Youssoufenes A2 and A3, Antibiotic Dimeric Cinnamoyl Lipids from the ΔdtlA Mutant of a Marine-Derived Streptomyces Strain
Abstract
:1. Introduction
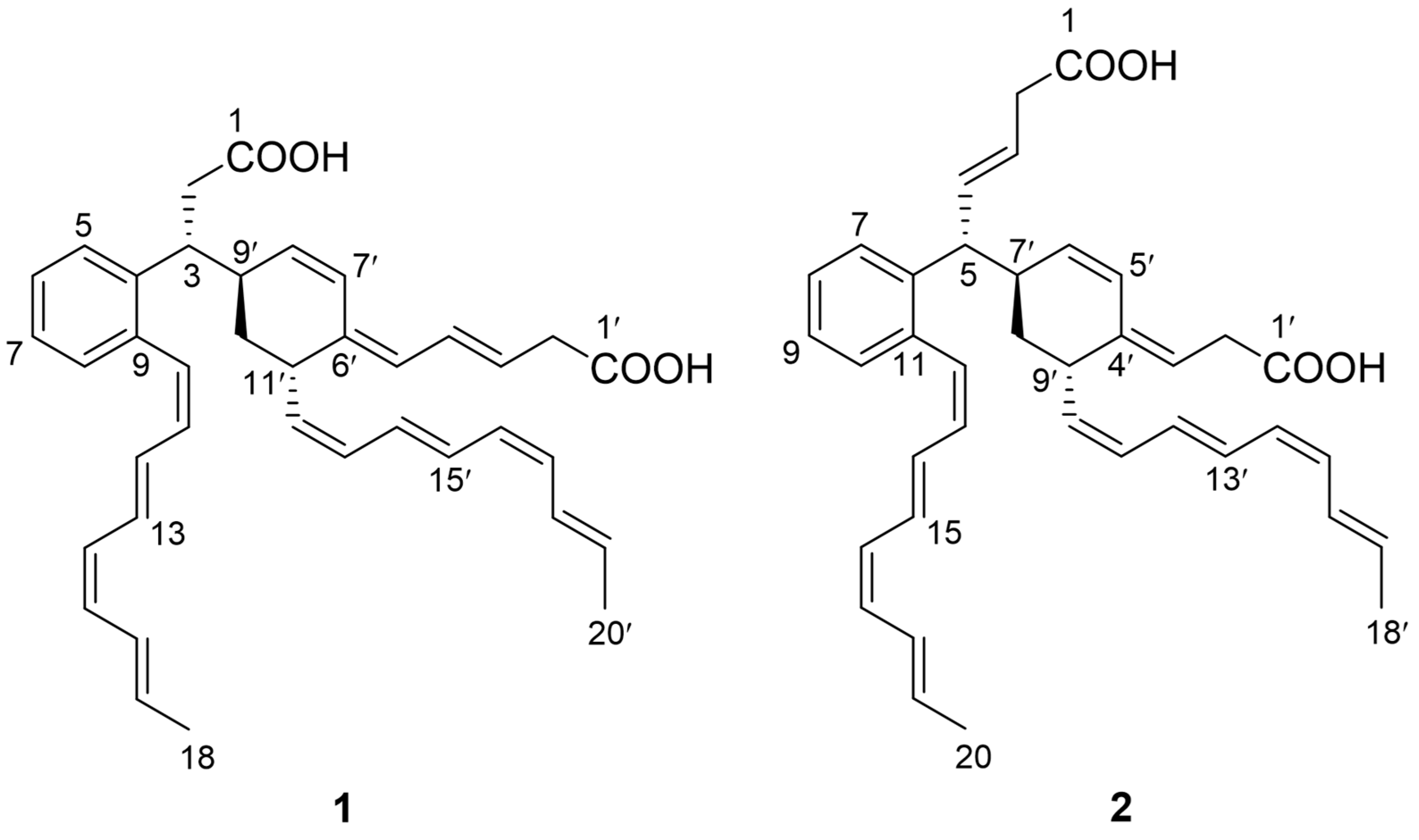
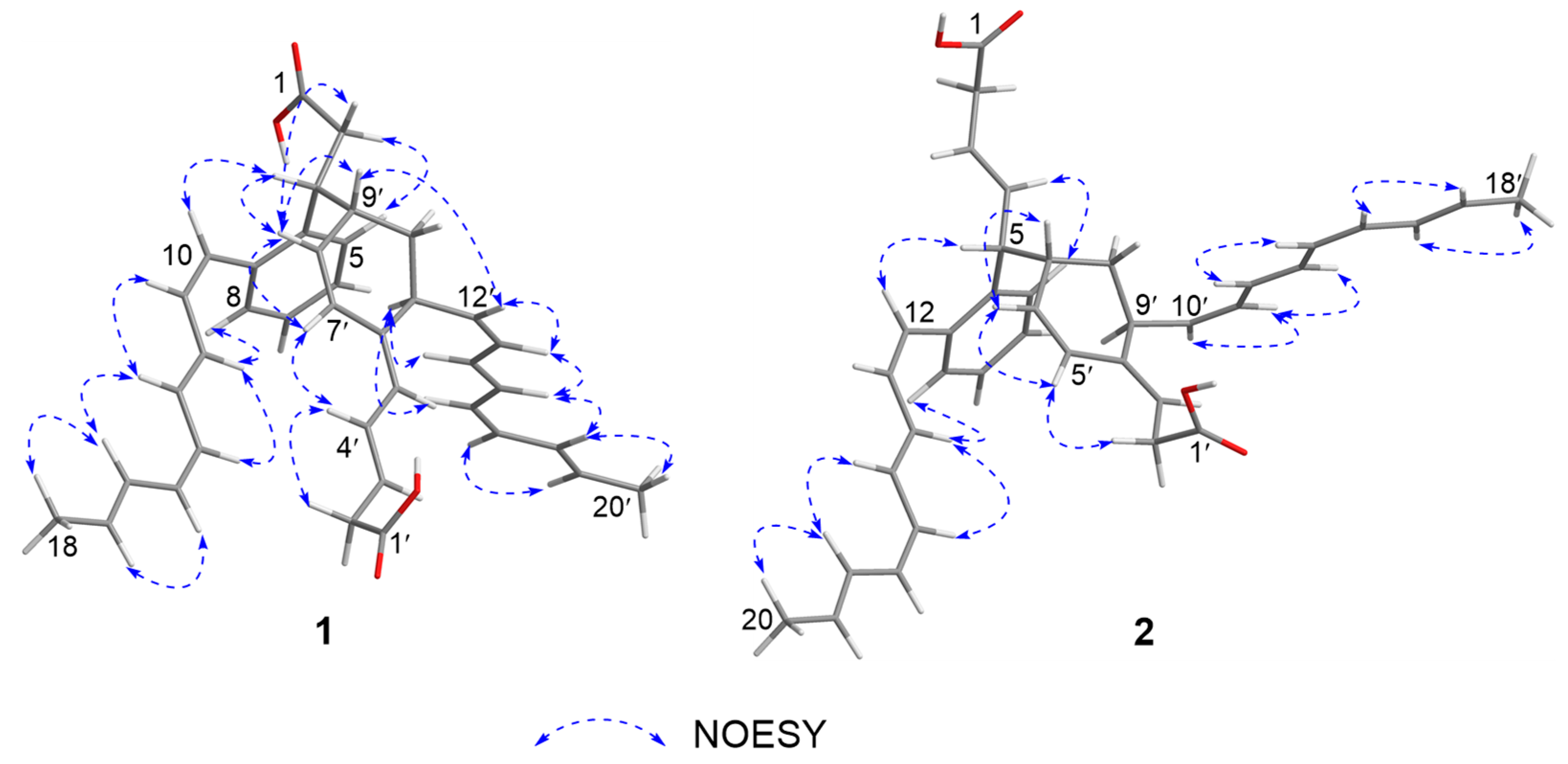
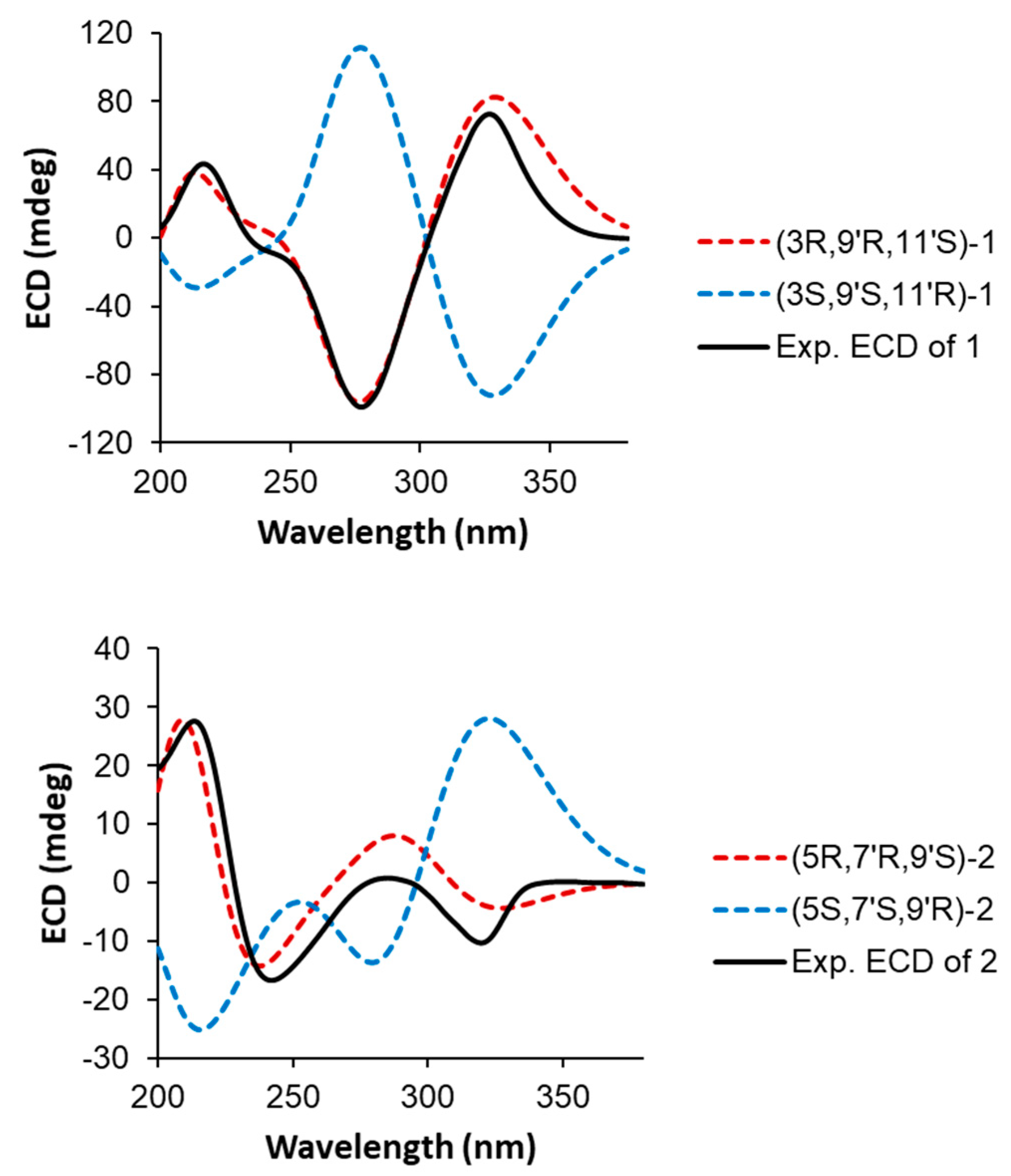
2. Results and Discussion
3. Materials and Methods
3.1. General Experimental Procedures
3.2. LC-MS-Based Production Analyses of the ΔdtlA Mutant Strain of S. youssoufiensis OUC6819
3.3. Fermentation, Extraction and Isolation of the Compounds
3.4. Computational Methods
3.5. Antibacterial Activity Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, H.; Liu, J.; Deng, Z.; Li, T.; Liu, Z.; Che, Q.; Li, W. Genetic Manipulation of an Aminotransferase Family Gene dtlA Activates Youssoufenes in Marine-Derived Streptomyces youssoufiensis. Org. Lett. 2020, 22, 729–733, Correction in Org. Lett. 2020, 22, 7773. [Google Scholar] [CrossRef] [PubMed]
- Bae, M.; Kim, H.; Moon, K.; Nam, S.-J.; Shin, J.; Oh, K.-B.; Oh, D.-C. Mohangamides A and B, New Dilactone-Tethered Pseudo-dimeric Peptides Inhibiting Candida albicans Isocitrate Lyase. Org. Lett. 2015, 17, 712–715. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Liu, C.; Zhang, B.; Guo, W.; Zhu, J.; Chang, C.-Y.; Zhao, E.; Jiao, R.; Tan, R.; Ge, H. Genome Mining and Biosynthesis of Kitacinnamycins as a STING Activator. Chem. Sci. 2019, 10, 4839–4846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bae, M.; Oh, J.; Bae, E.; Oh, J.; Hur, J.; Suh, Y.-G.; Lee, S.; Shin, J.; Oh, D.-C. WS9326H, an Antiangiogenic Pyrazolone-Bearing Peptide from an Intertidal Mudflat Actinomycete. Org. Lett. 2018, 20, 1999–2002. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Liu, Z.; Sun, C.; Shao, M.; Ma, J.; Wei, X.; Zhang, T.; Li, W.; Ju, J. Discovery and Biosynthesis of Atrovimycin, an Antitubercular and Antifungal Cyclodepsipeptide Featuring Vicinal-Dihydroxylated Cinnamic Acyl Chain. Org. Lett. 2019, 21, 2634–2638. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Yang, Z.; Zhang, C.; Liu, Z.; He, J.; Liu, Q.; Zhang, T.; Ju, J.; Ma, J. Genome Mining of Streptomyces atratus SCSIO ZH16: Discovery of Atratumycin and Identification of its Biosynthetic Gene Cluster. Org. Lett. 2019, 21, 1453–1457. [Google Scholar] [CrossRef] [PubMed]
- Ritzau, M.; Drautz, H.; Zähner, H.; Zeeck, A. Serpentene, a Novel Polyene Carboxylic Acid from Streptomyces. Liebigs An. Chem. 1993, 266, 433–435. [Google Scholar] [CrossRef]
- Ohlendorf, B.; Schulz, D.; Beese, P.; Erhard, A.; Schmaljohann, R.; Imhoff, J. Diacidene, a Polyene Dicarboxylic Acid from a Micromonospora Isolate from the German Wadden Sea. Naturforsch C J. Biosci. 2012, 67, 445–450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mehnaz, S.; Saleem, R.; Yameen, B.; Pianet, I.; Schnakenburg, G.; Pietraszkiewicz, H.; Valeriote, F.; Josten, M.; Sahl, H.-G.; Franzblau, S.; et al. Lahorenoic Acids A–C, Ortho-Dialkyl-Substituted Aromatic Acids from the Biocontrol Strain Pseudomonas aurantiaca PB-St2. J. Nat. Prod. 2013, 76, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Pohle, S.; Appelt, C.; Roux, M.; Fiedler, H.-P.; Süssmuth, R. Biosynthetic Gene Cluster of the Non-Ribosomally Synthesized Cyclodepsipeptide Skyllamycin: Deciphering Unprecedented Ways of Unusual Hydroxylation Reactions. J. Am. Chem. Soc. 2011, 133, 6194–6205. [Google Scholar] [CrossRef]
- Deng, Z.; Liu, J.; Li, T.; Li, H.; Liu, Z.; Dong, Y.; Li, W. An Unusual Type II Polyketide Synthase System Involved in Cinnamoyl Lipid Biosynthesis. Angew. Chem. 2021, 60, 153–158. [Google Scholar] [CrossRef]
- Che, Q.; Zhu, T.; Qi, X.; Mándi, A.; Kurtán, T.; Mo, X.; Li, D. Hybrid Isoprenoids from a Reeds Rhizosphere Soil Derived Actinomycete Streptomyces sp. CHQ-64. Org. Lett. 2012, 14, 3438–3441. [Google Scholar]
- Yao, T.; Liu, J.; Liu, Z.; Li, T.; Li, H.; Che, Q.; Zhu, T.; Li, D.; Li, W. Genome mining of cyclodipeptide synthases unravels unusual tRNA-dependent diketopiperazine-terpene biosynthetic machinery. Nat. Commun. 2018, 9, 4091. [Google Scholar]

| 1 | 2 | ||||
|---|---|---|---|---|---|
| Position | δC, type | δH (J in Hz) | Position | δC, type | δH (J in Hz) |
| 1 | b | - | 1 | b | - |
| 2 | 38.9, CH2 | 2.75, 2.62, m | 2 | b, CH2 | 2.95, m |
| 3 | 41.5, CH | 3.62, m | 3 | 125.3, CH | 5.66, m |
| 4 | 141.2, C | - | 4 | 134.1, CH | 5.74, m |
| 5 | 126.9, CH | 7.37, d (7.8) | 5 | 49.6, CH | 3.58, m |
| 6 | 126.9, CH | 7.27, t (7.5) | 6 | 141.4, C | - |
| 7 | 125.6, CH | 7.22, t (7.2) | 7 | 127.1, CH | 7.39, d (7.6) |
| 8 | 130.1, CH | 7.19, d (7.4) | 8 | 127.2, CH | 7.29, td (7.0, 0.9) |
| 9 | 137.4, C | - | 9 | 125.3, CH | 7.21, m |
| 10 | 129.9, CH | 6.83, d (11.3) | 10 | 129.9, CH | 7.19, m |
| 11 | 130.8, CH | 6.43, m | 11 | 136.5, C | - |
| 12 | 129.3, CH | 6.42, m | 12 | 129.3, CH | 6.60, m |
| 13 | 129.9, CH | 6.83, t (13.0) | 13 | 130.9, CH | 6.46, m |
| 14 | 127.1, CH | 5.84, t (10.8) | 14 | 129.1, CH | 6.33, m |
| 15 | 129.6, CH | 5.98, t (10.8) | 15 | 129.8, CH | 6.85, dd (14.2, 11.9) |
| 16 | 127.2, CH | 6.64, m | 16 | 127.0, CH | 5.83, m |
| 17 | 130.3, CH | 5.79, m | 17 | 129.5, CH | 5.97, m |
| 18 | 17.3, CH3 | 1.85, d (6.7) | 18 | 127.2, CH | 6.63, m |
| 1′ | a | - | 19 | 130.2, CH | 5.78, m |
| 2′ | 40.5, CH2 | 3.03, m | 20 | 17.3, CH3 | 1.84, d (6.7) |
| 3′ | 127.7, CH | 5.73, m | 1′ | a | - |
| 4′ | 127.4, CH | 6.48, dd (14.8, 11.9) | 2′ | 35.1, CH2 | 3.03, m |
| 5′ | 125.3, CH | 5.70, m | 3′ | 120.1, CH | 5.41, m |
| 6′ | 135.3, C | - | 4′ | 136.5, C | - |
| 7′ | 123.9, CH | 6.60, d (10.2) | 5′ | 123.1, CH | 6.33, m |
| 8′ | 131.2, CH | 5.71, t (10.9) | 6′ | 131.7, CH | 5.49, d (10.6, 9.1) |
| 9′ | 38.4, CH | 2.68, m | 7′ | 38.3, CH | 2.71, m |
| 10′ | 32.7, CH2 | 1.62, m 1.58, m | 8′ | 33.6, CH2 | 1.74, m 1.63, m |
| 11′ | 37.1 CH | 3.19, m | 9′ | 37.5, CH | 3.48, m |
| 12′ | 133.7, CH | 5.24, t (10.3) | 10′ | 134.1, CH | 5.41, m |
| 13′ | 128.4, CH | 6.05, t (10.3) | 11′ | 128.0, CH | 6.08, t (11.0) |
| 14′ | 128.1, CH | 6.30, dd (14.1, 12.0) | 12′ | 127.9, CH | 6.49, m |
| 15′ | 128.5, CH | 6.70, dd (14.6, 10.8) | 13′ | 128.4, CH | 6.71, m |
| 16′ | 127.1, CH | 5.96, m | 14′ | 127.2, CH | 5.97, m |
| 17′ | 129.7, CH | 5.98, t (10.8) | 15′ | 130.2, CH | 5.97, m |
| 18′ | 127.2, CH | 6.64, m | 16′ | 127.2, CH | 6.63, m |
| 19′ | 130.5, CH | 5.79, m | 17′ | 130.7, CH | 5.78, m |
| 20′ | 17.3, CH3 | 1.85, d (6.7) | 18′ | 17.3, CH3 | 1.84, d (6.7) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, J.; Li, H.; Liu, Z.; Li, T.; Xiao, F.; Li, W. Youssoufenes A2 and A3, Antibiotic Dimeric Cinnamoyl Lipids from the ΔdtlA Mutant of a Marine-Derived Streptomyces Strain. Mar. Drugs 2022, 20, 394. https://doi.org/10.3390/md20060394
Liu J, Li H, Liu Z, Li T, Xiao F, Li W. Youssoufenes A2 and A3, Antibiotic Dimeric Cinnamoyl Lipids from the ΔdtlA Mutant of a Marine-Derived Streptomyces Strain. Marine Drugs. 2022; 20(6):394. https://doi.org/10.3390/md20060394
Chicago/Turabian StyleLiu, Jing, Huayue Li, Zengzhi Liu, Tong Li, Fei Xiao, and Wenli Li. 2022. "Youssoufenes A2 and A3, Antibiotic Dimeric Cinnamoyl Lipids from the ΔdtlA Mutant of a Marine-Derived Streptomyces Strain" Marine Drugs 20, no. 6: 394. https://doi.org/10.3390/md20060394
APA StyleLiu, J., Li, H., Liu, Z., Li, T., Xiao, F., & Li, W. (2022). Youssoufenes A2 and A3, Antibiotic Dimeric Cinnamoyl Lipids from the ΔdtlA Mutant of a Marine-Derived Streptomyces Strain. Marine Drugs, 20(6), 394. https://doi.org/10.3390/md20060394






