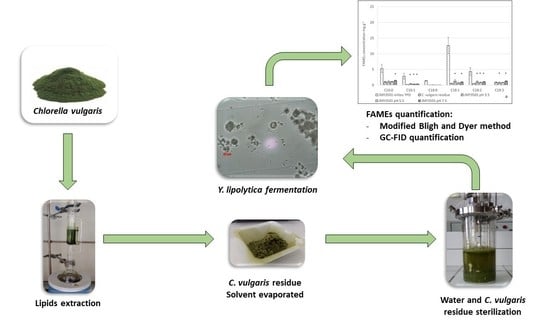
Evaluation of the Potential of Lipid-Extracted Chlorella vulgaris Residue for Yarrowia lipolytica Growth at Different pH Levels
Abstract
:1. Introduction
2. Results
2.1. Biochemical and Elemental Analysis of C. vulgaris Residue
2.2. Growth of Wild-Type and Mutant Y. lipolytica Strains in Lipid-Extracted C. vulgaris Residue and Control YPD Media
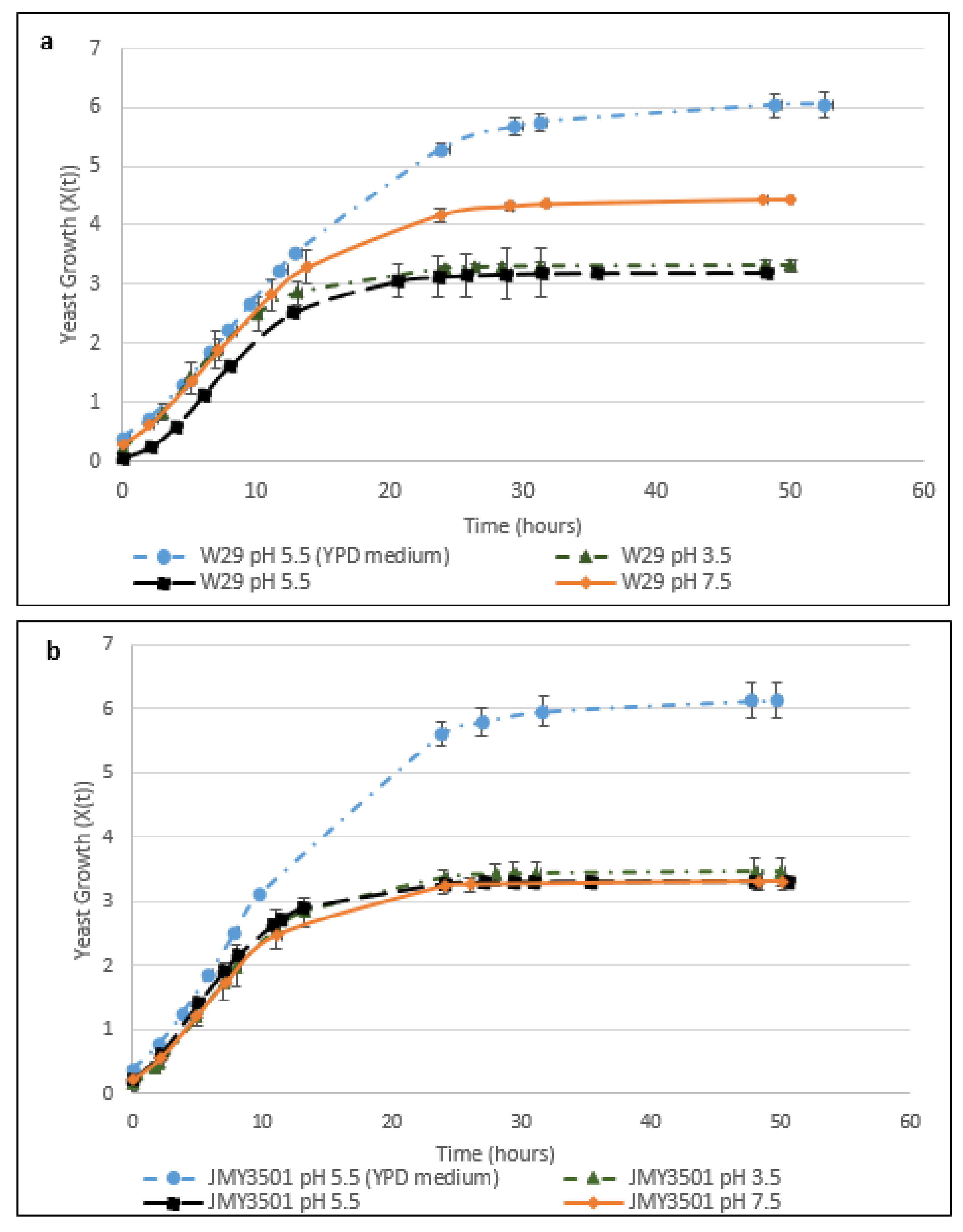
2.2.1. The Effect of pH on the Cell Growth of Y. lipolytica W29
2.2.2. The Effect of pH on the Cell Growth of JMY3501
2.3. Fatty Acid Profiles of Wild-Type and Mutant Y. lipolytica Strains
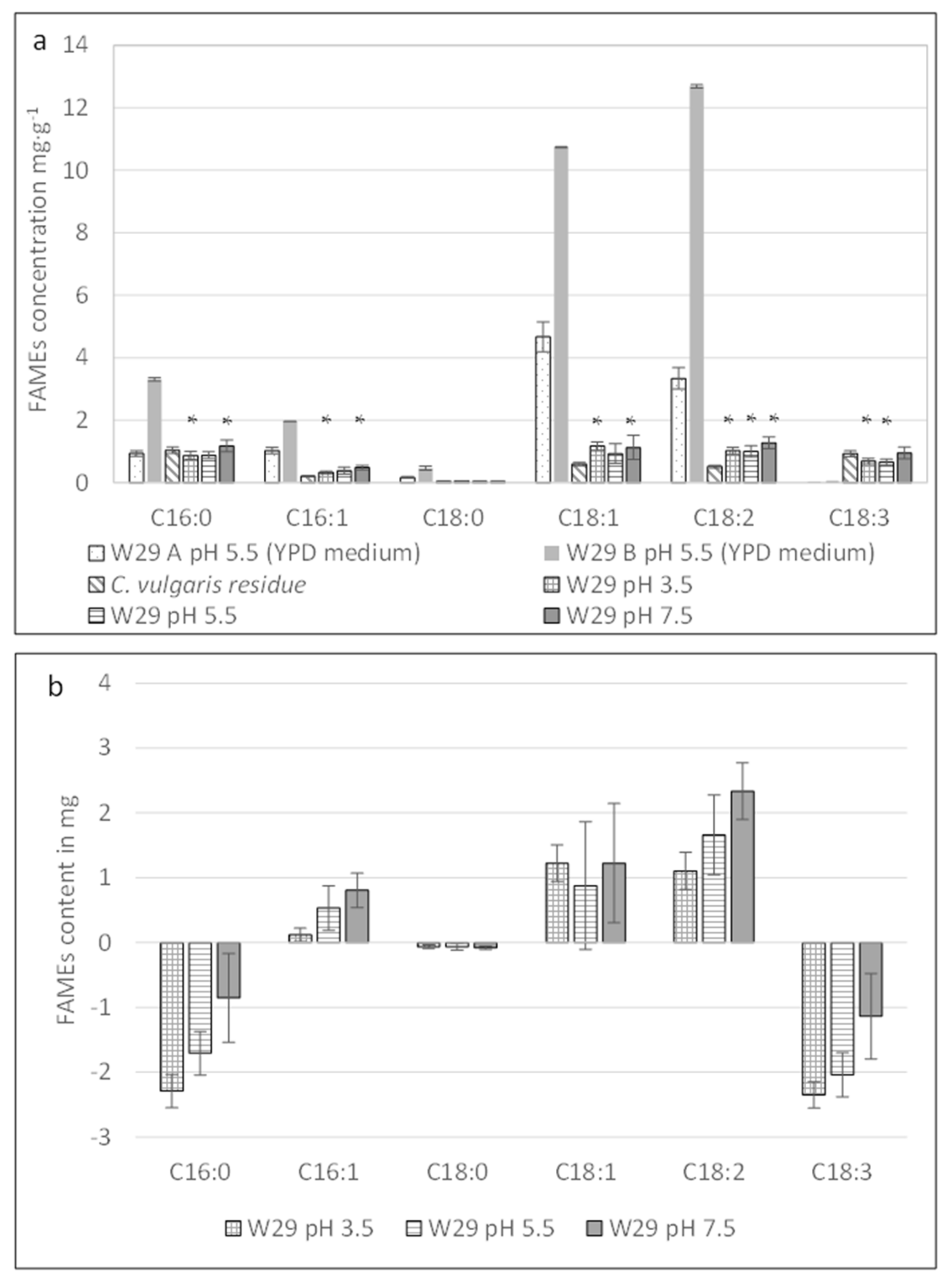
2.3.1. Effect of Different pH Levels on the Fatty Acid Profile of Y. lipolytica W29
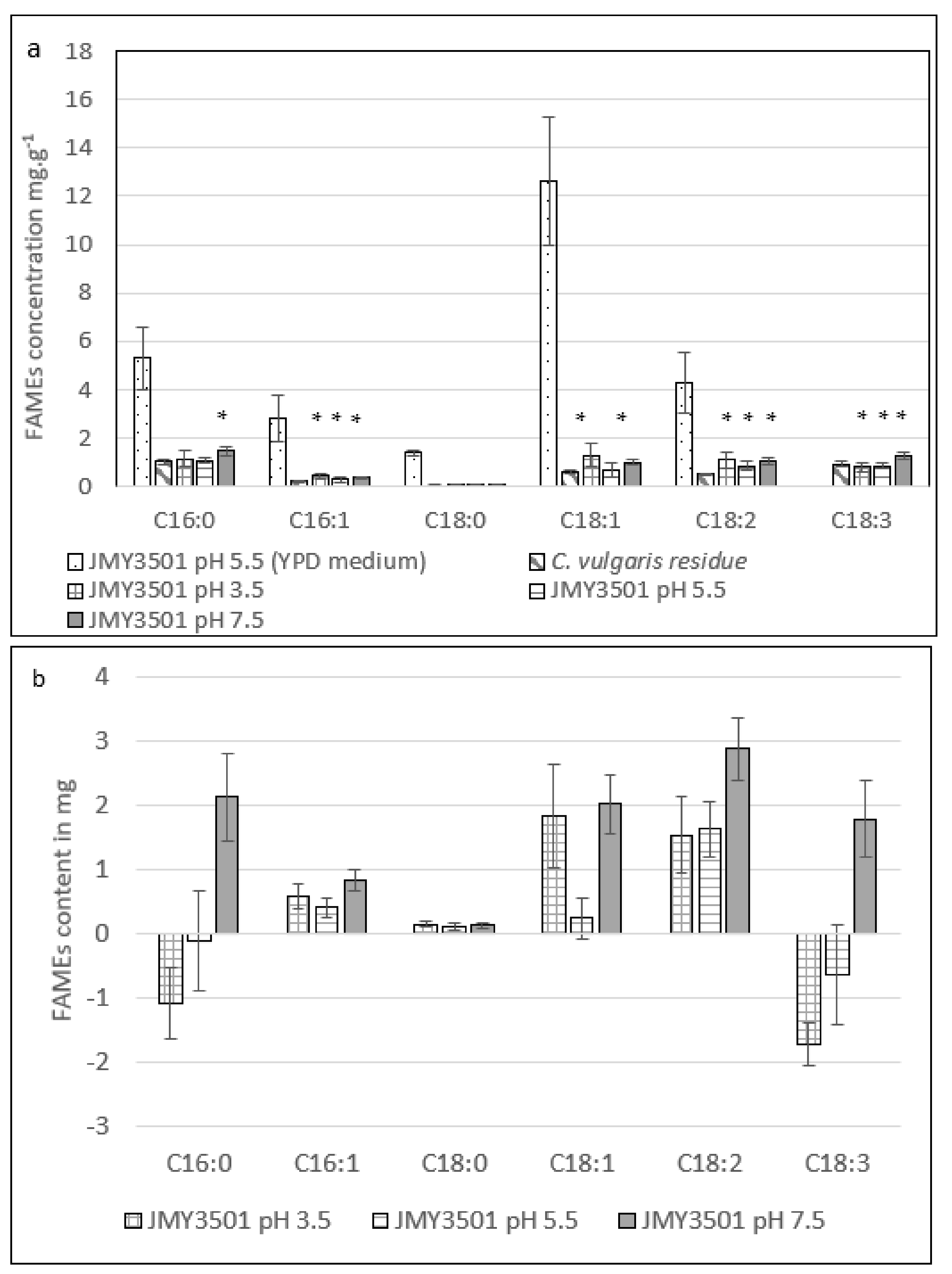
2.3.2. Effect of Different pH Levels on the Fatty Acid Profile of Y. lipolytica JMY3501
3. Discussion
4. Materials and Methods
4.1. Strains and Plasmid Genotype
4.2. Culture Conditions
4.3. Fermentation of C. vulgaris by Y. lipolytica
4.4. Analytical Procedure
4.4.1. Determination of Yeast Growth
4.4.2. Biochemical and Elementary Characterization of the Microalgae Strain after Lipid Extraction
4.4.3. Lipids Extraction
4.4.4. FAMEs Derivatization
4.4.5. FAMEs Gas Chromatography Analysis
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Safi, C.; Zebib, B.; Merah, O.; Pontalier, P.Y.; Vaca-Garcia, C. Morphology, composition, production, processing and applications of Chlorella vulgaris: A review. Renew. Sustain. Energy Rev. 2014, 35, 265–278. [Google Scholar] [CrossRef] [Green Version]
- De Morais, M.G.; da Silva Vaz, B.; de Morais, E.G.; Costa, J.A.V. Biologically Active Metabolites Synthesized by Microalgae. Biomed. Res. Int. 2015, 2015, 835761. [Google Scholar] [CrossRef] [Green Version]
- Falaise, C.; François, C.; Travers, M.-A.; Morga, B.; Haure, J.; Tremblay, R.; Turcotte, F.; Pasetto, P.; Gastineau, R.; Hardivillier, Y.; et al. Antimicrobial compounds from eukaryotic microalgae against human pathogens and diseases in aquaculture. Mar. Drugs 2016, 14, 159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, D.Y.Y.; Khoo, K.S.; Chew, K.W.; Tao, Y.; Ho, S.H.; Show, P.L. Potential utilization of bioproducts from microalgae for the quality enhancement of natural products. Bioresour. Technol. 2020, 304, 835761. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.U.; Cho, H.U.; Park, J.M.; Park, J.M.; Kim, Y.M. Enhanced microalgal biomass and lipid production from a consortium of indigenous microalgae and bacteria present in municipal wastewater under gradually mixotrophic culture conditions. Bioresour. Technol. 2017, 228, 290–297. [Google Scholar] [CrossRef] [PubMed]
- Zabed, H.M.; Akter, S.; Yun, J.; Zhang, G.; Awad, F.N.; Qi, X.; Sahu, J.N. Recent advances in biological pretreatment of microalgae and lignocellulosic biomass for biofuel production. Renew. Sustain. Energy Rev. 2019, 105, 105–128. [Google Scholar] [CrossRef]
- Ferreira, G.F.; Ríos Pinto, L.F.; Carvalho, P.O.; Coelho, M.B.; Eberlin, M.N.; Maciel Filho, R.; Fregolente, L.V. Biomass and lipid characterization of microalgae genera Botryococcus, Chlorella, and Desmodesmus aiming high-value fatty acid production. Biomass Convers. Biorefinery 2019, 11, 1675–1689. [Google Scholar] [CrossRef]
- Rumin, J.; Nicolau, E.; de Oliveira, R.G.; Fuentes-Grünewald, C.; Flynn, K.J.; Picot, L. A bibliometric analysis of microalgae research in the world, Europe, and the European Atlantic area. Mar. Drugs 2020, 18, 79. [Google Scholar] [CrossRef] [Green Version]
- Benemann, J. Microalgae for Biofuels and Animal Feeds. Energies 2013, 6, 5869–5886. [Google Scholar] [CrossRef] [Green Version]
- Araújo, R.; Calderón, F.V.; Sánchez López, J.; Azevedo, I.C.; Bruhn, A.; Fluch, S.; Tasende, M.G.; Ghaderiardakani, F.; IImjärv, T.; Laurans, M.; et al. Current Status of the Algae Production Industry in Europe: An Emerging Sector of the Blue Bioeconomy. Front. Mar. Sci. 2021. [Google Scholar] [CrossRef]
- Bhattacharya, M.; Goswami, S. Microalgae—A green multi-product biorefinery for future industrial prospects. Biocatal. Agric. Biotechnol. 2020, 25, 101580. [Google Scholar] [CrossRef]
- Younes, S.; Bracharz, F.; Awad, D.; Qoura, F.; Mehlmer, N.; Brueck, T. Microbial lipid production by oleaginous yeasts grown on Scenedesmus obtusiusculus microalgae biomass hydrolysate. Bioprocess Biosyst. Eng. 2020, 43, 1629–1638. [Google Scholar] [CrossRef] [PubMed]
- Seo, Y.H.; Han, S.; Han, J.I. Economic biodiesel production using algal residue as substrate of lipid producing yeast Cryptococcus curvatus. Renew. Energy 2014, 69, 473–478. [Google Scholar] [CrossRef]
- Yen, H.W.; Brune, D.E. Anaerobic co-digestion of algal sludge and waste paper to produce methane. Bioresour. Technol. 2007, 98, 130–134. [Google Scholar] [CrossRef]
- Jankowska, E.; Sahu, A.K.; Oleskowicz-Popiel, P. Biogas from microalgae: Review on microalgae’s cultivation, harvesting and pretreatment for anaerobic digestion. Renew. Sustain. Energy Rev. 2017, 75, 692–709. [Google Scholar] [CrossRef]
- Sun, M.L.; Madzak, C.; Liu, H.H.; Song, P.; Ren, L.J.; Huang, H.; Ji, X.J. Engineering Yarrowia lipolytica for efficient γ-linolenic acid production. Biochem. Eng. J. 2017, 117, 172–180. [Google Scholar] [CrossRef] [Green Version]
- Shahi, T.; Beheshti, B.; Zenouzi, A.; Almasi, M. Bio-oil production from residual biomass of microalgae after lipid extraction: The case of Dunaliella sp. Biocatal. Agric. Biotechnol. 2020, 23, 101494. [Google Scholar] [CrossRef]
- Huang, X.; Bai, S.; Liu, Z.; Hasunuma, T.; Kondo, A.; Ho, S.H. Fermentation of pigment-extracted microalgal residue using yeast cell-surface display: Direct high-density ethanol production with competitive life cycle impacts. Green Chem. 2020, 22, 153–162. [Google Scholar] [CrossRef]
- Kim, K.H.; Choi, I.S.; Kim, H.M.; Wi, S.G.; Bae, H.-J.J. Bioethanol production from the nutrient stress-induced microalga Chlorella vulgaris by enzymatic hydrolysis and immobilized yeast fermentation. Bioresour. Technol. 2014, 153, 47–54. [Google Scholar] [CrossRef]
- Lee, S.; Oh, Y.; Kim, D.; Kwon, D.; Lee, C.; Lee, J. Converting carbohydrates extracted from marine algae into ethanol using various ethanolic Escherichia coli strains. Appl. Biochem. Biotechnol. 2011, 164, 878–888. [Google Scholar] [CrossRef]
- Lee, O.K.; Oh, Y.-K.; Lee, E.Y. Bioethanol production from carbohydrate-enriched residual biomass obtained after lipid extraction of Chlorella sp. KR-1. Bioresour. Technol. 2015, 196, 22–27. [Google Scholar] [CrossRef]
- De Farias Silva, C.E.; Meneghello, D.; Bertucco, A. A systematic study regarding hydrolysis and ethanol fermentation from microalgal biomass. Biocatal. Agric. Biotechnol. 2018, 14, 172–182. [Google Scholar] [CrossRef]
- Htet, A.N.; Noguchi, M.; Ninomiya, K.; Tsuge, Y.; Kuroda, K.; Kajita, S.; Masai, E.; Katayama, Y.; Shikinaka, K.; Otsuka, Y.; et al. Application of microalgae hydrolysate as a fermentation medium for microbial production of 2-pyrone 4,6-dicarboxylic acid. J. Biosci. Bioeng. 2018, 125, 717–722. [Google Scholar] [CrossRef] [PubMed]
- Ratledge, C.; Wynn, J.P. The biochemistry and molecular biology of lipid accumulation in oleaginous microorganisms. Adv. Appl. Microbiol. 2002, 51, 1–52. [Google Scholar] [CrossRef] [PubMed]
- Groenewald, M.; Boekhout, T.; Neuvéglise, C.; Gaillardin, C.; van Dijck, P.W.M.; Wyss, M. Yarrowia lipolytica: Safety assessment of an oleaginous yeast with a great industrial potential. Crit. Rev. Microbiol. 2014, 40, 187–206. [Google Scholar] [CrossRef] [PubMed]
- Spagnuolo, M.; Hussain, M.S.; Gambill, L.; Blenner, M. Alternative substrate metabolism in Yarrowia lipolytica. Front. Microbiol. 2018, 9, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Thevenieau, F.; Beopoulos, A.; Desfougeres, T.; Sabirova, J.; Albertin, K.; Zinjarde, S.; Nicaud, J.-M. Uptake and Assimilation of Hydrophobic Substrates by the Oleaginous Yeast Yarrowia lipolytica. In Handbook of Hydrocarbon and Lipid Microbiology; Springer: Berlin/Heidelberg, Germany, 2010; pp. 1513–1527. [Google Scholar]
- Tezaki, S.; Iwama, R.; Kobayashi, S.; Shiwa, Y.; Yoshikawa, H.; Ohta, A.; Horiuchi, H.; Fukuda, R. Δ12-fatty acid desaturase is involved in growth at low temperature in yeast Yarrowia lipolytica. Biochem. Biophys. Res. Commun. 2017, 488, 165–170. [Google Scholar] [CrossRef]
- Krzyczkowska, J.; Kozłowska, M. Effect of Oils Extracted from Plant Seeds on the Growth and Lipolytic Activity of Yarrowia lipolytica Yeast. J. Am. Oil Chem. Soc. 2017, 94, 661–671. [Google Scholar] [CrossRef] [Green Version]
- Sestric, R.; Munch, G.; Cicek, N.; Sparling, R.; Levin, D.B. Growth and neutral lipid synthesis by Yarrowia lipolytica on various carbon substrates under nutrient-sufficient and nutrient-limited conditions. Bioresour. Technol. 2014, 164, 41–46. [Google Scholar] [CrossRef]
- Yang, S.; Wang, W.; Wei, H.; van Wychen, S.; Pienkos, P.T.; Zhang, M.; Himmel, M.E. Comparison of nitrogen depletion and repletion on lipid production in yeast and fungal species. Energies 2016, 9, 685. [Google Scholar] [CrossRef] [Green Version]
- Lopes, M.; Gomes, A.S.; Silva, C.M.; Belo, I. Microbial lipids and added value metabolites production by Yarrowia lipolytica from pork lard. J. Biotechnol. 2018, 265, 76–85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.; Jagtap, S.S.; Deewan, A.; Rao, C.V. pH selectively regulates citric acid and lipid production in Yarrowia lipolytica W29 during nitrogen-limited growth on glucose. J. Biotechnol. 2019, 290, 10–15. [Google Scholar] [CrossRef] [PubMed]
- Lazar, Z.; Dulermo, T.; Neuvéglise, C.; Crutz-Le Coq, A.M.; Nicaud, J.M. Hexokinase-A limiting factor in lipid production from fructose in Yarrowia lipolytica. Metab. Eng. 2014, 26, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Sekova, V.Y.; Dergacheva, D.I.; Isakova, E.P.; Gessler, N.N.; Tereshina, V.M.; Deryabina, Y.I. Soluble Sugar and Lipid Readjustments in the Yarrowia lipolytica Yeast at Various Temperatures and pH. Metabolites 2019, 9, 307. [Google Scholar] [CrossRef] [Green Version]
- Rashidi, B.; Trindade, L.M. Detailed biochemical and morphologic characteristics of the green microalga Neochloris oleoabundans cell wall. Algal Res. 2018, 35, 152–159. [Google Scholar] [CrossRef]
- Ochoa-Estopier, A.; Guillouet, S.E. D-stat culture for studying the metabolic shifts from oxidative metabolism to lipid accumulation and citric acid production in Yarrowia lipolytica. J. Biotechnol. 2014, 170, 35–41. [Google Scholar] [CrossRef]
- Timoumi, A.; Cléret, M.; Bideaux, C.; Guillouet, S.E.; Allouche, Y.; Molina-Jouve, C.; Fillaudeau, L.; Gorret, N. Dynamic behavior of Yarrowia lipolytica in response to pH perturbations: Dependence of the stress response on the culture mode. Appl. Microbiol. Biotechnol. 2017, 101, 351–366. [Google Scholar] [CrossRef]
- Azambuja, S.P.H.; Bonturi, N.; Miranda, E.A.; Gombert, A.K. Physiology and lipid accumulation capacity of different Yarrowia lipolytica and Rhodosporidium toruloides strains on glycerol. bioRxiv 2018, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez-Lopez, C.I.; Szabo, R.; Blanchin-Roland, S.; Gaillardin, C. Genetic control of extracellular protease synthesis in the yeast Yarrowia lipolytica. Genetics 2002, 160, 417–427. [Google Scholar] [CrossRef]
- Barth, G.; Gaillardin, C. Yarrowia lipolytica. In Nonconventional Yeasts in Biotechnology; Wolf, K., Ed.; Springer: Berlin/Heidelberg, Germany, 1996; pp. 313–388. [Google Scholar]
- Beopoulos, A.; Chardot, T.; Nicaud, J.M. Yarrowia lipolytica: A model and a tool to understand the mechanisms implicated in lipid accumulation. Biochimie 2009, 91, 692–696. [Google Scholar] [CrossRef]
- Wu, S.; Zhao, X.; Shen, H.; Wang, Q.; Zhao, Z.K. Microbial lipid production by Rhodosporidium toruloides under sulfate-limited conditions. Bioresour. Technol. 2011, 102, 1803–1807. [Google Scholar] [CrossRef] [PubMed]
- Mattanna, P.; Dallé da Rosa, P.; Gusso, A.P.; Richards, N.; Valente, P. Enhancement of microbial oil production by alpha-linolenic acid producing Yarrowia lipolytica strains QU22 and QU137. Food Sci. Biotechnol. 2014, 23, 1929–1934. [Google Scholar] [CrossRef]
- Cordova, L.T.; Alper, H.S. Production of α-linolenic acid in Yarrowia lipolytica using low-temperature fermentation. Appl. Microbiol. Biotechnol. 2018, 102, 8809–8816. [Google Scholar] [CrossRef] [PubMed]
- Gemperlein, K.; Dietrich, D.; Kohlstedt, M.; Zipf, G.; Bernauer, H.S.; Wittmann, C.; Wenzel, S.C.; Müller, R. Polyunsaturated fatty acid production by Yarrowia lipolytica employing designed myxobacterial PUFA synthases. Nat. Commun. 2019, 10, 4055. [Google Scholar] [CrossRef] [Green Version]
- Yan, F.X.; Dong, G.R.; Qiang, S.; Niu, Y.J.; Hu, C.Y.; Meng, Y.H. Overexpression of △12, △15-Desaturases for Enhanced Lipids Synthesis in Yarrowia lipolytica. Front. Microbiol. 2020, 11, 289. [Google Scholar] [CrossRef]
- Carsanba, E.; Papanikolaou, S.; Fickers, P.; Erten, H. Lipids by Yarrowia lipolytica Strains Cultivated on Glucose in Batch Cultures. Microorganisms 2020, 8, 1054. [Google Scholar] [CrossRef]
- Cui, J.; He, S.; Ji, X.; Lin, L.; Wei, Y.; Zhang, Q. Identification and characterization of a novel bifunctional Δ12/Δ15-fatty acid desaturase gene from Rhodosporidium kratochvilovae. Biotechnol. Lett. 2016, 38, 1155–1164. [Google Scholar] [CrossRef]
- Sayanova, O.; Haslam, R.; Guschina, I.; Lloyd, D.; Christie, W.W.; Harwood, J.L.; Napier, J.A. A bifunctional Δ12,Δ15-desaturase from Acanthamoeba castellanii directs the synthesis of highly unusual n-1 series unsaturated fatty acids. J. Biol. Chem. 2006, 281, 36533–36541. [Google Scholar] [CrossRef] [Green Version]
- Beopoulos, A.; Mrozova, Z.; Thevenieau, F.; Le Dall, M.T.; Hapala, I.; Papanikolaou, S.; Chardot, T.; Nicaud, J.M. Control of lipid accumulation in the yeast Yarrowia lipolytica. Appl. Environ. Microbiol. 2008, 74, 7779–7789. [Google Scholar] [CrossRef] [Green Version]
- Dulermo, T.; Tréton, B.; Beopoulos, A.; Gnankon, A.P.K.; Haddouche, R.; Nicaud, J.M. Characterization of the two intracellular lipases of Y. lipolytica encoded by TGL3 and TGL4 genes: New insights into the role of intracellular lipases and lipid body organisation. Biochim. Biophys. Acta—Mol. Cell Biol. Lipids 2013, 1831, 1486–1495. [Google Scholar] [CrossRef]
- Dulermo, T.; Nicaud, J.M. Involvement of the G3P shuttle and Β-oxidation pathway in the control of TAG synthesis and lipid accumulation in Yarrowia lipolytica. Metab. Eng. 2011, 13, 482–491. [Google Scholar] [CrossRef]
- Beopoulos, A.; Haddouche, R.; Kabran, P.; Dulermo, T.; Chardot, T.; Nicaud, J.M. Identification and characterization of DGA2, an acyltransferase of the DGAT1 acyl-CoA:diacylglycerol acyltransferase family in the oleaginous yeast Yarrowia lipolytica. New insights into the storage lipid metabolism of oleaginous yeasts. Appl. Microbiol. Biotechnol. 2012, 93, 1523–1537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryckebosch, E.; Muylaert, K.; Foubert, I. Optimization of an analytical procedure for extraction of lipids from microalgae. J. Am. Oil Chem. Soc. 2012, 89, 189–198. [Google Scholar] [CrossRef]
- Tjørve, K.M.C.; Tjørve, E. The use of Gompertz models in growth analyses, and new Gompertz-model approach: An addition to the Unified-Richards family. PLoS ONE 2017, 12, e0178691. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.L. Use of Dinitrosalicylic Acid Reagent for Determination of Reducing Sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Rice, E.W.; Baird, R.B.; Eaton, A.D. Standard Methods for the Examination of Water and Wastewater, 23th ed.; American Public Health Association: Washington, DC, USA; American Water Works Association: Washington, DC, USA, 2017. [Google Scholar]
- Sluiter, A.; Hames, B.; Ruiz, R.; Scarlata, C.; Sluiter, J.; Templeton, D. Determination of Ash in Biomass; NREL Laboratory Analytical Procedure (LAP): Golden, CO, USA, 2008; pp. 1–17. [Google Scholar]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef]
| Biochemical Compound | Content (Percentage of Dry Weight) |
| Proteins | 40.1 ± 5.3 |
| Reducing carbohydrates | 15.6 ± 2.1 |
| Fatty acids | 0.34 ± 0.02 |
| Elementary Composition | Content (Percentage of Dry Weight) |
| Total carbon | 34.9 ± 0.4 |
| Total nitrogen | 7.8 ± 0.2 |
| Ash | 11.2 ± 0.1 |
| Strains Medium | pH | Specific Growth Rate (h−1) | Cell Concentration (106 Cells·mL−1) | Generation Time (h−1) | Stationary Phase (h) |
|---|---|---|---|---|---|
| Y. lipolytica W29 YPD medium | 5.5 | 0.28 ± 0.03 | 1082.8 ± 4.61 | 2.50 ± 0.26 | 35 |
| Y. lipolytica W29 Lipid-extracted C. vulgaris residue | 3.5 | 0.28 ± 0.05 | 59.6 ± 8.1 | 2.56 ± 0.49 | 15 |
| 5.5 | 0.27 ± 0.06 | 200.8 ± 94.2 | 2.67 ± 0.59 | 15 | |
| 7.5 | 0.26 ± 0.01 | 235.4 ± 81.5 | 2.64 ± 0.09 | 25 | |
| Y. lipolytica JMY3501 YPD medium | 5.5 | 0.3238 ± 0.0004 | 1501.7 ± 33.4 | 2.14 ± 0.00 | 35 |
| Y. lipolytica JMY3501 Lipid-extracted C. vulgaris residue | 3.5 | 0.28 ± 0.08 | 77.1 ± 13.7 | 2.60 ± 0.76 | 15 |
| 5.5 | 0.28 ± 0.01 | 62.8 ± 18.3 | 2.49 ± 0.11 | 15 | |
| 7.5 | 0.26 ± 0.03 | 73.9 ± 6.6 | 2.69 ± 0.30 | 15 |
| Strains | pH | Media | Total Concentration of FAMEs mg.g−1 | Total Dry Biomass (g) | Total FAMEs (mg) |
|---|---|---|---|---|---|
| Lipid-extracted C. vulgaris residue | 7.5 | 3.3 ± 0.2 | 5.4 ± 0.9 | 17.8 ± 4.1 | |
| Y. lipolytica W29 | 5.5 | YPD | 25.4 ± 18.4 | 4.0 ± 0.4 | 101.6 ± 83.8 |
| Y. lipolytica W29 | 3.5 | Lipid-extracted C. vulgaris residue | 4.1 ± 0.5 | 3.8 ± 0.4 | 15.6 ± 3.3 |
| Y. lipolytica W29 | 5.5 | Lipid-extracted C. vulgaris residue | 3.9 ± 0.7 | 4.4 ± 0.3 | 17.1 ± 4.4 |
| Y. lipolytica W29 | 7.5 | Lipid-extracted C. vulgaris residue | 5.1 ± 0.7 | 4.0 ± 0.2 | 20.1 ± 3.9 |
| Y. lipolytica JMY3501 | 5.5 | YPD | 26.5 ± 6.1 | 4.4 ± 0.5 | 115.8 ± 39.8 |
| Y. lipolytica JMY3501 | 3.5 | Lipid-extracted C. vulgaris residue | 4.9 ± 1.5 | 4.1 ± 0.7 | 19.1 ± 9.4 |
| Y. lipolytica JMY3501 | 5.5 | Lipid-extracted C. vulgaris residue | 3.9 ± 0.6 | 5.0 ± 0.1 | 19.5 ± 3.5 |
| Y. lipolytica JMY3501 | 7.5 | Lipid-extracted C. vulgaris residue | 5.3 ± 0.6 | 5.26 ± 0.02 | 27.6 ± 1.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Delfau-Bonnet, G.; Imatoukene, N.; Clément, T.; Lopez, M.; Allais, F.; Hantson, A.-L. Evaluation of the Potential of Lipid-Extracted Chlorella vulgaris Residue for Yarrowia lipolytica Growth at Different pH Levels. Mar. Drugs 2022, 20, 264. https://doi.org/10.3390/md20040264
Delfau-Bonnet G, Imatoukene N, Clément T, Lopez M, Allais F, Hantson A-L. Evaluation of the Potential of Lipid-Extracted Chlorella vulgaris Residue for Yarrowia lipolytica Growth at Different pH Levels. Marine Drugs. 2022; 20(4):264. https://doi.org/10.3390/md20040264
Chicago/Turabian StyleDelfau-Bonnet, Guillaume, Nabila Imatoukene, Tiphaine Clément, Michel Lopez, Florent Allais, and Anne-Lise Hantson. 2022. "Evaluation of the Potential of Lipid-Extracted Chlorella vulgaris Residue for Yarrowia lipolytica Growth at Different pH Levels" Marine Drugs 20, no. 4: 264. https://doi.org/10.3390/md20040264
APA StyleDelfau-Bonnet, G., Imatoukene, N., Clément, T., Lopez, M., Allais, F., & Hantson, A.-L. (2022). Evaluation of the Potential of Lipid-Extracted Chlorella vulgaris Residue for Yarrowia lipolytica Growth at Different pH Levels. Marine Drugs, 20(4), 264. https://doi.org/10.3390/md20040264