Fish Skin Mucus Extracts: An Underexplored Source of Antimicrobial Agents
Abstract
1. Introduction
2. Fish Skin Mucus as a Promising Source of Antimicrobials
3. Composition of Fish Skin Mucus in Innate Immunity Antimicrobial Molecules
3.1. Antimicrobial Peptides (AMPs)
3.2. Proteins
3.3. Other Components
4. Antimicrobial Activity of Fish Skin Mucus
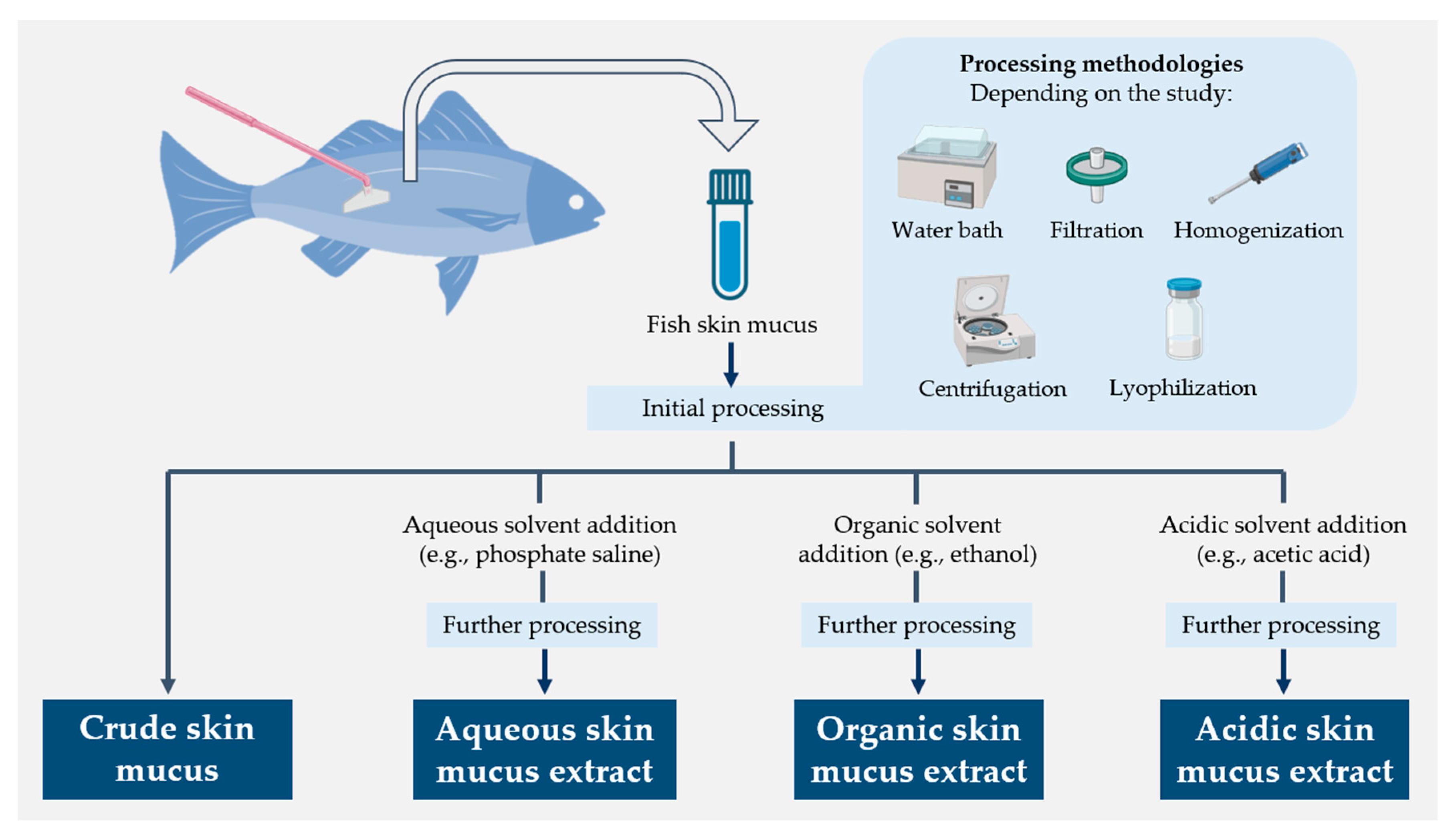
4.1. Antibacterial Activity of Fish Skin Mucus Extracts
4.1.1. Aqueous Extractions
4.1.2. Organic Extractions
4.1.3. Acidic Extractions
4.1.4. Crude Mucus
4.2. Antifungal Activity of Fish Skin Mucus Extracts
4.3. Antiviral Activity of Fish Skin Mucus
5. Omics Techniques as a Promising Tool in Fish Skin Mucus Research
6. Conclusions
7. Recommendations and Future Perspectives
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Spellberg, B.; Guidos, R.; Gilbert, D.; Bradley, J.; Boucher, H.W.; Scheld, W.M.; Bartlett, J.G.; Edwards, J., Jr.; Infectious Diseases Society of America. The epidemic of antibiotic-resistant infections: A call to action for the medical community from the Infectious Diseases Society of America. Clin. Infect. Dis. 2008, 46, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Perlin, D.S.; Rautemaa-Richardson, R.; Alastruey-Izquierdo, A. The global problem of antifungal resistance: Prevalence, mechanisms, and management. Lancet Infect. Dis. 2017, 17, e383–e392. [Google Scholar] [CrossRef]
- Mason, S.; Devincenzo, J.P.; Toovey, S.; Wu, J.Z.; Whitley, R.J. Comparison of antiviral resistance across acute and chronic viral infections. Antivir. Res. 2018, 158, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Butler, M.S.; Paterson, D.L. Antibiotics in the clinical pipeline in October 2019. J. Antibiot. 2020, 73, 329–364. [Google Scholar] [CrossRef] [PubMed]
- Bassetti, M.; Merelli, M.; Temperoni, C.; Astilean, A. New antibiotics for bad bugs: Where are we? Ann. Clin. Microbiol. Antimicrob. 2013, 12, 22. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, M.; Ćirić, A.; Stojković, D. Emerging antifungal targets and strategies. Int. J. Mol. Sci. 2022, 23, 2756. [Google Scholar] [CrossRef]
- Tompa, D.R.; Immanuel, A.; Srikanth, S.; Kadhirvel, S. Trends and strategies to combat viral infections: A review on FDA approved antiviral drugs. Int. J. Biol. Macromol. 2021, 172, 524–541. [Google Scholar] [CrossRef]
- Baker, R.E.; Mahmud, A.S.; Miller, I.F.; Rajeev, M.; Rasambainarivo, F.; Rice, B.L.; Takahashi, S.; Tatem, A.J.; Wagner, C.E.; Wang, L.-F. Infectious disease in an era of global change. Nat. Rev. Microbiol. 2022, 20, 193–205. [Google Scholar] [CrossRef]
- Pawlowski, A.C.; Johnson, J.W.; Wright, G.D. Evolving medicinal chemistry strategies in antibiotic discovery. Curr. Opin. Biotechnol. 2016, 42, 108–117. [Google Scholar] [CrossRef]
- Miethke, M.; Pieroni, M.; Weber, T.; Brönstrup, M.; Hammann, P.; Halby, L.; Arimondo, P.B.; Glaser, P.; Aigle, B.; Bode, H.B. Towards the sustainable discovery and development of new antibiotics. Nat. Rev. Chem. 2021, 5, 726–749. [Google Scholar] [CrossRef]
- Falco, A.; Adamek, M.; Pereiro, P.; Hoole, D.; Encinar, J.A.; Novoa, B.; Mallavia, R. The Immune System of Marine Organisms as Source for Drugs against Infectious Diseases. Mar. Drugs 2022, 20, 363. [Google Scholar] [CrossRef]
- Breitbart, M.; Rohwer, F. Here a virus, there a virus, everywhere the same virus? Trends Microbiol. 2005, 13, 278–284. [Google Scholar] [CrossRef]
- Hawksworth, D.L.; Lücking, R. Fungal diversity revisited: 2.2 to 3.8 million species. Microbiol. Spectr. 2017, 5, 4. [Google Scholar] [CrossRef] [PubMed]
- Flemming, H.-C.; Wuertz, S. Bacteria and archaea on Earth and their abundance in biofilms. Nat. Rev. Microbiol. 2019, 17, 247–260. [Google Scholar] [CrossRef]
- Durand, G.A.; Raoult, D.; Dubourg, G. Antibiotic discovery: History, methods and perspectives. Int. J. Antimicrob. Agents 2019, 53, 371–382. [Google Scholar] [CrossRef]
- Spížek, J.; Sigler, K.; Řezanka, T.; Demain, A. Biogenesis of antibiotics—Viewing its history and glimpses of the future. Folia Microbiol. 2016, 61, 347–358. [Google Scholar] [CrossRef]
- Oloke, J. Activity pattern of natural and synthetic antibacterial agents among hospital isolates. Microbios 2000, 102, 175–181. [Google Scholar] [PubMed]
- Pancu, D.F.; Scurtu, A.; Macasoi, I.G.; Marti, D.; Mioc, M.; Soica, C.; Coricovac, D.; Horhat, D.; Poenaru, M.; Dehelean, C. Antibiotics: Conventional therapy and natural compounds with antibacterial activity—A pharmaco-toxicological screening. Antibiotics 2021, 10, 401. [Google Scholar] [CrossRef]
- Bar-On, Y.M.; Phillips, R.; Milo, R. The biomass distribution on Earth. Proc. Natl. Acad. Sci. USA 2018, 115, 6506–6511. [Google Scholar] [CrossRef]
- Forterre, P.; Prangishvili, D. The origin of viruses. Res. Microbiol. 2009, 160, 466–472. [Google Scholar] [CrossRef] [PubMed]
- Mojzsis, S.J.; Arrhenius, G.; McKeegan, K.; Harrison, T.; Nutman, A.; Friend, C. Evidence for life on Earth before 3,800 million years ago. Nature 1996, 384, 55–59. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.Y.-C.; Kumar, S.; Hedges, S.B. Divergence time estimates for the early history of animal phyla and the origin of plants, animals and fungi. Proc. R. Soc. Lond. Ser. B Biol. Sci. 1999, 266, 163–171. [Google Scholar] [CrossRef] [PubMed]
- Richter, D.; Grün, R.; Joannes-Boyau, R.; Steele, T.E.; Amani, F.; Rué, M.; Fernandes, P.; Raynal, J.-P.; Geraads, D.; Ben-Ncer, A. The age of the hominin fossils from Jebel Irhoud, Morocco, and the origins of the Middle Stone Age. Nature 2017, 546, 293–296. [Google Scholar] [CrossRef] [PubMed]
- Murphy, W.J.; Eizirik, E.; Johnson, W.E.; Zhang, Y.P.; Ryder, O.A.; O’Brien, S.J. Molecular phylogenetics and the origins of placental mammals. Nature 2001, 409, 614–618. [Google Scholar] [CrossRef] [PubMed]
- Sallan, L.C. Major issues in the origins of ray-finned fish (A. ctinopterygii) biodiversity. Biol. Rev. 2014, 89, 950–971. [Google Scholar] [CrossRef]
- Suttle, C.A. Marine viruses—Major players in the global ecosystem. Nat. Rev. Microbiol. 2007, 5, 801–812. [Google Scholar] [CrossRef]
- Madsen, E.L. Microorganisms and their roles in fundamental biogeochemical cycles. Curr. Opin. Biotechnol. 2011, 22, 456–464. [Google Scholar] [CrossRef]
- Hagström, A.k.; Pommier, T.; Rohwer, F.; Simu, K.; Stolte, W.; Svensson, D.; Zweifel, U.L. Use of 16S ribosomal DNA for delineation of marine bacterioplankton species. Appl. Environ. Microbiol. 2002, 68, 3628–3633. [Google Scholar] [CrossRef]
- Margulis, L.; Chapman, M.J. Kingdoms and Domains: An Illustrated Guide to the Phyla of Life on Earth; Academic Press: Cambridge, MA, USA, 2009. [Google Scholar]
- Kong, D.-X.; Jiang, Y.-Y.; Zhang, H.-Y. Marine natural products as sources of novel scaffolds: Achievement and concern. Drug Discov. Today 2010, 15, 884–886. [Google Scholar] [CrossRef]
- Harizani, M.; Ioannou, E.; Roussis, V. The Laurencia paradox: An endless source of chemodiversity. Prog. Chem. Org. Nat. Prod. 2016, 102, 91–252. [Google Scholar]
- Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2019, 36, 122–173. [Google Scholar] [CrossRef] [PubMed]
- Ntie-Kang, F.; Svozil, D. An enumeration of natural products from microbial, marine and terrestrial sources. Phys. Sci. Rev. 2020, 5. [Google Scholar] [CrossRef]
- Principe, P.P.; Fisher, W.S. Spatial distribution of collections yielding marine natural products. J. Nat. Prod. 2018, 81, 2307–2320. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, S.; Adholeya, A.; Deshmukh, S.K. The pharmacological potential of non-ribosomal peptides from marine sponge and tunicates. Front. Pharmacol. 2016, 7, 333. [Google Scholar] [CrossRef] [PubMed]
- Brinchmann, M.F. Immune relevant molecules identified in the skin mucus of fish using -omics technologies. Mol. BioSyst. 2016, 12, 2056–2063. [Google Scholar] [CrossRef] [PubMed]
- Reverter, M.; Tapissier-Bontemps, N.; Lecchini, D.; Banaigs, B.; Sasal, P. Biological and Ecological Roles of External Fish Mucus: A Review. Fishes 2018, 3, 41. [Google Scholar] [CrossRef]
- Tiralongo, F.; Messina, G.; Lombardo, B.M.; Longhitano, L.; Li Volti, G.; Tibullo, D. Skin mucus of marine fish as a source for the development of antimicrobial agents. Front. Mar. Sci. 2020, 7, 760. [Google Scholar] [CrossRef]
- Nelson, J.S.; Grande, T.C.; Wilson, M.V. Fishes of the World; John Wiley & Sons: Hoboken, NJ, USA, 2016. [Google Scholar]
- Martin, A.P. Increasing genomic complexity by gene duplication and the origin of vertebrates. Am. Nat. 1999, 154, 111–128. [Google Scholar] [CrossRef]
- Hurley, I.A.; Mueller, R.L.; Dunn, K.A.; Schmidt, E.J.; Friedman, M.; Ho, R.K.; Prince, V.E.; Yang, Z.; Thomas, M.G.; Coates, M.I. A new time-scale for ray-finned fish evolution. Proc. R. Soc. B Biol. Sci. 2007, 274, 489–498. [Google Scholar] [CrossRef]
- Ravi, V.; Venkatesh, B. Rapidly evolving fish genomes and teleost diversity. Curr. Opin. Genet. Dev. 2008, 18, 544–550. [Google Scholar] [CrossRef] [PubMed]
- Azam, F.; Malfatti, F. Microbial structuring of marine ecosystems. Nat. Rev. Microbiol. 2007, 5, 782–791. [Google Scholar] [CrossRef] [PubMed]
- Bergh, Ø.; Børsheim, K.Y.; Bratbak, G.; Heldal, M. High abundance of viruses found in aquatic environments. Nature 1989, 340, 467–468. [Google Scholar] [CrossRef] [PubMed]
- Ducklow, H. Bacterial production and biomass in the oceans. Microb. Ecol. Oceans 2000, 1, 85–120. [Google Scholar]
- Grossart, H.-P.; Van den Wyngaert, S.; Kagami, M.; Wurzbacher, C.; Cunliffe, M.; Rojas-Jimenez, K. Fungi in aquatic ecosystems. Nat. Rev. Microbiol. 2019, 17, 339–354. [Google Scholar] [CrossRef] [PubMed]
- Castro, R.; Tafalla, C. Overview of fish immunity. In Mucosal Health in Aquaculture; Elsevier: Amsterdam, The Netherlands, 2015; pp. 3–54. [Google Scholar]
- Ponnappan, N.; Budagavi, D.P.; Yadav, B.K.; Chugh, A. Membrane-active peptides from marine organisms-antimicrobials, cell-penetrating peptides and Peptide toxins: Applications and prospects. Probiotics Antimicrob. Proteins 2015, 7, 75–89. [Google Scholar] [CrossRef] [PubMed]
- Litman, G.W.; Rast, J.P.; Fugmann, S.D. The origins of vertebrate adaptive immunity. Nat. Rev. Immunol. 2010, 10, 543–553. [Google Scholar] [CrossRef]
- Esteban, M.Á.; Cerezuela, R. Fish mucosal immunity: Skin. In Mucosal Health in Aquaculture; Elsevier: Amsterdam, The Netherlands, 2015; pp. 67–92. [Google Scholar]
- Salinas, I. The mucosal immune system of teleost fish. Biology 2015, 4, 525–539. [Google Scholar] [CrossRef]
- Ellis, A. Innate host defense mechanisms of fish against viruses and bacteria. Dev. Comp. Immunol. 2001, 25, 827–839. [Google Scholar] [CrossRef]
- Gomez, D.; Sunyer, J.O.; Salinas, I. The mucosal immune system of fish: The evolution of tolerating commensals while fighting pathogens. Fish Shellfish Immunol. 2013, 35, 1729–1739. [Google Scholar] [CrossRef]
- Otero-Gonzáiez, A.J.; Magalhaes, B.S.; Garcia-Villarino, M.; López-Abarrategui, C.; Sousa, D.A.; Dias, S.C.; Franco, O.L. Antimicrobial peptides from marine invertebrates as a new frontier for microbial infection control. FASEB J. 2010, 24, 1320–1334. [Google Scholar] [CrossRef]
- Abdelmohsen, U.R.; Balasubramanian, S.; Oelschlaeger, T.A.; Grkovic, T.; Pham, N.B.; Quinn, R.J.; Hentschel, U. Potential of marine natural products against drug-resistant fungal, viral, and parasitic infections. Lancet Infect. Dis. 2017, 17, e30–e41. [Google Scholar] [CrossRef] [PubMed]
- Tassanakajon, A.; Rimphanitchayakit, V.; Visetnan, S.; Amparyup, P.; Somboonwiwat, K.; Charoensapsri, W.; Tang, S. Shrimp humoral responses against pathogens: Antimicrobial peptides and melanization. Dev. Comp. Immunol. 2018, 80, 81–93. [Google Scholar] [CrossRef]
- Shephard, K.L. Functions for fish mucus. Rev. Fish Biol. Fish. 1994, 4, 401–429. [Google Scholar] [CrossRef]
- Esteban, M.Á. An Overview of the Immunological Defenses in Fish Skin. ISRN Immunol. 2012, 2012, 853470. [Google Scholar] [CrossRef]
- Shephard, K.L. Mucus on the epidermis of fish and its influence on drug delivery. Adv. Drug Deliv. Rev. 1993, 11, 403–417. [Google Scholar] [CrossRef]
- Liu, X.; Wu, H.; Liu, Q.; Wang, Q.; Xiao, J.; Zhang, Y. Skin-injured Zebrafish, Danio rerio, are more Susceptible to Vibrio anguillarum Infection. J. World Aquac. Soc. 2015, 46, 301–310. [Google Scholar] [CrossRef]
- Adamek, M.; Matras, M.; Rebl, A.; Stachnik, M.; Falco, A.; Bauer, J.; Miebach, A.-C.; Teitge, F.; Jung-Schroers, V.; Abdullah, M. Don’t Let It Get Under Your Skin!—Vaccination Protects the Skin Barrier of Common Carp From Disruption Caused by Cyprinid Herpesvirus 3. Front. Immunol. 2022, 13, 787021. [Google Scholar] [CrossRef]
- Dash, S.; Das, S.K.; Samal, J.; Thatoi, H.N. Epidermal mucus, a major determinant in fish health: A review. Iran. J. Vet. Res. 2018, 19, 72–81. [Google Scholar]
- Hussain, A.; Sachan, S.G. Fish Epidermal Mucus as a Source of Diverse Therapeutical Compounds. Int. J. Pept. Res. Ther. 2023, 29, 36. [Google Scholar] [CrossRef]
- Rakers, S.; Niklasson, L.; Steinhagen, D.; Kruse, C.; Schauber, J.; Sundell, K.; Paus, R. Antimicrobial peptides (AMPs) from fish epidermis: Perspectives for investigative dermatology. J. Investig. Dermatol. 2013, 133, 1140–1149. [Google Scholar] [CrossRef] [PubMed]
- Mahlapuu, M.; Håkansson, J.; Ringstad, L.; Björn, C. Antimicrobial peptides: An emerging category of therapeutic agents. Front. Cell. Infect. Microbiol. 2016, 6, 194. [Google Scholar] [CrossRef] [PubMed]
- Hancock, R.E.; Haney, E.F.; Gill, E.E. The immunology of host defence peptides: Beyond antimicrobial activity. Nat. Rev. Immunol. 2016, 16, 321. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, B.O.; Wu, Z.; Nuding, S.; Groscurth, S.; Marcinowski, M.; Beisner, J.; Buchner, J.; Schaller, M.; Stange, E.F.; Wehkamp, J. Reduction of disulphide bonds unmasks potent antimicrobial activity of human β-defensin 1. Nature 2011, 469, 419–423. [Google Scholar] [CrossRef] [PubMed]
- Haag, A.F.; Kerscher, B.; Dall’Angelo, S.; Sani, M.; Longhi, R.; Baloban, M.; Wilson, H.M.; Mergaert, P.; Zanda, M.; Ferguson, G.P. Role of cysteine residues and disulfide bonds in the activity of a legume root nodule-specific, cysteine-rich peptide. J. Biol. Chem. 2012, 287, 10791–10798. [Google Scholar] [CrossRef]
- Falco, A.; Ortega-Villaizan, M.; Chico, V.; Brocal, I.; Perez, L.; Coll, J.M.; Estepa, A. Antimicrobial peptides as model molecules for the development of novel antiviral agents in aquaculture. Mini Rev. Med. Chem. 2009, 9, 1159–1164. [Google Scholar] [CrossRef]
- Masso-Silva, J.A.; Diamond, G. Antimicrobial peptides from fish. Pharmaceuticals 2014, 7, 265–310. [Google Scholar] [CrossRef]
- Huan, Y.; Kong, Q.; Mou, H.; Yi, H. Antimicrobial peptides: Classification, design, application and research progress in multiple fields. Front. Microbiol. 2020, 2559. [Google Scholar] [CrossRef]
- Fernandes, J.M.O.; Kemp, G.D.; Molle, M.G.; Smith, V.J. Anti-microbial properties of histone H2A from skin secretions of rainbow trout, Oncorhynchus mykiss. Biochem. J. 2002, 368, 611–620. [Google Scholar] [CrossRef]
- Birkemo, G.A.; Lüders, T.; Andersen, Ø.; Nes, I.F.; Nissen-Meyer, J. Hipposin, a histone-derived antimicrobial peptide in Atlantic halibut (Hippoglossus hippoglossus L.). Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2003, 1646, 207–215. [Google Scholar] [CrossRef]
- Park, I.Y.; Park, C.B.; Kim, M.S.; Kim, S.C. Parasin I, an antimicrobial peptide derived from histone H2A in the catfish, Parasilurus asotus. FEBS Lett. 1998, 437, 258–262. [Google Scholar] [CrossRef]
- Cho, J.H.; Park, I.Y.; Kim, H.S.; Lee, W.T.; Kim, M.S.; Kim, S.C. Cathepsin D produces antimicrobial peptide parasin I from histone H2A in the skin mucosa of fish. FASEB J. 2002, 16, 429–431. [Google Scholar] [CrossRef]
- Bergsson, G.; Agerberth, B.; Jörnvall, H.; Gudmundsson, G.H. Isolation and identification of antimicrobial components from the epidermal mucus of Atlantic cod (Gadus morhua). FEBS J. 2005, 272, 4960–4969. [Google Scholar] [CrossRef]
- Fernandes, J.M.O.; Molle, G.; Kemp, G.D.; Smith, V.J. Isolation and characterisation of oncorhyncin II, a histone H1-derived antimicrobial peptide from skin secretions of rainbow trout, Oncorhynchus mykiss. Dev. Comp. Immunol. 2004, 28, 127–138. [Google Scholar] [CrossRef]
- Lüders, T.; Birkemo, G.A.; Nissen-Meyer, J.; Andersen, Ø.; Nes, I.F. Proline conformation-dependent antimicrobial activity of a proline-rich histone h1 N-terminal Peptide fragment isolated from the skin mucus of Atlantic salmon. Antimicrob. Agents Chemother. 2005, 49, 2399–2406. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, S.; Ross, N.W.; MacKinnon, S.L. Myxinidin, A Novel Antimicrobial Peptide from the Epidermal Mucus of Hagfish, Myxine glutinosa L. Mar. Biotechnol. 2009, 11, 748. [Google Scholar] [CrossRef]
- Fernandes, J.M.O.; Saint, N.; Kemp, G.D.; Smith, V.J. Oncorhyncin III: A potent antimicrobial peptide derived from the non-histone chromosomal protein H6 of rainbow trout, Oncorhynchus mykiss. Biochem. J. 2003, 373, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Su, Y. Isolation and identification of pelteobagrin, a novel antimicrobial peptide from the skin mucus of yellow catfish (Pelteobagrus fulvidraco). Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2011, 158, 149–154. [Google Scholar] [CrossRef]
- Lazarovici, P.; Primor, N.; Loew, L.M. Purification and pore-forming activity of two hydrophobic polypeptides from the secretion of the Red Sea Moses sole (Pardachirus marmoratus). J. Biol. Chem. 1986, 261, 16704–16713. [Google Scholar] [CrossRef] [PubMed]
- Ruangsri, J.; Salger, S.A.; Caipang, C.M.A.; Kiron, V.; Fernandes, J.M.O. Differential expression and biological activity of two piscidin paralogues and a novel splice variant in Atlantic cod (Gadus morhua L.). Fish Shellfish Immunol. 2012, 32, 396–406. [Google Scholar] [CrossRef]
- Cole, A.M.; Weis, P.; Diamond, G. Isolation and Characterization of Pleurocidin, an Antimicrobial Peptide in the Skin Secretions of Winter Flounder. J. Biol. Chem. 1997, 272, 12008–12013. [Google Scholar] [CrossRef]
- Fernandes, J.M.O.; Smith, V.J. A novel antimicrobial function for a ribosomal peptide from rainbow trout skin. Biochem. Biophys. Res. Commun. 2002, 296, 167–171. [Google Scholar] [CrossRef] [PubMed]
- Robinette, D.; Wada, S.; Arroll, T.; Levy, M.G.; Miller, W.L.; Noga, E.J. Antimicrobial activity in the skin of the channel catfish Ictalurus punctatus: Characterization of broad-spectrum histone-like antimicrobial proteins. Cell. Mol. Life Sci. CMLS 1998, 54, 467–475. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Park, C.B.; Kim, M.S.; Kim, S.C. cDNA cloning and characterization of buforin I, an antimicrobial peptide: A cleavage product of histone H2A. Biochem. Biophys. Res. Commun. 1996, 229, 381–387. [Google Scholar] [CrossRef] [PubMed]
- Valero, Y.; Chaves-Pozo, E.; Meseguer, J.; Esteban, M.; Cuesta, A. Biologial Role of Fish Antimicrobial Peptides. In Antimicrobial Peptides; Nova Science Publishers, Inc.: Hauppauge, NY, USA, 2013; pp. 31–60. [Google Scholar]
- Oren, Z.; Shai, Y. A Class of Highly Potent Antibacterial Peptides Derived from Pardaxin, A Pore-Forming Peptide Isolated from Moses Sole Fish Pardachirus marmoratus. Eur. J. Biochem. 1996, 237, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Bae, J.-S.; Jung, J.-M.; An, C.M.; Kim, J.-W.; Hwang, S.D.; Kwon, M.-G.; Park, M.-A.; Kim, M.-C.; Park, C.-I. Piscidin: Antimicrobial peptide of rock bream, Oplegnathus fasciatus. Fish Shellfish Immunol. 2016, 51, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Ebran, N.; Julien, S.; Orange, N.; Auperin, B.; Molle, G. Isolation and characterization of novel glycoproteins from fish epidermal mucus: Correlation between their pore-forming properties and their antibacterial activities. Biochim. Biophys. Acta (BBA)-Biomembr. 2000, 1467, 271–280. [Google Scholar] [CrossRef]
- Easy, R.H.; Trippel, E.A.; Burt, M.D.B.; Cone, D.K. Identification of transferrin in Atlantic cod Gadus morhua epidermal mucus. J. Fish Biol. 2012, 81, 2059–2063. [Google Scholar] [CrossRef]
- Ræder, I.L.U.; Paulsen, S.M.; Smalås, A.O.; Willassen, N.P. Effect of fish skin mucus on the soluble proteome of Vibrio salmonicida analysed by 2-D gel electrophoresis and tandem mass spectrometry. Microb. Pathog. 2007, 42, 36–45. [Google Scholar] [CrossRef]
- Santoso, H.B.; Suhartono, E.; Yunita, R.; Biyatmoko, D. Epidermal mucus as a potential biological matrix for fish health analysis. Egypt. J. Aquat. Biol. Fish. 2020, 24, 361–382. [Google Scholar] [CrossRef]
- Raposo, C.D.; Canelas, A.B.; Barros, M.T. Human Lectins, Their Carbohydrate Affinities and Where to Find Them. Biomolecules 2021, 11, 188. [Google Scholar] [CrossRef]
- Suzuki, Y.; Tasumi, S.; Tsutsui, S.; Okamoto, M.; Suetake, H. Molecular diversity of skin mucus lectins in fish. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2003, 136, 723–730. [Google Scholar] [CrossRef]
- Chong, K.; Joshi, S.; Jin, L.T.; Shu-Chien, A.C. Proteomics profiling of epidermal mucus secretion of a cichlid (Symphysodon aequifasciata) demonstrating parental care behavior. Proteomics 2006, 6, 2251–2258. [Google Scholar] [CrossRef]
- Cordero, H.; Brinchmann, M.F.; Cuesta, A.; Meseguer, J.; Esteban, M.A. Skin mucus proteome map of European sea bass (Dicentrarchus labrax). Proteomics 2015, 15, 4007–4020. [Google Scholar] [CrossRef] [PubMed]
- Rajan, B.; Fernandes, J.M.O.; Caipang, C.M.A.; Kiron, V.; Rombout, J.H.W.M.; Brinchmann, M.F. Proteome reference map of the skin mucus of Atlantic cod (Gadus morhua) revealing immune competent molecules. Fish Shellfish Immunol. 2011, 31, 224–231. [Google Scholar] [CrossRef] [PubMed]
- Tsutsui, S.; Tasumi, S.; Suetake, H.; Suzuki, Y. Lectins Homologous to Those of Monocotyledonous Plants in the Skin Mucus and Intestine of Pufferfish, Fugu rubripes. J. Biol. Chem. 2003, 278, 20882–20889. [Google Scholar] [CrossRef] [PubMed]
- Tsutsui, S.; Yamaguchi, M.; Hirasawa, A.; Nakamura, O.; Watanabe, T. Common Skate (Raja kenojei) Secretes Pentraxin into the Cutaneous Secretion: The First Skin Mucus Lectin in Cartilaginous Fish*. J. Biochem. 2009, 146, 295–306. [Google Scholar] [CrossRef]
- Patel, D.M.; Brinchmann, M.F. Skin mucus proteins of lumpsucker (Cyclopterus lumpus). Biochem. Biophys. Rep. 2017, 9, 217–225. [Google Scholar] [CrossRef]
- Gewurz, H.; Zhang, X.-H.; Lint, T.F. Structure and function of the pentraxins. Curr. Opin. Immunol. 1995, 7, 54–64. [Google Scholar] [CrossRef]
- Ramos, F.; Smith, A.C. The C-reactive protein (CRP) test for the detection of early disease in fishes. Aquaculture 1978, 14, 261–266. [Google Scholar] [CrossRef]
- Lazado, C.C.; Skov, P.V. Secretory Proteins in the Skin Mucus of Nile Tilapia (Oreochromis niloticus) are Modulated Temporally by Photoperiod and Bacterial Endotoxin Cues. Fishes 2019, 4, 57. [Google Scholar] [CrossRef]
- Nigam, A.K.; Kumari, U.; Mittal, S.; Mittal, A.K. Comparative analysis of innate immune parameters of the skin mucous secretions from certain freshwater teleosts, inhabiting different ecological niches. Fish Physiol. Biochem. 2012, 38, 1245–1256. [Google Scholar] [CrossRef]
- Ren, T.; Koshio, S.; Ishikawa, M.; Yokoyama, S.; Micheal, F.R.; Uyan, O.; Tung, H.T. Influence of dietary vitamin C and bovine lactoferrin on blood chemistry and non-specific immune responses of Japanese eel, Anguilla japonica. Aquaculture 2007, 267, 31–37. [Google Scholar] [CrossRef]
- Subramanian, S.; MacKinnon, S.L.; Ross, N.W. A comparative study on innate immune parameters in the epidermal mucus of various fish species. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2007, 148, 256–263. [Google Scholar] [CrossRef] [PubMed]
- Hjelmeland, K.; Christie, M.; Raa, J. Skin mucus protease from rainbow trout, Salmo gairdneri Richardson, and its biological significance. J. Fish Biol. 1983, 23, 13–22. [Google Scholar] [CrossRef]
- Aranishi, F.; Nakane, M. Epidermal proteases of the Japanese eel. Fish Physiol. Biochem. 1997, 16, 471–478. [Google Scholar] [CrossRef]
- Aranishi, F.; Nakane, M. Epidermal Proteinases in the European Eel. Physiol. Zool. 1997, 70, 563–570. [Google Scholar] [CrossRef]
- Firth, K.; Johnson, S.; Ross, N. Characterisation of proteases in the skin mucus of Atlantic Salmon (Salmo salar) infected with the Salmon louse (Lepeophtheirus salmonis) and in whole-body louse homogenate. J. Parasitol. 2001, 86, 1199–1205. [Google Scholar] [CrossRef]
- Jurado, J.; Fuentes-Almagro, C.A.; Guardiola, F.A.; Cuesta, A.; Esteban, M.Á.; Prieto-Álamo, M.-J. Proteomic profile of the skin mucus of farmed gilthead seabream (Sparus aurata). J. Proteom. 2015, 120, 21–34. [Google Scholar] [CrossRef] [PubMed]
- Molle, V.; Campagna, S.; Bessin, Y.; Ebran, N.; Saint, N.; Molle, G. First evidence of the pore-forming properties of a keratin from skin mucus of rainbow trout (Oncorhynchus mykiss, formerly Salmo gairdneri). Biochem. J. 2008, 411, 33–40. [Google Scholar] [CrossRef]
- Hayes, M.J.; Rescher, U.; Gerke, V.; Moss, S.E. Annexin–actin interactions. Traffic 2004, 5, 571–576. [Google Scholar] [CrossRef]
- Cordero, H.; Brinchmann, M.F.; Cuesta, A.; Esteban, M.A. Chronic wounds alter the proteome profile in skin mucus of farmed gilthead seabream. BMC Genom. 2017, 18, 939. [Google Scholar] [CrossRef]
- Easy, R.H.; Ross, N.W. Changes in Atlantic salmon Salmo salar mucus components following short- and long-term handling stress. J. Fish Biol. 2010, 77, 1616–1631. [Google Scholar] [CrossRef]
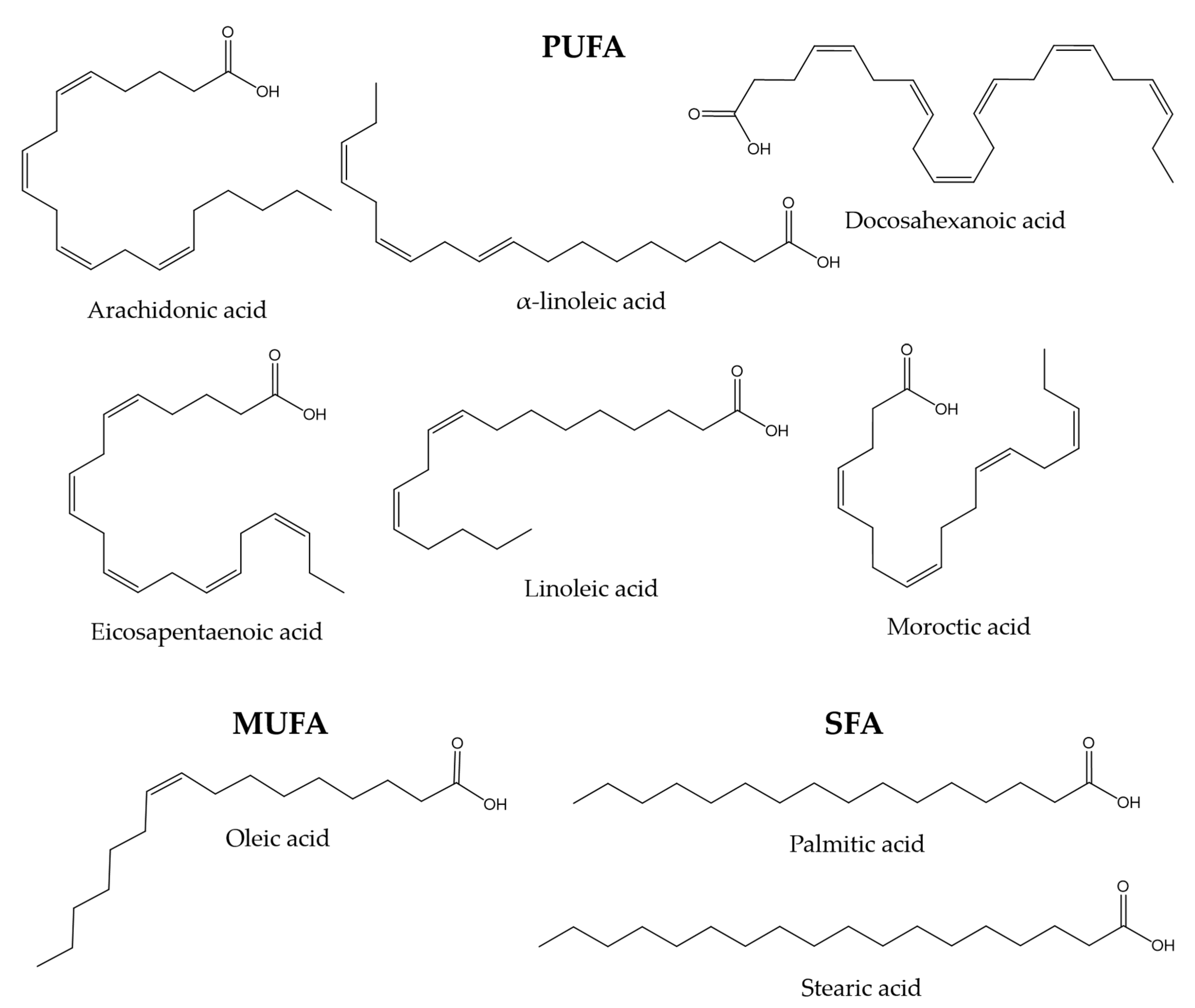
- Balasubramanian, S.; Gunasekaran, G. Fatty acids and amino acids composition in skin epidermal mucus of selected fresh water fish mugil cephalus. World J. Pharm. Pharm. Sci. 2015, 4, 1275–1287. [Google Scholar]
- Torrecillas, S.; Montero, D.; Domínguez, D.; Robaina, L.; Izquierdo, M. Skin Mucus Fatty Acid Composition of Gilthead Sea Bream (Sparus Aurata): A Descriptive Study in Fish Fed Low and High Fish Meal Diets. Fishes 2019, 4, 15. [Google Scholar] [CrossRef]
- Lewis, R.W. Fish cutaneous mucus: A new source of skin surface lipid. Lipids 1970, 5, 947–949. [Google Scholar] [CrossRef]
- Ekman, D.R.; Skelton, D.M.; Davis, J.M.; Villeneuve, D.L.; Cavallin, J.E.; Schroeder, A.; Jensen, K.M.; Ankley, G.T.; Collette, T.W. Metabolite Profiling of Fish Skin Mucus: A Novel Approach for Minimally-Invasive Environmental Exposure Monitoring and Surveillance. Environ. Sci. Technol. 2015, 49, 3091–3100. [Google Scholar] [CrossRef]
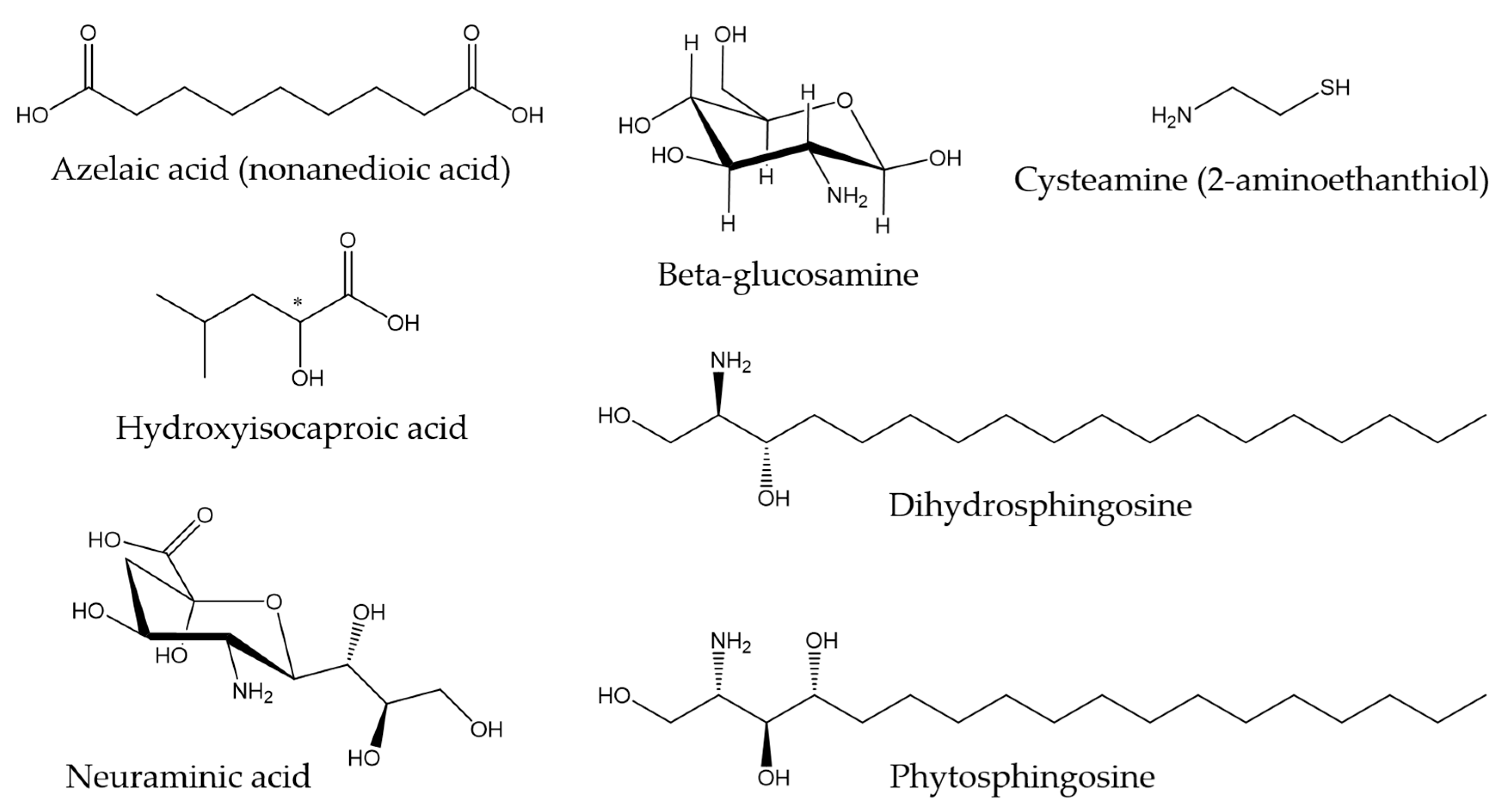
- Holland, K.; Bojar, R. Antimicrobial effects of azelaic acid. J. Dermatol. Treat. 1993, 4, S8–S11. [Google Scholar] [CrossRef]
- Sakko, M.; Moore, C.; Novak-Frazer, L.; Rautemaa, V.; Sorsa, T.; Hietala, P.; Järvinen, A.; Bowyer, P.; Tjäderhane, L.; Rautemaa, R. 2-hydroxyisocaproic acid is fungicidal for Candida and Aspergillus species. Mycoses 2014, 57, 214–221. [Google Scholar] [CrossRef] [PubMed]
- Patel, M.; Ashraf, M.S.; Siddiqui, A.J.; Ashraf, S.A.; Sachidanandan, M.; Snoussi, M.; Adnan, M.; Hadi, S. Profiling and Role of Bioactive Molecules from Puntius sophore (Freshwater/Brackish Fish) Skin Mucus with Its Potent Antibacterial, Antiadhesion, and Antibiofilm Activities. Biomolecules 2020, 10, 920. [Google Scholar] [CrossRef]
- Lee, Y.; Bilung, L.M.; Sulaiman, B.; Chong, Y.L. The antibacterial activity of fish skin mucus with various extraction solvents and their in-vitro evaluation methods. Int. Aquat. Res. 2020, 12, 1–21. [Google Scholar]
- Wang, H.; Tang, W.; Zhang, R.; Ding, S. Analysis of enzyme activity, antibacterial activity, antiparasitic activity and physico-chemical stability of skin mucus derived from Amphiprion clarkii. Fish Shellfish Immunol. 2019, 86, 653–661. [Google Scholar] [CrossRef]
- Al-Rasheed, A.; Handool, K.O.; Garba, B.; Noordin, M.M.; Bejo, S.K.; Kamal, F.M.; Daud, H.H.M. Crude extracts of epidermal mucus and epidermis of climbing perch Anabas testudineus and its antibacterial and hemolytic activities. Egypt. J. Aquat. Res. 2018, 44, 125–129. [Google Scholar] [CrossRef]
- Sanahuja, I.; Fernández-Alacid, L.; Ordóñez-Grande, B.; Sánchez-Nuño, S.; Ramos, A.; Araujo, R.M.; Ibarz, A. Comparison of several non-specific skin mucus immune defences in three piscine species of aquaculture interest. Fish Shellfish Immunol. 2019, 89, 428–436. [Google Scholar] [CrossRef]
- Leng, W.; Wu, X.; Xiong, Z.; Shi, T.; Sun, Q.; Yuan, L.; Gao, R. Study on antibacterial properties of mucus extract of snakehead (Channa argus) against Escherichia coli and its application in chilled fish fillets preservation. LWT 2022, 167, 113840. [Google Scholar] [CrossRef]
- Dhanaraj, M.; Haniffa, M.A.; Arun Singh, S.V.; Ramakrishnan, C.; Manikandaraja, D.; James Milton, M. Antibacterial Activity of Skin and Intestinal Mucus of Five Different Freshwater Fish Species Viz., Channa striatus, C. micropeltes, C. marulius, C. Punctatus and C. gachua. Malays. J. Sci. 2009, 28, 256–262. [Google Scholar] [CrossRef]
- Wei, O.Y.; Xavier, R.; Marimuthu, K. Screening of antibacterial activity of mucus extract of snakehead fish, Channa striatus. Eur. Rev. Med. Pharmacol. Sci. 2010, 14, 675–681. [Google Scholar]
- Ramesh, B.K. Assessment of antimicrobial peptides from mucus of fish. Indian J. Clin. Biochem. 2013, 1, 5–8. [Google Scholar]
- Haniffa, M.A.; Viswanathan, S.; Jancy, D.; Poomari, K.; Manik, S. Antibacterial studies of fish mucus from two marketed air-breathing fishes-Channa striatus and Heteropneustes fossilis. Int. Res. J. Microbiol. 2014, 5, 22–27. [Google Scholar]
- Guardiola, F.A.; Cuesta, A.; Abellán, E.; Meseguer, J.; Esteban, M.A. Comparative analysis of the humoral immunity of skin mucus from several marine teleost fish. Fish Shellfish Immunol. 2014, 40, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Caruso, G.; Maricchiolo, G.; Genovese, L.; De Pasquale, F.; Caruso, R.; Denaro, M.G.; Delia, S.; Laganà, P. Comparative Study of Antibacterial and Haemolytic Activities in Sea Bass, European Eel and Blackspot Seabream. Open Mar. Biol. J. 2014, 8, 10–16. [Google Scholar]
- Magariños, B.; Pazos, F.G.; Santos, Y.; Romalde, J.L.; Toranzo, A.E. Response of Pasteurella piscicida and Flexibacter maritimus to skin mucus of marine fish. Dis. Aquat. Org. 1995, 21, 103–108. [Google Scholar] [CrossRef]
- Manikantan, G.; Lyla, S.; Khan, S.A.; Vijayanand, P.; Edward, G.; Jothi, G. Bioactive potency of epidermal mucus extracts from greasy grouper, Epinephelus tauvina (Forsskal, 1775). J. Coast. Life Med. 2016, 4, 510–520. [Google Scholar] [CrossRef]
- Hellio, C.; Pons, A.M.; Beaupoil, C.; Bourgougnon, N.; Gal, Y.L. Antibacterial, antifungal and cytotoxic activities of extracts from fish epidermis and epidermal mucus. Int. J. Antimicrob. Agents 2002, 20, 214–219. [Google Scholar] [CrossRef] [PubMed]
- Katra, N.; Hisar, O.; Karatas, S.; Turgay, E.; Sarvan, C.; KATRA, N. In vitro antimicrobial activities of extracts from ballan wrasse (Labrus bergylta) skin mucus. Mar. Sci. Technol. Bull. 2016, 5, 13–15. [Google Scholar]
- Subramanian, S.; Ross, N.W.; MacKinnon, S.L. Comparison of antimicrobial activity in the epidermal mucus extracts of fish. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2008, 150, 85–92. [Google Scholar] [CrossRef]
- García-Marciano, M.; Apún-Molina, J.P.; Sainz-Hernández, J.C.; Santamaría-Miranda, A.; Medina-Godoy, S.; Aguiñaga-Cruz, J.A. Antibacterial activity evaluation of the nile tilapia Oreochromis niloticus (Linnaeus, 1758) skin mucus, against vibrio bacteria affecting the white shrimp Penaeus vannamei. Lat. Am. J. Aquat. Res. 2019, 47, 580–585. [Google Scholar] [CrossRef]
- Lirio, G.A.C.; De Leon, J.A.A.; Villafuerte, A.G. Antimicrobial Activity of Epidermal Mucus from Top Aquaculture Fish Species against Medically-Important Pathogens. Walailak J. Sci. Technol. 2018, 16, 329–340. [Google Scholar] [CrossRef]
- Wibowo, A.; Fadjar, M. Utilization of Tilapia Mucus to Inhibit Vibrio harveyi on Vannamei (Litopenaeus vannamei). J. Life Sci. Biomed. 2015, 5, 141–148. [Google Scholar]
- Elavarasi, K.; Ranjini, S.; Rajagopal, T.; Rameshkumar, G.; Ponmanickam, P. Bactericidal proteins of skin mucus and skin extracts from fresh water fishes, Clarias batrachus and Tilapia mossambicus. Thai J. Pharm. Sci. 2013, 37, 194–200. [Google Scholar]
- Mahadevan, G.; Mohan, K.; Vinoth, J.; Ravi, V. Biotic potential of mucus extracts of giant mudskipper Periophthalmodon schlosseri (Pallas, 1770) from Pichavaram, southeast coast of India. J. Basic Appl. Zool. 2019, 80, 13. [Google Scholar] [CrossRef]
- Pethkar, M.R.; Lokhande, M.V. Antifungal activity of skin mucus of three cultivable fish species. Int. J. Zool. Stud. 2017, 2, 1–3. [Google Scholar]
- Manivasagan, P.; Annamalai, N.; Ashokkumar, S.; Sampathkumar, P. Studies on the proteinaceous gel secretion from the skin of the catfish, Arius maculatus (Thunberg, 1792). Afr. J. Biotechnol. 2009, 8, 7125–7129. [Google Scholar]
- Patil, R.; Ingole, J.; Sathe, T.; Jadhav, A. Antibacterial activity of fish mucus from Clarias batrachus (Linn.) against selected microbes. Biolife 2015, 3, 788–791. [Google Scholar]
- Loganathan, K.; Muniyan, M.; Prakash, A.A.; Raja, P.S.; Prakash, M. Studies on the Role of Mucus from Clarias batrachus (Linn) Against Selected Microbes. Int. J. Pharm. Appl. 2011, 2, 202–206. [Google Scholar]
- Kumari, U.; Nigam, A.K.; Mittal, S.; Mittal, A.K. Antibacterial properties of the skin mucus of the freshwater fishes, Rita rita and Channa punctatus. Eur. Rev. Med. Pharmacol. Sci. 2011, 15, 781–786. [Google Scholar]
- Subhashini, S.; Lavanya, J.; Jain, S.; Agihotri, T. Screening of Antibacterial and Cytotoxic Activity of Extracts from Epidermis and Epidermal Mucus of Barbonymus Schwanenfeldii (Tinfoil Barb Fish). Int. J. Res. Eng. Technol. 2013, 2, 492–497. [Google Scholar] [CrossRef]
- Islam, M.M.; Hossain, M.M.M.; Islam, M.S.; Khondoker, S.; Khatun, M.A. Competitive antibacterial activity of two Indian major carps and two Chinese carps fish mucus against common pathogenic bacteria at aquaculture pond. Int. J. Fish. Aquat. Stud. 2014, 2, 158–162. [Google Scholar] [CrossRef]
- Balasubramanian, S.; Rani, P.B.; Prakash, A.A.; Prakash, M.; Senthilraja, P.; Gunasekaran, G. Antimicrobial properties of skin mucus from four freshwater cultivable Fishes (Catla catla, Hypophthalmichthys molitrix, Labeo rohita and Ctenopharyngodon idella). Afr. J. Microbiol. Res. 2011, 6, 5110–5120. [Google Scholar]
- Nigam, A.K.; Kumari, U.; Mittal, S.; Mittal, A.K. Evaluation of antibacterial activity and innate immune components in skin mucus of Indian major carp, Cirrhinus mrigala. Aquac. Res. 2017, 48, 407–418. [Google Scholar] [CrossRef]
- Kuppulakshmi, C.; Prakash, M.; Gunasekaran, G.; Manimegalai, G.; Sarojini, S. Antibacterial properties of fish mucus from Channa punctatus and Cirrhinus mrigala. Eur. Rev. Med. Pharmacol. Sci. 2008, 12, 149–153. [Google Scholar]
- Kumari, S.; Tyor, A.K.; Bhatnagar, A. Evaluation of the antibacterial activity of skin mucus of three carp species. Int. Aquat. Res. 2019, 11, 225–239. [Google Scholar] [CrossRef]
- Tyor, A.; Kumari, S. Biochemical characterization and antibacterial properties of fish skin mucus of fresh water fish, Hypophthalmichthys nobilis. Int. J. Pharm. Pharm. Sci. 2016, 8, 132–136. [Google Scholar]
- Fuochi, V.; Li Volti, G.; Camiolo, G.; Tiralongo, F.; Giallongo, C.; Distefano, A.; Petronio Petronio, G.; Barbagallo, I.; Viola, M.; Furneri, P.M.; et al. Antimicrobial and Anti-Proliferative Effects of Skin Mucus Derived from Dasyatis pastinaca (Linnaeus, 1758). Mar. Drugs 2017, 15, 342. [Google Scholar] [CrossRef]
- Hisar, O.; Hisar, S.A.; Uyanik, M.H.; Sahin, T.; Cakir, F.; Yilmaz, S. In vitro antimicrobial and antifungal activities of aqueous skin mucus from rainbow trout (Oncorhynchus mykiss) on human pathogens. Mar. Sci. Technol. Bull. 2014, 3, 19–22. [Google Scholar]
- Guardiola, F.A.; Cuartero, M.; del Mar Collado-González, M.; Díaz Baños, F.G.; Cuesta, A.; Moriñigo, M.Á.; Esteban, M.Á. Terminal carbohydrates abundance, immune related enzymes, bactericidal activity and physico-chemical parameters of the Senegalese sole (Solea senegalensis, Kaup) skin mucus. Fish Shellfish Immunol. 2017, 60, 483–491. [Google Scholar] [CrossRef]
- Cho, J.H.; Park, I.Y.; Kim, M.S.; Kim, S.C. Matrix metalloproteinase 2 is involved in the regulation of the antimicrobial peptide parasin I production in catfish skin mucosa. FEBS Lett. 2002, 531, 459–463. [Google Scholar] [CrossRef]
- Yount, N.Y.; Bayer, A.S.; Xiong, Y.Q.; Yeaman, M.R. Advances in antimicrobial peptide immunobiology. Pept. Sci. Orig. Res. Biomol. 2006, 84, 435–458. [Google Scholar] [CrossRef]
- Afkarian, M.; Bhasin, M.; Dillon, S.T.; Guerrero, M.C.; Nelson, R.G.; Knowler, W.C.; Thadhani, R.; Libermann, T.A. Optimizing a proteomics platform for urine biomarker discovery. Mol. Cell. Proteom. 2010, 9, 2195–2204. [Google Scholar] [CrossRef]
- Ming, L.; Xiaoling, P.; Yan, L.; Lili, W.; Qi, W.; Xiyong, Y.; Boyao, W.; Ning, H. Purification of antimicrobial factors from human cervical mucus. Hum. Reprod. 2007, 22, 1810–1815. [Google Scholar] [CrossRef]
- Fischer, C.L. Antimicrobial Activity of Host-Derived Lipids. Antibiotics 2020, 9, 75. [Google Scholar] [CrossRef]
- Al-Arifa, N.; Mughal, M.; Hanif, A.; Batool, A. Effect of alkaline pH on Biozctive Molecules of Epidermal mucus from Labeo rohita. Turk. J. Biochem. 2011, 36, 29–34. [Google Scholar]
- Ikram, M.; Ridzwan, B.H. A preliminary screening of antifungal activities from skin mucus extract of Malaysian local swamp eel (Monopterus albus). Int. Res. J. Pharm. 2013, 3, 1–8. [Google Scholar]
- Nishiyama, Y.; Nakaoka, C.; Hiratani, T.; Abe, S.; Uchida, K.; Yamaguchi, H. Synergy of lysozyme and lanoconazole on the morphology of Candida albicans. J. Electron Microsc. 2001, 50, 41–49. [Google Scholar] [CrossRef]
- Sowa-Jasiłek, A.; Zdybicka-Barabas, A.; Stączek, S.; Wydrych, J.; Skrzypiec, K.; Mak, P.; Deryło, K.; Tchórzewski, M.; Cytryńska, M. Galleria mellonella lysozyme induces apoptotic changes in Candida albicans cells. Microbiol. Res. 2016, 193, 121–131. [Google Scholar] [CrossRef]
- Zarei, M.; Aminzadeh, S.; Zolgharnein, H.; Safahieh, A.; Daliri, M.; Noghabi, K. Characterization of A Chitinase with Antifungal Activity from a Native Serratia. Braz. J. Microbiol. 2011, 42, 1017–1029. [Google Scholar] [CrossRef]
- Raj, V.S.; Fournier, G.; Rakus, K.; Ronsmans, M.; Ouyang, P.; Michel, B.; Delforges, C.; Costes, B.; Farnir, F.; Leroy, B. Skin mucus of Cyprinus carpio inhibits cyprinid herpesvirus 3 binding to epidermal cells. Vet. Res. 2011, 42, 1–10. [Google Scholar] [CrossRef]
- Valero, Y.; Arizcun, M.; Cortes, J.; Ramirez-Cepeda, F.; Guzman, F.; Mercado, L.; Esteban, M.Á.; Chaves-Pozo, E.; Cuesta, A. NK-lysin, dicentracin and hepcidin antimicrobial peptides in European sea bass. Ontogenetic development and modulation in juveniles by nodavirus. Dev. Comp. Immunol. 2020, 103, 103516. [Google Scholar] [CrossRef]
- Falco, A.; Medina-Gali, R.M.; Poveda, J.A.; Bello-Perez, M.; Novoa, B.; Encinar, J.A. Antiviral activity of a Turbot (Scophthalmus maximus) NK-lysin peptide by inhibition of low-pH virus-induced membrane fusion. Mar. Drugs 2019, 17, 87. [Google Scholar] [CrossRef]
- Casadei, E.; Wang, T.; Zou, J.; Vecino, J.L.G.; Wadsworth, S.; Secombes, C.J. Characterization of three novel β-defensin antimicrobial peptides in rainbow trout (Oncorhynchus mykiss). Mol. Immunol. 2009, 46, 3358–3366. [Google Scholar] [CrossRef]
- Falco, A.; Chico, V.; Marroqui, L.; Perez, L.; Coll, J.; Estepa, A. Expression and antiviral activity of a β-defensin-like peptide identified in the rainbow trout (Oncorhynchus mykiss) EST sequences. Mol. Immunol. 2008, 45, 757–765. [Google Scholar] [CrossRef]
- Mai, W.-j.; Wang, W.-n. Protection of blue shrimp (Litopenaeus stylirostris) against the White Spot Syndrome Virus (WSSV) when injected with shrimp lysozyme. Fish Shellfish Immunol. 2010, 28, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Guo, N.; Chen, S.; Guo, X.; Liu, X.; Ye, S.; Chai, Q.; Wang, Y.; Liu, B.; He, Q. Antiviral activity of Piscidin 1 against pseudorabies virus both in vitro and in vivo. Virol. J. 2019, 16, 95. [Google Scholar] [CrossRef]
- Rodrigues, P.M.; Silva, T.S.; Dias, J.; Jessen, F. PROTEOMICS in aquaculture: Applications and trends. J. Proteom. 2012, 75, 4325–4345. [Google Scholar] [CrossRef] [PubMed]
- Ao, J.; Mu, Y.; Xiang, L.-X.; Fan, D.; Feng, M.; Zhang, S.; Shi, Q.; Zhu, L.-Y.; Li, T.; Ding, Y.; et al. Genome sequencing of the perciform fish Larimichthys crocea provides insights into molecular and genetic mechanisms of stress adaptation. PLoS Genet. 2015, 11, e1005118. [Google Scholar] [CrossRef]
- Carda-Diéguez, M.; Ghai, R.; Rodríguez-Valera, F.; Amaro, C. Wild eel microbiome reveals that skin mucus of fish could be a natural niche for aquatic mucosal pathogen evolution. Microbiome 2017, 5, 162. [Google Scholar] [CrossRef] [PubMed]
- Greer, J.B.; Andrzejczyk, N.E.; Mager, E.M.; Stieglitz, J.D.; Benetti, D.; Grosell, M.; Schlenk, D. Whole-Transcriptome Sequencing of Epidermal Mucus as a Novel Method for Oil Exposure Assessment in Juvenile Mahi-Mahi (Coryphaena hippurus). Environ. Sci. Technol. Lett. 2019, 6, 538–544. [Google Scholar] [CrossRef]
- Parida, S.; Mohapatra, A.; Kar, B.; Mohanty, J.; Sahoo, P.K. Transcriptional analysis of immune-relevant genes in the mucus of Labeo rohita, experimentally infected with Argulus siamensis. Acta Parasitol. 2018, 63, 125–133. [Google Scholar] [CrossRef]
- Nissa, M.U.; Pinto, N.; Parkar, H.; Goswami, M.; Srivastava, S. Proteomics in fisheries and aquaculture: An approach for food security. Food Control 2021, 127, 108125. [Google Scholar] [CrossRef]
- Minniti, G.; Rød Sandve, S.; Padra, J.T.; Heldal Hagen, L.; Lindén, S.; Pope, P.B.; Arntzen, M.Ø.; Vaaje-Kolstad, G. The Farmed Atlantic Salmon (Salmo salar) Skin-Mucus Proteome and Its Nutrient Potential for the Resident Bacterial Community. Genes 2019, 10, 515. [Google Scholar] [CrossRef]
- Liu, H.-h.; Sun, Q.; Jiang, Y.-t.; Fan, M.-h.; Wang, J.-x.; Liao, Z. In-depth proteomic analysis of Boleophthalmus pectinirostris skin mucus. J. Proteom. 2019, 200, 74–89. [Google Scholar] [CrossRef]
- Chong, K.; Sock Ying, T.; Foo, J.; Toong Jin, L.; Chong, A. Characterisation of proteins in epidermal mucus of discus fish (Symphysodon spp.) during parental phase. Aquaculture 2005, 249, 469–476. [Google Scholar] [CrossRef]
- Rajan, B.; Lokesh, J.; Kiron, V.; Brinchmann, M.F. Differentially expressed proteins in the skin mucus of Atlantic cod (Gadus morhua) upon natural infection with Vibrio anguillarum. BMC Vet. Res. 2013, 9, 103. [Google Scholar] [CrossRef]
- Xiong, Y.; Dan, C.; Ren, F.; Su, Z.; Zhang, Y.; Mei, J. Proteomic profiling of yellow catfish (Pelteobagrus fulvidraco) skin mucus identifies differentially-expressed proteins in response to Edwardsiella ictaluri infection. Fish Shellfish Immunol. 2020, 100, 98–108. [Google Scholar] [CrossRef]
- Fernández-Montero, Á.; Torrecillas, S.; Montero, D.; Acosta, F.; Prieto-Álamo, M.-J.; Abril, N.; Jurado, J. Proteomic profile and protease activity in the skin mucus of greater amberjack (Seriola dumerili) infected with the ectoparasite Neobenedenia girellae—An immunological approach. Fish Shellfish Immunol. 2021, 110, 100–115. [Google Scholar] [CrossRef]
- Pérez-Sánchez, J.; Terova, G.; Simó-Mirabet, P.; Rimoldi, S.; Folkedal, O.; Calduch-Giner, J.A.; Olsen, R.E.; Sitjà-Bobadilla, A. Skin Mucus of Gilthead Sea Bream (Sparus aurata L.). Protein Mapping and Regulation in Chronically Stressed Fish. Front. Physiol. 2017, 8, 34. [Google Scholar] [CrossRef]
- Fæste, C.K.; Tartor, H.; Moen, A.; Kristoffersen, A.B.; Dhanasiri, A.K.S.; Anonsen, J.H.; Furmanek, T.; Grove, S. Proteomic profiling of salmon skin mucus for the comparison of sampling methods. J. Chromatogr. B 2020, 1138, 121965. [Google Scholar] [CrossRef] [PubMed]
- Goodacre, R.; Vaidyanathan, S.; Dunn, W.B.; Harrigan, G.G.; Kell, D.B. Metabolomics by numbers: Acquiring and understanding global metabolite data. Trends Biotechnol. 2004, 22, 245–252. [Google Scholar] [CrossRef] [PubMed]
- Samuelsson, L.M.; Larsson, D.G.J. Contributions from metabolomics to fish research. Mol. BioSyst. 2008, 4, 974–979. [Google Scholar] [CrossRef] [PubMed]
- Wen, B.; Zhou, J.-Q.; Gao, J.-Z.; Chen, H.-R.; Shen, Y.-Q.; Chen, Z.-Z. Sex-dependent changes in the skin mucus metabolome of discus fish (Symphysodon haraldi) during biparental care. J. Proteom. 2020, 221, 103784. [Google Scholar] [CrossRef]
- Ivanova, L.; Tartor, H.; Grove, S.; Kristoffersen, A.B.; Uhlig, S. Workflow for the Targeted and Untargeted Detection of Small Metabolites in Fish Skin Mucus. Fishes 2018, 3, 21. [Google Scholar] [CrossRef]
- Reyes-López, F.E.; Ibarz, A.; Ordóñez-Grande, B.; Vallejos-Vidal, E.; Andree, K.B.; Balasch, J.C.; Fernández-Alacid, L.; Sanahuja, I.; Sánchez-Nuño, S.; Firmino, J.P.; et al. Skin Multi-Omics-Based Interactome Analysis: Integrating the Tissue and Mucus Exuded Layer for a Comprehensive Understanding of the Teleost Mucosa Functionality as Model of Study. Front. Immunol. 2021, 11, 613824. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Parente, J.; Harris, S.M.; Woods, D.E.; Hancock, R.E.; Falla, T.J. Antimicrobial peptide therapeutics for cystic fibrosis. Antimicrob. Agents Chemother. 2005, 49, 2921–2927. [Google Scholar] [CrossRef] [PubMed]
- Rai, M.; Pandit, R.; Gaikwad, S.; Kövics, G. Antimicrobial peptides as natural bio-preservative to enhance the shelf-life of food. J. Food Sci. Technol. 2016, 53, 3381–3394. [Google Scholar] [CrossRef] [PubMed]
| Variable 1 | Virus | Bacteria | Fungi | Human | Livestock | Fish |
|---|---|---|---|---|---|---|
| Number [19] | 1031 | 1030 | 1027 | 1010 | 1010 | 1015 |
| Mass, Gt C [19] | 0.2 | 70 | 12 | 0.06 | 0.1 | 0.7 |
| Time, years | 3.8 × 109 [20] 2 | 3.8 × 109 [21] | 1.5 × 109 [22] | 3 × 105 [23] | 84 × 106 [24] | 5 × 108 [25] |
| Family | AMP | Species | Ref. |
|---|---|---|---|
| Histone 2A * | N-acetylated Histone 2A | Oncorhynchus mykiss | [72] |
| Hipposin | Hippoglossus hippoglossus | [73] | |
| Parasin I | Parasilurus asotus | [74,75] | |
| Histone 2B * | Histone H2B | Gadus morhua | [76] |
| Histone H1 * | Oncorhyncin II | O. mykiss | [77] |
| SAMP H1 | Salmo salar | [78] | |
| Myxinidin | Myxinidin | Myxine glutinosa | [79] |
| Non-histone chromosomal protein H6 * | Oncorhyncin III | O. mykiss | [80] |
| Pelteobagrin | Pelteobagrin | Pelteobagrus fulvidraco | [81] |
| Pardaxin | Pardaxin I, II | Pardachirus marmoratus | [82] |
| Piscidin | Piscidin 1, 2, 2β | G. morhua | [83] |
| Pleurocidin | Pleurocidin | Pleuronectes americanus | [84] |
| Ribosomal proteins * | 40S Ribosomal protein S30 | O. mykiss | [85] |
| 60S Ribosomal protein L35, L36A, L40 | G. morhua | [76] |
| Extraction | Aqueous | Organic | Acidic | Crude | Ref. | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Solvent | W | AB | PS | TBS | ET | DCM | AA | TFA | |||||||||||
| Gram | + | − | + | − | + | − | + | − | + | − | + | − | + | − | + | − | + | − | |
| Perciformes order | |||||||||||||||||||
| Amphiprion clarkii | [126] | ||||||||||||||||||
| Anabas testudineus | [127] | ||||||||||||||||||
| Argyrosomus regius | [128] | ||||||||||||||||||
| Channa argus | [129] | ||||||||||||||||||
| Channa marulius | [130] | ||||||||||||||||||
| Channa micropeltes | [130] | ||||||||||||||||||
| Channa striata | [130,131,132,133] | ||||||||||||||||||
| Dentex dentex | [134] | ||||||||||||||||||
| Dicentrarchus labrax | [128,134,135,136] | ||||||||||||||||||
| Epinephelus marginatus | [134] | ||||||||||||||||||
| Epinephelus tauvina | [137] | ||||||||||||||||||
| Labrus bergylta | [138,139] | ||||||||||||||||||
| Morone saxatilis | [140] | ||||||||||||||||||
| Oreochromis niloticus | [141,142,143] | ||||||||||||||||||
| O. mossambicus | [144] | ||||||||||||||||||
| Pagellus bogaraveo | [135] | ||||||||||||||||||
| P. schlosseri | [145] | ||||||||||||||||||
| Sparus aurata | [128,134,136] | ||||||||||||||||||
| Umbrina cirrosa | [134] | ||||||||||||||||||
| Anguilliformes order | |||||||||||||||||||
| Anguilla anguilla | [135,146] | ||||||||||||||||||
| Siluriformes order | |||||||||||||||||||
| Arius maculatus | [147] | ||||||||||||||||||
| Clarias batrachus | [142,144,148,149] | ||||||||||||||||||
| Heteropneustes fossilis | [133] | ||||||||||||||||||
| Rita rita | [150] | ||||||||||||||||||
| Cypriniformes order | |||||||||||||||||||
| B. schwanenfeldii | [151] | ||||||||||||||||||
| Catla catla | [146,152,153] | ||||||||||||||||||
| Cirrhinus mrigala | [146,154,155] | ||||||||||||||||||
| Ctenopharyngodon idella | [152,153,156] | ||||||||||||||||||
| Cyprinus carpio | [140,156] | ||||||||||||||||||
| H. nobilis | [152,153,156,157] | ||||||||||||||||||
| Labeo rohita | [152,153] | ||||||||||||||||||
| Puntius sophore | [124] | ||||||||||||||||||
| Anabantiformes order | |||||||||||||||||||
| Channa gachua | [130] | ||||||||||||||||||
| Channa punctatus | [130,150,155] | ||||||||||||||||||
| Myliobatiformes order | |||||||||||||||||||
| Dasyatis pastinaca | [158] | ||||||||||||||||||
| Gadiformes order | |||||||||||||||||||
| Gadus morhua | [76,138] | ||||||||||||||||||
| M. aeglefinus | [140] | ||||||||||||||||||
| Pollachius virens | [138] | ||||||||||||||||||
| Myxiniformes order | |||||||||||||||||||
| Myxine glutinosa | [140] | ||||||||||||||||||
| Salmoniformes order | |||||||||||||||||||
| Oncorhynchus mykiss | [159] | ||||||||||||||||||
| Salvelinus alpinus | [140] | ||||||||||||||||||
| Salvelinus fontinalis | [140] | ||||||||||||||||||
| Pleuronectiformes order | |||||||||||||||||||
| Platichthys flesus | [138] | ||||||||||||||||||
| Scophthalmus rhombus | [138] | ||||||||||||||||||
| Scophthalmus maximus | [136] | ||||||||||||||||||
| Solea senegalensis | [160] | ||||||||||||||||||
| Solea solea | [138] | ||||||||||||||||||
 no effect;
no effect;  low or variable inhibition;
low or variable inhibition;  strong inhibition. W: water; AB: ammonium bicarbonate; PS: phosphate saline; TBS: Tris-buffered saline; ET: ethanol; DCM: dichloromethane; AA: acetic acid; TFA: trifluoroacetic acid. Fish family and species full names: Anabantidae: Anabas testudineus Bloch; Anguillidae: Anguilla anguilla L.; Ariidae: Arius maculatus Thunberg; Bagridae: Rita rita Hamilton; Channidae: Channa argus Cantor, Channa gachua Hamilton, Channa marulius Hamilton, Channa micropeltes G. Cuvier, Channa punctatus Bloch, Channa striata Bloch; Cichlidae: Oreochromis mossambicus W. K. H. Peters, Oreochromis niloticus L., Clarias batrachus L.; Cyprinidae: Barbonymus schwanenfeldii Bleeker, Catla catla Hamilton, Cirrhinus mrigala Hamilton, Ctenopharyngodon idella Valenciennes, Cyprinus carpio L., Hypophthalmichthys nobilis Richardson, Labeo rohita Hamilton, Puntius sophore Hamilton; Dasyatidae: Dasyatis pastinaca L.; Gadidae: Gadus morhua L., Melanogrammus aeglefinus L., Pollachius virens L.; Heteropneustidae: Heteropneustes fossilis Bloch; Labridae: Labrus bergylta Ascanius; Moronidae: Dicentrarchus labrax L., Morone saxatilis Walbaum; Myxinidae: Myxine glutinosa L.; Oxudercidae: Periophthalmodon schlosseri Pallas; Pleuronectidae: Platichthys flesus L.; Pomacentridae: Amphiprion clarkii Bennet; Salmonidae: Oncorhynchus mykiss Walbaum, Salvelinus alpinus L., Salvelinus fontinalis Mitchill; Sciaenidae: Argyrosomus regius Asso, Umbrina cirrosa L.; Scophthalmidae: Scophthalmus maximus L., Scophthalmus rhombus L.; Serranidae: Epinephelus marginatus Lowe, Epinephelus tauvina Forsskål; Soleidae: Solea senegalensis Kaup, Solea solea L.; Sparidae: Dentex dentex L., Pagellus bogaraveo Brünnich, Sparus aurata L.
strong inhibition. W: water; AB: ammonium bicarbonate; PS: phosphate saline; TBS: Tris-buffered saline; ET: ethanol; DCM: dichloromethane; AA: acetic acid; TFA: trifluoroacetic acid. Fish family and species full names: Anabantidae: Anabas testudineus Bloch; Anguillidae: Anguilla anguilla L.; Ariidae: Arius maculatus Thunberg; Bagridae: Rita rita Hamilton; Channidae: Channa argus Cantor, Channa gachua Hamilton, Channa marulius Hamilton, Channa micropeltes G. Cuvier, Channa punctatus Bloch, Channa striata Bloch; Cichlidae: Oreochromis mossambicus W. K. H. Peters, Oreochromis niloticus L., Clarias batrachus L.; Cyprinidae: Barbonymus schwanenfeldii Bleeker, Catla catla Hamilton, Cirrhinus mrigala Hamilton, Ctenopharyngodon idella Valenciennes, Cyprinus carpio L., Hypophthalmichthys nobilis Richardson, Labeo rohita Hamilton, Puntius sophore Hamilton; Dasyatidae: Dasyatis pastinaca L.; Gadidae: Gadus morhua L., Melanogrammus aeglefinus L., Pollachius virens L.; Heteropneustidae: Heteropneustes fossilis Bloch; Labridae: Labrus bergylta Ascanius; Moronidae: Dicentrarchus labrax L., Morone saxatilis Walbaum; Myxinidae: Myxine glutinosa L.; Oxudercidae: Periophthalmodon schlosseri Pallas; Pleuronectidae: Platichthys flesus L.; Pomacentridae: Amphiprion clarkii Bennet; Salmonidae: Oncorhynchus mykiss Walbaum, Salvelinus alpinus L., Salvelinus fontinalis Mitchill; Sciaenidae: Argyrosomus regius Asso, Umbrina cirrosa L.; Scophthalmidae: Scophthalmus maximus L., Scophthalmus rhombus L.; Serranidae: Epinephelus marginatus Lowe, Epinephelus tauvina Forsskål; Soleidae: Solea senegalensis Kaup, Solea solea L.; Sparidae: Dentex dentex L., Pagellus bogaraveo Brünnich, Sparus aurata L.| Fish Species (Family) | Extraction 1 | Sensitive Fungi | Non-Sensitive Fungi | Antimicrobial Assay 2 | Reference |
|---|---|---|---|---|---|
| Anguilla anguilla L. (Anguillidae) | C | Aspergillus awamori, Colletotrichum falcatum, Fusarium oxysporum | DD | [146] | |
| Catla catla Hamilton (Cyprinidae) | PS | Aspergillus flavus, Aspergillus niger, Candida albicans, Mucor globosus, Rhizopus arrhizus | DD | [153] | |
| C | A. awamori, C. falcatum, F. oxysporum | DD | [146] | ||
| Cirrhinus mrigala Hamilton (Cyprinidae) | C | A. awamori, C. falcatum, F. oxysporum | DD | [146] | |
| Clarias batrachus L. (Clariidae) | PS | A. niger, Aspergillus nidulans, Fusarium moniliforme, C. albicans, Trichoderma koningi | DD | [149] | |
| Ctenopharyngodon idella Valenciennes (Cyprinidae) | PS | A. flavus, C. albicans, M. globosus, R. arrhizus | A. niger | DD | [153] |
| Cyprinus carpio L. (Cyprinidae) | PS, AA, DCM | C. albicans | BD | [140] | |
| Dasyatis pastinaca L. (Dasyatidae) | C | C. albicans, Candida glabrata, C. tropicalis | BD | [158] | |
| Gadus morhua L. (Gadidae) | ACN + 1% TFA | C. albicans | AWD, BD | [76] | |
| W, DCM | C. albicans, Candida brusei, C. tropicalis, Issatchenkia orientalis, Saccharomyces cerevisiae | AWD, BD | [138] | ||
| Hypophthalmichthys molitrix Valenciennes (Cyprinidae) | PS | A. flavus, A. niger, M. globosus | C. albicans, R. arrhizus | DD | [153] |
| Labeo rohita Hamilton (Cyprinidae) | NaOH, NaOH + TH | C. albicans | AWD | [166] | |
| PS | A. flavus, A. niger, C. albicans, M. globosus, R. arrhizus | DD | [153] | ||
| Melanogrammus aeglefinus L. (Gadidae) | AB, AA, ET, DCM | C. albicans | BD | [140] | |
| Monopterus albus Zuiew (Synbranchidae) | C, W, PS | C. albicans, Candida krusei, Cryptococcus neoformans, Fusarium sp. | DD | [167] | |
| Morone saxatilis Walbaum (Moronidae) | AB, AA, ET, DCM | C. albicans | BD | [140] | |
| Myxine glutinosa L. (Myxinidae) | AB, AA, ET, DCM | C. albicans | BD | [140] | |
| Oncorhynchus mykiss Walbaum (Salmonidae) | C | C. albicans, Candida parapsilosis | DD | [159] | |
| Periophthalmodon schlosseri Pallas (Gobiidae) | PS, ET | C. albicans, A. flavus, Mucor sp., Trichoderma longibriachtin | DD, BD | [145] | |
| Salvelinus alpinus L. (Salmonidae) | AB, AA, ET, DCM | C. albicans | BD | [140] | |
| Salvelinus fontinalis Mitchill (Salmonidae) | AB, AA, ET, DCM | C. albicans | BD | [140] | |
| Scophtalamus rhombus L. (Scophthalmidae) | W, ET, DCM | C. tropicalis, S. cerevisiae | C. brusei, C. albicans, I. orientalis | AWD, BD | [138] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Díaz-Puertas, R.; Adamek, M.; Mallavia, R.; Falco, A. Fish Skin Mucus Extracts: An Underexplored Source of Antimicrobial Agents. Mar. Drugs 2023, 21, 350. https://doi.org/10.3390/md21060350
Díaz-Puertas R, Adamek M, Mallavia R, Falco A. Fish Skin Mucus Extracts: An Underexplored Source of Antimicrobial Agents. Marine Drugs. 2023; 21(6):350. https://doi.org/10.3390/md21060350
Chicago/Turabian StyleDíaz-Puertas, Rocío, Mikolaj Adamek, Ricardo Mallavia, and Alberto Falco. 2023. "Fish Skin Mucus Extracts: An Underexplored Source of Antimicrobial Agents" Marine Drugs 21, no. 6: 350. https://doi.org/10.3390/md21060350
APA StyleDíaz-Puertas, R., Adamek, M., Mallavia, R., & Falco, A. (2023). Fish Skin Mucus Extracts: An Underexplored Source of Antimicrobial Agents. Marine Drugs, 21(6), 350. https://doi.org/10.3390/md21060350







