Scholar] [CrossRef]
- Ma, W.J.; Wang, C.; Kothandapani, J.; Luzentales-Simpson, M.; Menzies, S.C.; Bescucci, D.M.; Lange, M.E.; Fraser, A.S.C.; Gusse, J.F.; House, K.E.; et al. Bespoke plant glycoconjugates for gut microbiota-mediated drug targeting. Science 2025, 388, 1410–1416. [Google Scholar] [CrossRef]
- Koropatkin, N.M.; Cameron, E.A.; Martens, E.C. How glycan metabolism shapes the human gut microbiota. Nat. Rev. Genet. 2012, 10, 323–335. [Google Scholar] [CrossRef]
- Tannock, G.W. Modulating the gut microbiota of humans by dietary intervention with plant glycans. Appl. Environ. Microbiol. 2021, 87, e02757-20. [Google Scholar] [CrossRef]
- Zhernakova, A.; Kurilshikov, A.; Bonder, M.J.; Tigchelaar, E.F.; Schirmer, M.; Vatanen, T.; Mujagic, Z.; Vila, A.V.; Falony, G.; Vieira-Silva, S.; et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science 2016, 352, 565–569. [Google Scholar] [CrossRef] [PubMed]
- Tufail, M.A.; Schmitz, R.A. Exploring the probiotic potential of Bacteroides spp. within one health paradigm. Probiotics Antimicrob. Proteins 2025, 17, 681–704. [Google Scholar] [CrossRef] [PubMed]
- Tan, H.; Zhai, Q.; Chen, W. Investigations of Bacteroides spp. towards next-generation probiotics. Food Res. Int. 2019, 116, 637–644. [Google Scholar] [CrossRef] [PubMed]
- De Filippis, F.; Esposito, A.; Ercolini, D. Outlook on next-generation probiotics from the human gut. Cell. Mol. Life Sci. 2022, 79, 76. [Google Scholar] [CrossRef] [PubMed]
- Rawat, P.S.; Seyed, H.A.S.; Meng, X.; Liu, W. Utilization of glycosaminoglycans by the human gut microbiota: Participating bacteria and their enzymatic machineries. Gut Microbes 2022, 14, 2068367. [Google Scholar] [CrossRef] [PubMed]
- Brown, H.A.; Koropatkin, N.M. Host glycan utilization within the Bacteroidetes Sus-like paradigm. Glycobiology 2021, 31, 697–706. [Google Scholar] [CrossRef]
- Bolam, D.N.; Koropatkin, N.M. Glycan recognition by the Bacteroidetes Sus-like systems. Curr. Opin. Struct. Biol. 2012, 22, 563–569. [Google Scholar] [CrossRef]
- Despres, J.; Forano, E.; Lepercq, P.; Comtet-Marre, S.; Jubelin, G.; Yeoman, C.J.; Miller, M.E.; Fields, C.J.; Terrapon, N.; Le Bourvellec, C.; et al. Unraveling the pectinolytic function of Bacteroides xylanisolvens using a RNA-seq approach and mutagenesis. BMC Genom. 2016, 17, 147. [Google Scholar] [CrossRef]
- Rakoff-Nahoum, S.; Foster, K.R.; Comstock, L.E. The evolution of cooperation within the gut microbiota. Nat. Cell Biol. 2016, 533, 255–259. [Google Scholar] [CrossRef]
- Culp, E.J.; Goodman, A.L. Cross-feeding in the gut microbiome: Ecology and mechanisms. Cell Host Microbe 2023, 31, 485–499. [Google Scholar] [CrossRef]
- Fang, Z.; Ma, M.; Wang, Y.; Dai, W.; Shang, Q.; Yu, G. Degradation and fermentation of hyaluronic acid by Bacteroides spp. from the human gut microbiota. Carbohydr. Polym. 2024, 334, 122074. [Google Scholar] [CrossRef]
- Dubois, M.; Gilles, K.; Hamilton, J.; Rebers, P.; Smith, F. A colorimetric method for the determination of sugars. Nature 1951, 168, 167. [Google Scholar] [CrossRef]
- Dubois, M.; Gilles, K.A.; Hamilton, J.K.; Rebers, P.A.; Smith, F. Colorimetric method for determination of sugars and related substances. Anal. Chem. 1956, 28, 350–356. [Google Scholar] [CrossRef]
- Drula, E.; Garron, M.-L.; Dogan, S.; Lombard, V.; Henrissat, B.; Terrapon, N. The carbohydrate-active enzyme database: Functions and literature. Nucleic Acids Res. 2022, 50, D571–D577. [Google Scholar] [CrossRef] [PubMed]
- Henrissat, B.; Davies, G. Structural and sequence-based classification of glycoside hydrolases. Curr. Opin. Struct. Biol. 1997, 7, 637–644. [Google Scholar] [CrossRef] [PubMed]
- Coutinho, P.M.; Deleury, E.; Davies, G.J.; Henrissat, B. An evolving hierarchical family classification for glycosyltransferases. J. Mol. Biol. 2003, 328, 307–317. [Google Scholar] [CrossRef] [PubMed]
- Lombard, V.; Bernard, T.; Rancurel, C.; Brumer, H.; Coutinho, P.M.; Henrissat, B. A hierarchical classification of polysaccharide lyases for glycogenomics. Biochem. J. 2010, 432, 437–444. [Google Scholar] [CrossRef]
- Boraston, A.B.; Bolam, D.N.; Gilbert, H.J.; Davies, G.J. Carbohydrate-binding modules: Fine-tuning polysaccharide recognition. Biochem. J. 2004, 382, 769–781. [Google Scholar] [CrossRef]
- Levasseur, A.; Drula, E.; Lombard, V.; Coutinho, P.M.; Henrissat, B. Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes. Biotechnol. Biofuels 2013, 6, 41. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, N.; Li, M.; Yuan, X.; Fu, T.; Lv, Y.; Shang, Q. Selective Utilization of Polyguluronate by the Human Gut Bacteroides Species. Mar. Drugs 2025, 23, 348. https://doi.org/10.3390/md23090348
Liu N, Li M, Yuan X, Fu T, Lv Y, Shang Q. Selective Utilization of Polyguluronate by the Human Gut Bacteroides Species. Marine Drugs. 2025; 23(9):348. https://doi.org/10.3390/md23090348
Chicago/Turabian StyleLiu, Nuo, Ming Li, Xiangting Yuan, Tianyu Fu, Youjing Lv, and Qingsen Shang. 2025. "Selective Utilization of Polyguluronate by the Human Gut Bacteroides Species" Marine Drugs 23, no. 9: 348. https://doi.org/10.3390/md23090348
APA StyleLiu, N., Li, M., Yuan, X., Fu, T., Lv, Y., & Shang, Q. (2025). Selective Utilization of Polyguluronate by the Human Gut Bacteroides Species. Marine Drugs, 23(9), 348. https://doi.org/10.3390/md23090348




