Total Phenolic Levels, In Vitro Antioxidant Properties, and Fatty Acid Profile of Two Microalgae, Tetraselmis marina Strain IMA043 and Naviculoid Diatom Strain IMA053, Isolated from the North Adriatic Sea
Abstract
1. Introduction
2. Results and Discussion
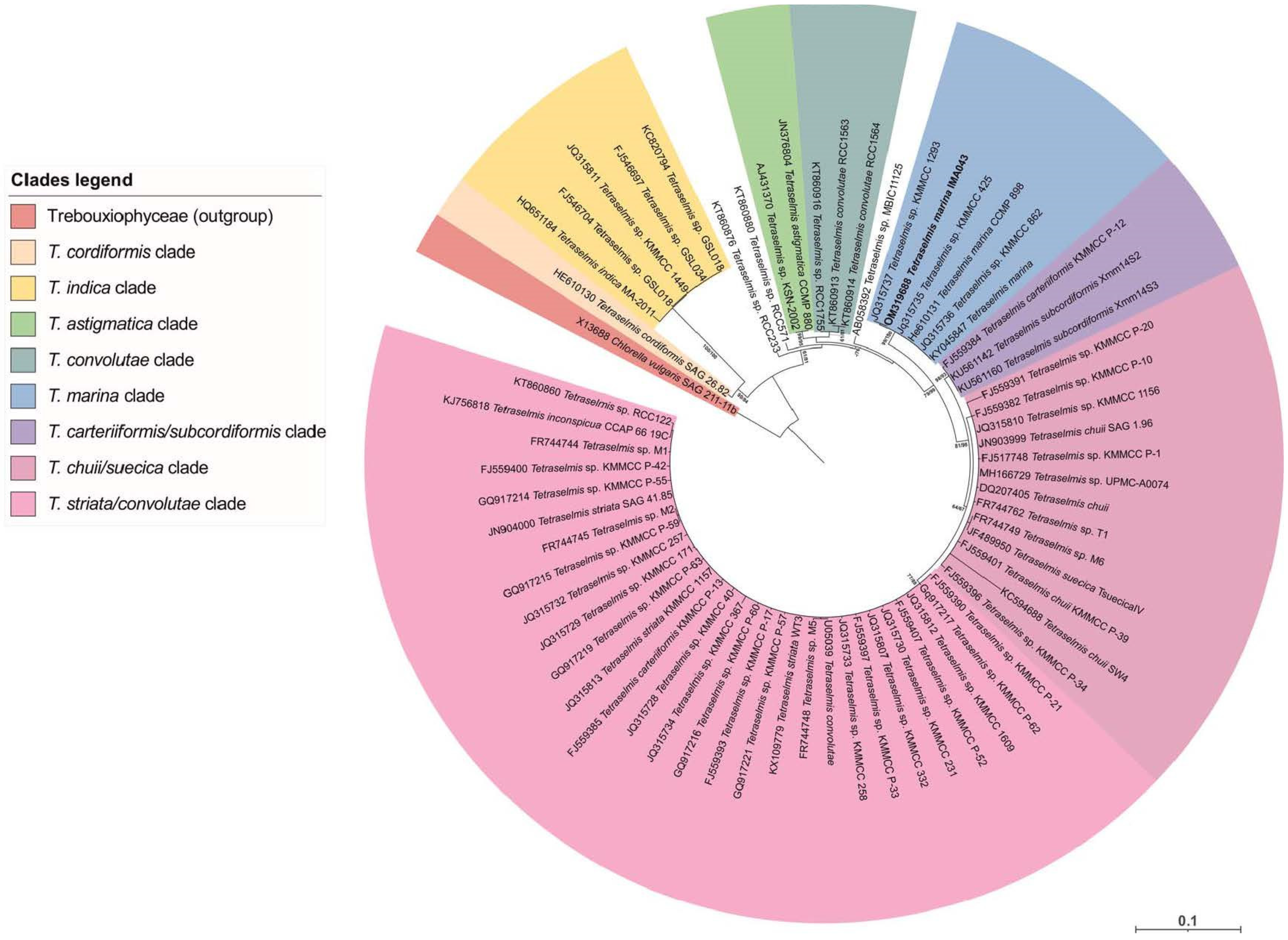
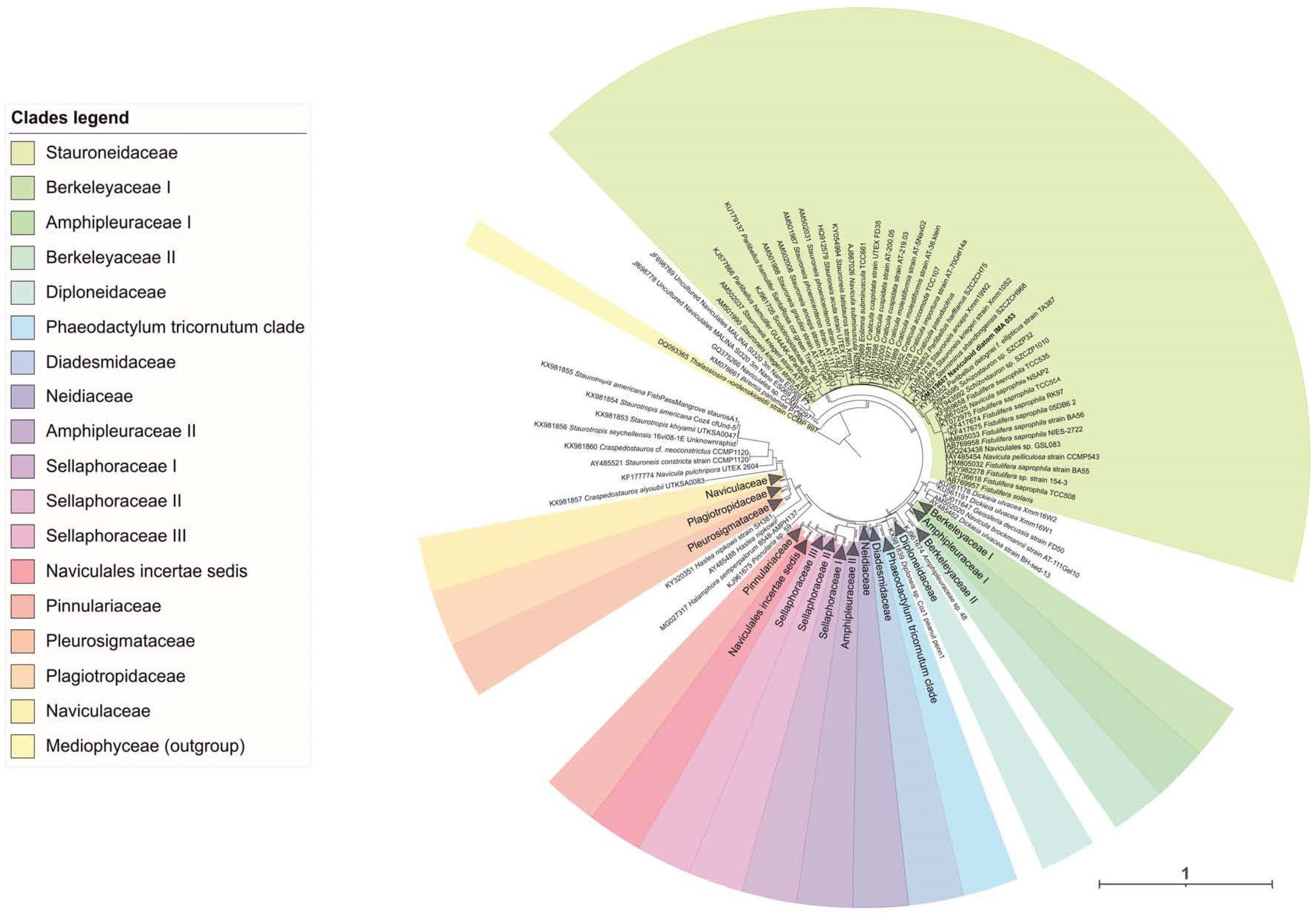
2.1. Molecular Analyses
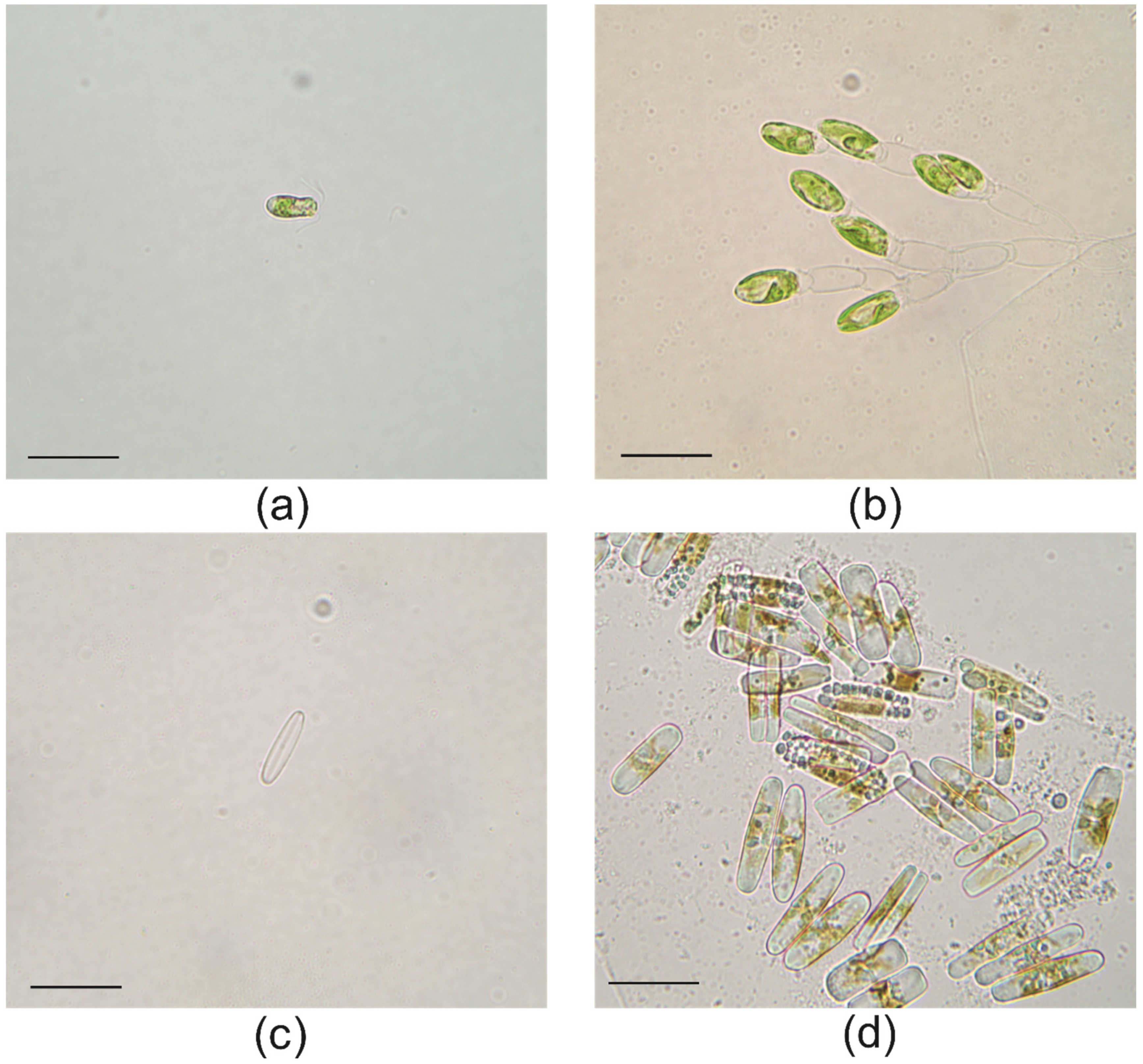
2.2. Light Microscopy
2.3. Total Phenolics Content (TPC) of the Extracts
2.4. In Vitro Antioxidant Properties
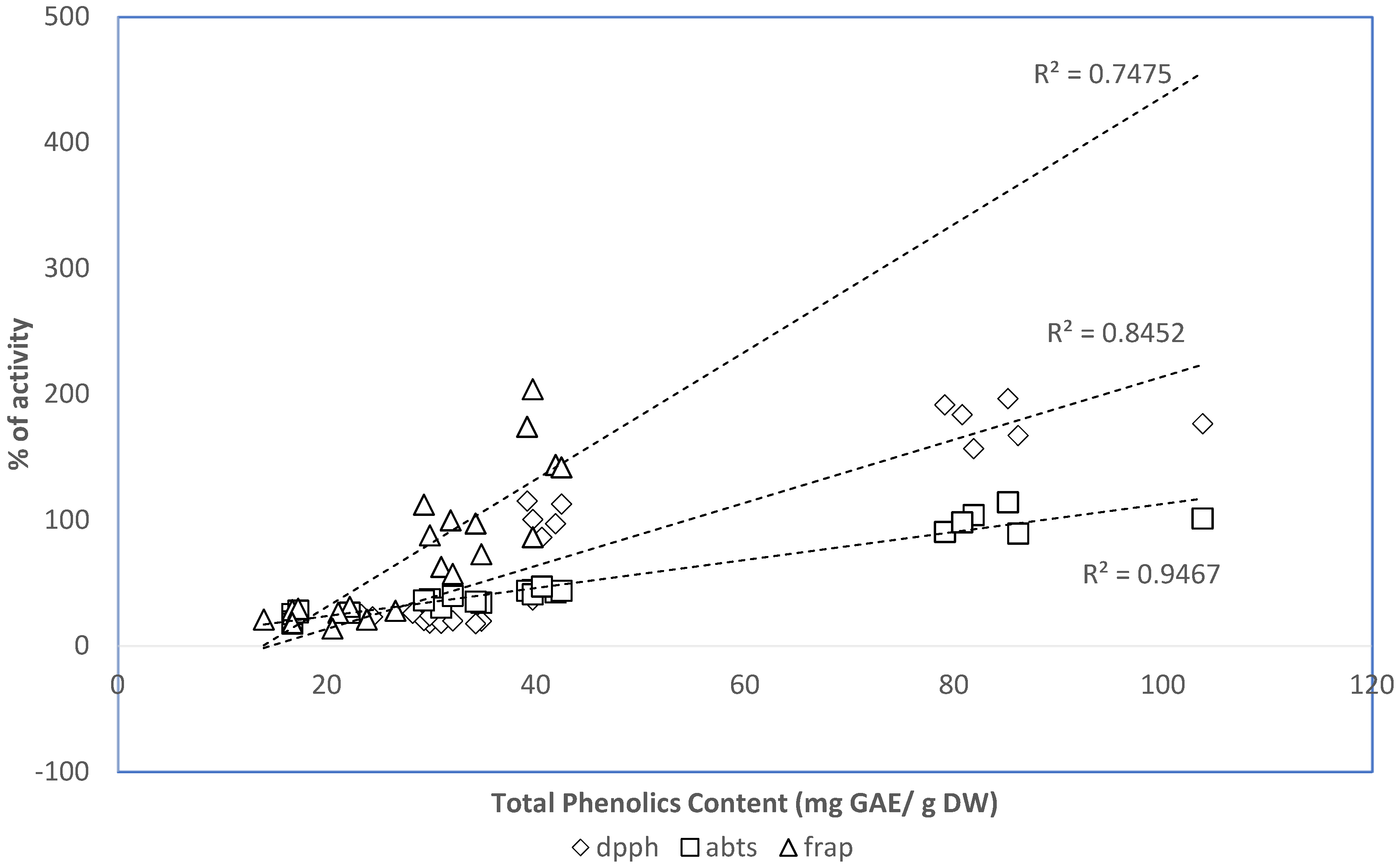
2.5. Relation between Phenolic Contents of the Extracts and the In Vitro Antioxidant Properties
2.6. FAMEs Profile
3. Materials and Methods
3.1. Chemicals
3.2. Sampling, Strain Isolation and Culture Set Up
3.3. Genetic Analyses
3.4. Light Microscopy
3.5. Preparation of the Extracts
3.6. Total Phenolics Content (TPC) of the Extracts
3.7. In Vitro Antioxidant Properties
3.7.1. Radical-Based Assays: RSA on DPPH and ABTS
3.7.2. Metal-Based Assays: Ferric Reducing Activity Power (FRAP) and Metal Chelation of Iron and Copper
3.8. Evaluation of the FA Profile of the Biomass
3.8.1. FAME Preparation
3.8.2. Determination of FAMEs Profile by GC–MS
3.9. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Guedes, A.C.; Amaro, H.M.; Malcata, F.X. Microalgae as sources of carotenoids. Mar. Drugs 2011, 9, 625–644. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.Y.Y.; Khoo, K.S.; Chew, K.W.; Tao, Y.; Ho, S.H.; Show, P.L. Potential utilization of bioproducts from microalgae for the quality enhancement of natural products. Bioresour. Technol. 2020, 304, 122997. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Lopez, C.; Pereira, A.G.; Lourenço-Lopes, C.; Garcia-Oliveira, P.; Cassani, L.; Fraga-Corral, M.; Prieto, M.A.; Simal-Gandara, J. Main bioactive phenolic compounds in marine algae and their mechanisms of action supporting potential health benefits. Food Chem. 2021, 341, 128262. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Gu, X.; Yang, D.; Xu, S.; Wang, S.; Chen, X.; Wang, Z. Bioactive substances and potentiality of marine microalgae. Food Sci. Nutr. 2021, 9, 5279–5292. [Google Scholar] [CrossRef] [PubMed]
- Pulz, O.; Gross, W. Valuable products from biotechnology of microalgae. Appl. Microbiol. Biotechnol. 2004, 65, 635–648. [Google Scholar] [CrossRef] [PubMed]
- Fernández, F.G.A.; Reis, A.; Wijffels, R.H.; Barbosa, M.; Verdelho, V.; Llamas, B. The role of microalgae in the bioeconomy. New Biotechnol. 2021, 61, 99–107. [Google Scholar] [CrossRef]
- Priyadarshani, I.; Rath, B. Commercial and industrial applications of micro algae—A review. J. Algal Biomass Util. 2012, 3, 89–100. [Google Scholar]
- Bohutskyi, P.; Liu, K.; Nasr, L.K.; Byers, N.; Rosenberg, J.N.; Oyler, G.A.; Betenbaugh, M.J.; Bouwer, E.J. Bioprospecting of microalgae for integrated biomass production and phytoremediation of unsterilized wastewater and anaerobic digestion centrate. Appl. Microbiol. Biotechnol. 2015, 99, 6139–6154. [Google Scholar] [CrossRef]
- Rindi, F.; Braga, J.C.; Martin, S.; Peña, V.; Le Gall, L.; Caragnano, A.; Aguirre, J. Coralline Algae in a Changing Mediterranean Sea: How Can We Predict Their Future, if We Do Not Know Their Present? Front. Mar. Sci. 2019, 6, 723. [Google Scholar] [CrossRef]
- Dahmen-Ben Moussa, I.; Chtourou, H.; Karray, F.; Sayadi, S.; Dhouib, A. Nitrogen or phosphorus repletion strategies for enhancing lipid or carotenoid production from Tetraselmis marina. Bioresour. Technol. 2017, 238, 325–332. [Google Scholar] [CrossRef]
- Cameron, H.; Mata, M.T.; Riquelme, C. The effect of heavy metals on the viability of Tetraselmis marina AC16-MESO and an evaluation of the potential use of this microalga in bioremediation. PeerJ 2018, 6, e5295. [Google Scholar] [CrossRef] [PubMed]
- Fimbres-Olivarría, D.; López-Elías, J.A.; Carvajal-Millán, E.; Márquez-Escalante, J.A.; Martínez-Córdova, L.R.; Miranda-Baeza, A.; Enríquez-Ocaña, F.; Valdéz-Holguín, J.E.; Brown-Bojórquez, F. Navicula sp. Sulfated Polysaccharide Gels Induced by Fe(III): Rheology and Microstructure. Int. J. Mol. Sci. 2016, 17, 1238. [Google Scholar] [CrossRef] [PubMed]
- Guiry, M.D.; Guiry, G.M. Algaebase. Available online: https://www.algaebase.org/about/ (accessed on 3 January 2022).
- Kim, Y.S.; Li, X.F.; Kang, K.H.; Ryu, B.M.; Kim, S.K. Stigmasterol isolated from marine microalgae Navicula incerta induces apoptosis in human hepatoma HepG2 cells. BMB Rep. 2014, 47, 433–438. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, M.; Sugiyama, H.; Maeda, Y.; Sato, R.; Tanaka, T.; Matsunaga, T. Marine diatom, Navicula sp. strain JPCC DA0580 and marine green alga, Chlorella sp. strain NKG400014 as potential sources for biodiesel production. Appl. Biochem. Biotechnol. 2010, 161, 483–490. [Google Scholar] [CrossRef] [PubMed]
- Muto, M.; Tanaka, M.; Liang, Y.; Yoshino, T.; Matsumoto, M.; Tanaka, T. Enhancement of glycerol metabolism in the oleaginous marine diatom Fistulifera solaris JPCC DA0580 to improve triacylglycerol productivity. Biotechnol. Biofuels 2015, 8, 4. [Google Scholar] [CrossRef] [PubMed]
- Chung, K.T.; Wong, T.Y.; Wei, C.I.; Huang, Y.W.; Lin, Y. Tannins and human health: A review. Crit. Rev. Food Sci. Nutr. 1998, 38, 421–464. [Google Scholar] [CrossRef]
- Li, H.B.; Cheng, K.W.; Wong, C.C.; Fan, K.W.; Chen, F.; Jiang, Y. Evaluation of antioxidant capacity and total phenolic content of different fractions of selected microalgae. Food Chem. 2007, 102, 771–776. [Google Scholar] [CrossRef]
- Haminiuk, C.W.I.; Plata-Oviedo, M.S.V.; de Mattos, G.; Carpes, S.T.; Branco, I.G. Extraction and quantification of phenolic acids and flavonols from Eugenia pyriformis using different solvents. J. Food Sci. Technol. 2012, 51, 2862–2866. [Google Scholar] [CrossRef]
- Lee, S.-H.; Karawita, R.; Affan, A.; Lee, J.-B.; Lee, K.-W.; Lee, B.-J.; Kim, D.-W.; Jeon, Y.-J. Potential of Benthic Diatoms Achnanthes longipes, Amphora coffeaeformisand Navicula sp. (Bacillariophyceae) as Antioxidant Sources. ALGAE 2009, 24, 47–55. [Google Scholar] [CrossRef]
- Custódio, L.; Justo, T.; Silvestre, L.; Barradas, A.; Duarte, C.V.; Pereira, H.; Barreira, L.; Rauter, A.P.; Alberício, F.; Varela, J. Microalgae of different phyla display antioxidant, metal chelating and acetylcholinesterase inhibitory activities. Food Chem. 2012, 131, 134–140. [Google Scholar] [CrossRef]
- Goiris, K.; Muylaert, K.; Fraeye, I.; Foubert, I.; De Brabanter, J.; De Cooman, L. Antioxidant potential of microalgae in relation to their phenolic and carotenoid content. J. Appl. Phycol. 2012, 24, 1477–1486. [Google Scholar] [CrossRef]
- Archer, L.; McGee, D.; Parkes, R.; Paskuliakova, A.; McCoy, G.R.; Adamo, G.; Cusimano, A.; Bongiovanni, A.; Gillespie, E.; Touzet, N. Antioxidant Bioprospecting in Microalgae: Characterisation of the Potential of Two Marine Heterokonts from Irish Waters. Appl. Biochem. Biotechnol. 2021, 193, 981–997. [Google Scholar] [CrossRef] [PubMed]
- Bhosale, P. Environmental and cultural stimulants in the production of carotenoids from microorganisms. Appl. Microbiol. Biotechnol. 2004, 63, 351–361. [Google Scholar] [CrossRef]
- Kepekçi, R.A.; Saygideger, S.D. Enhancement of phenolic compound production in Spirulina platensis by two-step batch mode cultivation. J. Appl. Phycol. 2012, 24, 897–905. [Google Scholar] [CrossRef]
- Queimada, A.J.; Mota, F.L.; Pinho, S.P.; Macedo, E.A. Solubilities of Biologically Active Phenolic Compounds: Measurements and Modeling. J. Phys. Chem. B 2009, 113, 3469–3476. [Google Scholar] [CrossRef]
- Pan, W.; Xu, H.; Cui, Y.; Song, D.; Feng, Y.Q. Improved liquid–liquid–liquid microextraction method and its application to analysis of four phenolic compounds in water samples. J. Chromatogr. A 2008, 1203, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Hajimahmoodi, M.; Faramarzi, M.A.; Mohammadi, N.; Soltani, N.; Oveisi, M.R.; Nafissi-Varcheh, N. Evaluation of antioxidant properties and total phenolic contents of some strains of microalgae. J. Appl. Phycol. 2010, 22, 43–50. [Google Scholar] [CrossRef]
- Oreopoulou, V.; Tzia, C. Utilization of Plant By-Products for the Recovery of Proteins, Dietary Fibers, Antioxidants, and Colorants. In Utilization of by-Products and Treatment of Waste in the Food Industry; Springer: Boston, MA, USA, 2007; pp. 209–232. [Google Scholar] [CrossRef]
- Dring, M.J. Stress Resistance and Disease Resistance in Seaweeds: The Role of Reactive Oxygen Metabolism. Adv. Bot. Res. 2005, 43, 175–207. [Google Scholar] [CrossRef]
- Cirulis, J.T.; Scott, J.A.; Ross, G.M. Management of oxidative stress by microalgae. Can. J. Physiol. Pharmacol. 2013, 91, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Scaglioni, P.T.; Badiale-Furlong, E. Can Microalgae Act as Source of Preservatives in Food Chain? J. Food Sci. Eng. 2017, 7, 283–296. [Google Scholar] [CrossRef][Green Version]
- Ulloa, G.; Otero, A.; Sánchez, M.; Sineiro, J.; Núñez, M.J.; Fábregas, J. Effect of Mg, Si, and Sr on growth and antioxidant activity of the marine microalga Tetraselmis suecica. J. Appl. Phycol. 2011, 24, 1229–1236. [Google Scholar] [CrossRef]
- Banskota, A.H.; Sperker, S.; Stefanova, R.; McGinn, P.J.; O’Leary, S.J.B. Antioxidant properties and lipid composition of selected microalgae. J. Appl. Phycol. 2018, 31, 309–318. [Google Scholar] [CrossRef]
- Natrah, F.M.I.; Yusoff, F.M.; Shariff, M.; Abas, F.; Mariana, N.S. Screening of Malaysian indigenous microalgae for antioxidant properties and nutritional value. J. Appl. Phycol. 2007, 19, 711–718. [Google Scholar] [CrossRef]
- Custódio, L.; Soares, F.; Pereira, H.; Barreira, L.; Vizetto-Duarte, C.; Rodrigues, M.J.; Rauter, A.P.; Alberício, F.; Varela, J. Fatty acid composition and biological activities of Isochrysis galbana T-ISO, Tetraselmis sp. and Scenedesmus sp.: Possible application in the pharmaceutical and functional food industries. J. Appl. Phycol. 2014, 26, 151–161. [Google Scholar] [CrossRef]
- Foo, S.C.; Yusoff, F.M.; Ismail, M.; Basri, M.; Khong, N.M.H.; Chan, K.W.; Yau, S.K. Efficient solvent extraction of antioxidant-rich extract from a tropical diatom, Chaetoceros calcitrans (Paulsen) Takano 1968. Asian Pac. J. Trop. Biomed. 2015, 5, 834–840. [Google Scholar] [CrossRef]
- Affan, A.; Karawita, R.; Jeon, Y.J.; Lee, J.B. Growth characteristics and antioxidant properties of the benthic diatom Navicula incerta (Bacillariophyceae) from Jeju Island, Korea1. J. Phycol. 2007, 43, 823–832. [Google Scholar] [CrossRef]
- Karthikeyan, P. In vitro Antioxidant Activity of Marine Diatoms. IOSR J. Environ. Sci. Toxicol. Food Technol. 2013, 5, 32–37. [Google Scholar] [CrossRef]
- Ordoñez, A.A.L.; Gomez, J.D.; Vattuone, M.A.; Isla, M.I. Antioxidant activities of Sechium edule (Jacq.) Swartz extracts. Food Chem. 2006, 97, 452–458. [Google Scholar] [CrossRef]
- Zengin, G.; Uysal, A.; Aktumsek, A.; Mocan, A.; Mollica, A.; Locatelli, M.; Custodio, L.; Neng, N.R.; Nogueira, J.M.F.; Aumeeruddy-Elalfi, Z.; et al. Euphorbia denticulata Lam.: A promising source of phyto-pharmaceuticals for the development of novel functional formulations. Biomed. Pharmacother. 2017, 87, 27–36. [Google Scholar] [CrossRef]
- Halliwell, B. Antioxidants in human health and disease. Annu. Rev. Nutr. 1996, 16, 33–50. [Google Scholar] [CrossRef]
- Valko, M.; Rhodes, C.J.; Moncol, J.; Izakovic, M.; Mazur, M. Free radicals, metals and antioxidants in oxidative stress-induced cancer. Chem. Biol. Interact. 2006, 160, 1–40. [Google Scholar] [CrossRef] [PubMed]
- Plaza, M.; Santoyo, S.; Jaime, L.; García-Blairsy Reina, G.; Herrero, M.; Señoráns, F.J.; Ibáñez, E. Screening for bioactive compounds from algae. J. Pharm. Biomed. Anal. 2010, 51, 450–455. [Google Scholar] [CrossRef] [PubMed]
- Kelman, D.; Posner, E.K.; McDermid, K.J.; Tabandera, N.K.; Wright, P.R.; Wright, A.D. Antioxidant Activity of Hawaiian Marine Algae. Mar. Drugs 2012, 10, 403–416. [Google Scholar] [CrossRef] [PubMed]
- Djeridane, A.; Yousfi, M.; Nadjemi, B.; Boutassouna, D.; Stocker, P.; Vidal, N. Antioxidant activity of some algerian medicinal plants extracts containing phenolic compounds. Food Chem. 2006, 97, 654–660. [Google Scholar] [CrossRef]
- Maadane, A.; Merghoub, N.; Ainane, T.; El Arroussi, H.; Benhima, R.; Amzazi, S.; Bakri, Y.; Wahby, I. Antioxidant activity of some Moroccan marine microalgae: Pufa profiles, carotenoids and phenolic content. J. Biotechnol. 2015, 215, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, C.; Pereira, H.; Franca, J.; Matos, J.; Monteiro, I.; Pousão-Ferreira, P.; Gomes, A.; Barreira, L.; Varela, J.; Neng, N.; et al. Lipid composition and some bioactivities of 3 newly isolated microalgae (Tetraselmis sp. IMP3, Tetraselmis sp. CTP4, and Skeletonema sp.). Aquac. Int. 2020, 28, 711–727. [Google Scholar] [CrossRef]
- Parkes, R.; Archer, L.; Gee, D.M.; Smyth, T.J.; Gillespie, E.; Touzet, N. Differential responses in EPA and fucoxanthin production by the marine diatom Stauroneis sp. under varying cultivation conditions. Biotechnol. Prog. 2021, 37, e3197. [Google Scholar] [CrossRef]
- Guzmán, H.M.; de la Valido, A.J.; Duarte, L.C.; Presmanes, K.F. Analysis of interspecific variation in relative fatty acid composition: Use of flow cytometry to estimate unsaturation index and relative polyunsaturated fatty acid content in microalgae. J. Appl. Phycol. 2010, 23, 7–15. [Google Scholar] [CrossRef]
- Renaud, S.M.; Thinh, L.V.; Lambrinidis, G.; Parry, D.L. Effect of temperature on growth, chemical composition and fatty acid composition of tropical Australian microalgae grown in batch cultures. Aquaculture 2002, 211, 195–214. [Google Scholar] [CrossRef]
- Teoh, M.L.; Chu, W.L.; Marchant, H.; Phang, S.M. Influence of culture temperature on the growth, biochemical composition and fatty acid profiles of six Antarctic microalgae. J. Appl. Phycol. 2004, 16, 421–430. [Google Scholar] [CrossRef]
- Najdenski, H.M.; Gigova, L.G.; Iliev, I.I.; Pilarski, P.S.; Lukavský, J.; Tsvetkova, I.V.; Ninova, M.S.; Kussovski, V.K. Antibacterial and antifungal activities of selected microalgae and cyanobacteria. Int. J. Food Sci. Technol. 2013, 48, 1533–1540. [Google Scholar] [CrossRef]
- Davoodbasha, M.A.; Edachery, B.; Nooruddin, T.; Lee, S.Y.; Kim, J.W. An evidence of C16 fatty acid methyl esters extracted from microalga for effective antimicrobial and antioxidant property. Microb. Pathog. 2018, 115, 233–238. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Gao, B.; Huang, L.; Su, M.; Dai, C.; Zhang, C. Evaluation of oleaginous eustigmatophycean microalgae as potential biorefinery feedstock for the production of palmitoleic acid and biodiesel. Bioresour. Technol. 2018, 270, 30–37. [Google Scholar] [CrossRef] [PubMed]
- De Souza, C.O.; Vannice, G.K.; Neto, J.C.R.; Calder, P.C. Is Palmitoleic Acid a Plausible Nonpharmacological Strategy to Prevent or Control Chronic Metabolic and Inflammatory Disorders? Mol. Nutr. Food Res. 2018, 62, 1700504. [Google Scholar] [CrossRef]
- Tutunchi, H.; Ostadrahimi, A.; Saghafi-Asl, M. The Effects of Diets Enriched in Monounsaturated Oleic Acid on the Management and Prevention of Obesity: A Systematic Review of Human Intervention Studies. Adv. Nutr. 2020, 11, 864–877. [Google Scholar] [CrossRef] [PubMed]
- Deshmukh, S.; Kumar, R.; Bala, K. Microalgae biodiesel: A review on oil extraction, fatty acid composition, properties and effect on engine performance and emissions. Fuel Process. Technol. 2019, 191, 232–247. [Google Scholar] [CrossRef]
- Xu, H.; Miao, X.; Wu, Q. High quality biodiesel production from a microalga Chlorella protothecoides by heterotrophic growth in fermenters. J. Biotechnol. 2006, 126, 499–507. [Google Scholar] [CrossRef]
- Trentin, R.; Custódio, L.; Rodrigues, M.J.; Moschin, E.; Sciuto, K.; da Silva, J.P.; Moro, I. Exploring Ulva australis Areschoug for possible biotechnological applications: In vitro antioxidant and enzymatic inhibitory properties, and fatty acids contents. Algal Res. 2020, 50, 101980. [Google Scholar] [CrossRef]
- Li, Q.; Du, W.; Liu, D. Perspectives of microbial oils for biodiesel production. Appl. Microbiol. Biotechnol. 2008, 80, 749–756. [Google Scholar] [CrossRef]
- Chisti, Y. Biodiesel from microalgae. Biotechnol. Adv. 2007, 25, 294–306. [Google Scholar] [CrossRef]
- Huang, M.; Xin, X.; Shi, H.; Lin, Y.; Ling, H.; Ge, J. Identification of six novel microalgal strains and characterization of its potential for development of high-value compounds. S. Afr. J. Bot. 2022, 147, 1–7. [Google Scholar] [CrossRef]
- Rodolfi, L.; Zittelli, G.C.; Bassi, N.; Padovani, G.; Biondi, N.; Bonini, G.; Tredici, M.R. Microalgae for oil: Strain selection, induction of lipid synthesis and outdoor mass cultivation in a low-cost photobioreactor. Biotechnol. Bioeng. 2009, 102, 100–112. [Google Scholar] [CrossRef] [PubMed]
- Abou-Shanab, R.A.I.; Hwang, J.H.; Cho, Y.; Min, B.; Jeon, B.H. Characterization of microalgal species isolated from fresh-water bodies as a potential source for biodiesel production. Appl. Energy 2011, 88, 3300–3306. [Google Scholar] [CrossRef]
- Guillard, R.R.L. Culture of Phytoplankton for Feeding Marine Invertebrates. In Culture of Marine Invertebrate Animals; Springer: Boston, MA, USA, 1975; pp. 29–60. [Google Scholar] [CrossRef]
- Edgcomb, V.P.; Kysela, D.T.; Teske, A.; de Vera Gomez, A.; Sogin, M.L.; Edgcomb, V.P.; Kysela, D.T.; Teske, A.; de Vera Gomez, A.; Sogin, M.L. Benthic eukaryotic diversity in the Guaymas Basin hydrothermal vent environment. Proc. Natl. Acad. Sci. USA 2002, 99, 7658–7662. [Google Scholar] [CrossRef]
- Medlin, L.; Elwood, H.J.; Stickel, S.; Sogin, M.L. The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene 1988, 71, 491–499. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Trifinopoulos, J.; Nguyen, L.T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Minh, B.Q.; Nguyen, M.A.T.; Von Haeseler, A. Ultrafast Approximation for Phylogenetic Bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Velioglu, Y.S.; Mazza, G.; Gao, L.; Oomah, B.D. Antioxidant Activity and Total Phenolics in Selected Fruits, Vegetables, and Grain Products. J. Agric. Food Chem. 1998, 46, 4113–4117. [Google Scholar] [CrossRef]
- Brand-Williams, W.; Cuvelier, M.E.; Berset, C. Use of a Free Radical Method to Evaluate Antioxidant Activity. LWT-Food Sci. Technol. 1995, 28, 25–30. [Google Scholar] [CrossRef]
- Custódio, L.; Patarra, J.; Alberício, F.; da Rosa Neng, N.; Nogueira, J.M.F.; Romano, A. Phenolic composition, antioxidant potential and in vitro inhibitory activity of leaves and acorns of Quercus suber on key enzymes relevant for hyperglycemia and Alzheimer’s disease. Ind. Crops Prod. 2015, 64, 45–51. [Google Scholar] [CrossRef]
- Re, R.; Pellegrini, N.; Proteggente, A.; Pannala, A.; Yang, M.; Rice-Evans, C. Antioxidant activity applying an improved ABTS radical cation decolorization assay. Free Radic. Biol. Med. 1999, 26, 1231–1237. [Google Scholar] [CrossRef]
- Megías, C.; Pastor-Cavada, E.; Torres-Fuentes, C.; Girón-Calle, J.; Alaiz, M.; Juan, R.; Pastor, J.; Vioque, J. Chelating, antioxidant and antiproliferative activity of Vicia sativa polyphenol extracts. Eur. Food Res. Technol. 2009, 230, 353–359. [Google Scholar] [CrossRef]
- Lepage, G.; Roy, C.C. Improved recovery of fatty acid through direct transesterification without prior extraction or purification. J. Lipid Res. 1984, 25, 1391–1396. [Google Scholar] [CrossRef]
- Pereira, H.; Barreira, L.; Figueiredo, F.; Custódio, L.; Vizetto-Duarte, C.; Polo, C.; Rešek, E.; Engelen, A.; Varela, J. Polyunsaturated Fatty Acids of Marine Macroalgae: Potential for Nutritional and Pharmaceutical Applications. Mar. Drugs 2012, 10, 1920–1935. [Google Scholar] [CrossRef] [PubMed]
| Extract | TPC | |
|---|---|---|
| T. marina (IMA043) | Water | 20.45 ± 1.87 d |
| Methanol | 25.19 ± 1.26 cd | |
| DCM | 86.14 ± 4.52 a | |
| Naviculoid diatom (IMA053) | Water | 17.80 ± 0.88 d |
| Methanol | 31.85 ± 0.92 c | |
| DCM | 40.58 ± 0.66 b |
| Extract | ABTS | DPPH | |||||
|---|---|---|---|---|---|---|---|
| 1 mg/mL | 5 mg/mL | 10 mg/mL | 1 mg/mL | 5 mg/mL | 10 mg/mL | ||
| Naviculoid diatom (IMA053) | Methanol | n.a. | 23.37 ± 0.58 c | 35.54 ± 1.28 b | n.a. | 12.53 ± 0.44 c | 18.91 ± 0.46 c |
| Water | n.a. | 11.06 ± 1.27 d | 23.51 ± 1.81 c | n.a. | n.a. | n.a. | |
| DCM | 10.09 ± 0.53 b | 46.91 ± 0.42 b | 43.83 ± 0.84 b | 18.25 ± 6.08 b | 41.55 ± 2.47 b | 91.38 ± 11.80 b | |
| T. marina (IMA043) | Methanol | n.a. | n.a. | n.a. | n.a. | 17.29 ± 0.35 c | 24.94 ± 0.71 c |
| Water | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | |
| DCM | 24.66 ± 0.32 a | 88.19 ± 2.41 a | 99.65 ± 3.76 a | 40.96 ± 0.94 a | 103.43 ± 7.12 a | 178.75 ± 6.13 a | |
| BHT * | 94.77 ± 0.04 | 80.07 ± 0.72 | |||||
| Extract | FRAP | |||
|---|---|---|---|---|
| 1 mg/mL | 5 mg/mL | 10 mg/mL | ||
| Naviculoid diatom (IMA053) | Methanol | 22.01 ± 0.68 c | 60.68 ± 3.54 ab | 70.23 ± 5.47 b |
| Water | n.a. | 23.29 ± 4.24 b | 26.75 ± 1.73 c | |
| DCM | 46.35 ± 8.26 ab | 135.10 ± 45.63 a | 150.10 ± 17.85 a | |
| T. marina (IMA043) | Methanol | 36.17 ± 2.04 bc | 92.28 ± 4.13 ab | 103.17 ± 3.26 b |
| Water | n.a. | 19.56 ± 2.15 b | 21.47 ± 2.14 c | |
| DCM | 67.16 ± 3.46 a | 40.02 ± 5.62 ab | n.a. | |
| Extract | ICA | CCA | |||||
|---|---|---|---|---|---|---|---|
| 1 mg/mL | 5 mg/mL | 10 mg/mL | 1 mg/mL | 5 mg/mL | 10 mg/mL | ||
| Naviculoid diatom (IMA053) | Methanol | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. |
| Water | n.a. | 83.43 ± 2.22 a | 92.05 ± 3.54 a | 13.92 ± 2.39 b | 29.23 ± 5.53 a | 35.31 ± 3.89 b | |
| DCM | n.a. | 49.18 ± 3.77 b | 63.57 ± 5.34 b | n.a. | 21.11 ± 8.01 a | 67.48 ± 14.68a | |
| T. marina (IMA043) | Methanol | 34.12 ± 3.00 | n.a. | n.a. | n.a. | n.a. | n.a. |
| Water | n.a. | 59.25 ± 4.23 b | 68.00 ± 6.13 b | 21.00 ± 2.88 a | 25.92 ± 3.41 a | 28.87 ± 1.57 b | |
| DCM | n.a. | n.a. | n.a. | n.a. | n.a. | 11.58 ± 1.49 c | |
| EDTA * | 72.28 ± 2.79 | 89.69 ± 0.55 | |||||
| Fatty Acid | Common Name | T. marina (IMA043) | Naviculoid Diatom (IMA053) |
|---|---|---|---|
| ∑SFA | 38.28 ± 2.00 | 45.70 ± 1.28 | |
| (C14:0) | Methyl myristate | 0.03 ± 0.08 | 3.15 ± 0.08 |
| (C15:0) | Methyl pentadecanoate | n.d. | 1.50 ± 0.96 |
| (C16:0) | Methyl palmitate | 37.74 ± 1.99 | 40.68 ± 0.83 |
| (C18:0) | Methyl stearate | 0.42 ± 0.12 | 0.37 ± 0.11 |
| (C24:0) | Methyl lignocerate | 0.09 ± 0.09 | n.d. |
| ∑MUFA | 45.41 ± 1.75 | 49.81 ± 1.00 | |
| (C16:1) | Methyl palmitoleate | 0.71 ± 0.20 | 47.25 ± 0.92 |
| (C18:1n9c) | cis-9-Oleic acid methyl ester | 37.52 ± 0.49 | 0.51 ± 0.15 |
| (C18:1n9t) | trans-9-Elaidic acid methyl ester | 3.17 ± 1.23 | 1.03 ± 0.29 |
| (C20:1) | Methyl cis-11-eicosenoate | 4.01 ± 1.12 | n.d. |
| (C22:1n9) | Methyl erucate | n.d. | 0.60 ± 0.18 |
| (C24:1n9) | Methyl nervonate | n.d. | 0.42 ± 0.13 |
| ∑PUFA | 16.31 ± 1.64 | 4.57 ± 0.47 | |
| (C19:3n3) | Methyl linolenate | 2.38 ± 1.43 | n.d. |
| (C18:2n6c) | Methyl linoleate | 11.01 ± 0.29 | 0.08 ± 0.08 |
| (C20:4n6) | cis-5,8,11,14-Eicosatetraenoic acid methyl ester | n.d. | 1.06 ± 0.34 |
| (C20:5n3) | cis-5,8,11,14,17-Eicosapentaenoic acid methyl ester | 2.50 ± 0.72 | 3.43 ± 0.32 |
| (C20:3n3) | cis-11,14,17-Eicosatrienoic acid methyl ester | 0.41 ± 0.22 | n.d. |
| (C20:2) | cis-11,14-Eicosadienoic acid methyl ester | 0.02 ± 0.03 | n.d. |
| ∑n-3 | 5.28 ± 1.61 | 3.43 ± 0.32 | |
| ∑n-6 | 11.01 ± 0.29 | 1.14 ± 0.35 | |
| ∑n-6/∑n-3 | 2.08 | 0.33 | |
| PUFA/SFA | 0.43 | 0.10 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trentin, R.; Custódio, L.; Rodrigues, M.J.; Moschin, E.; Sciuto, K.; da Silva, J.P.; Moro, I. Total Phenolic Levels, In Vitro Antioxidant Properties, and Fatty Acid Profile of Two Microalgae, Tetraselmis marina Strain IMA043 and Naviculoid Diatom Strain IMA053, Isolated from the North Adriatic Sea. Mar. Drugs 2022, 20, 207. https://doi.org/10.3390/md20030207
Trentin R, Custódio L, Rodrigues MJ, Moschin E, Sciuto K, da Silva JP, Moro I. Total Phenolic Levels, In Vitro Antioxidant Properties, and Fatty Acid Profile of Two Microalgae, Tetraselmis marina Strain IMA043 and Naviculoid Diatom Strain IMA053, Isolated from the North Adriatic Sea. Marine Drugs. 2022; 20(3):207. https://doi.org/10.3390/md20030207
Chicago/Turabian StyleTrentin, Riccardo, Luísa Custódio, Maria João Rodrigues, Emanuela Moschin, Katia Sciuto, José Paulo da Silva, and Isabella Moro. 2022. "Total Phenolic Levels, In Vitro Antioxidant Properties, and Fatty Acid Profile of Two Microalgae, Tetraselmis marina Strain IMA043 and Naviculoid Diatom Strain IMA053, Isolated from the North Adriatic Sea" Marine Drugs 20, no. 3: 207. https://doi.org/10.3390/md20030207
APA StyleTrentin, R., Custódio, L., Rodrigues, M. J., Moschin, E., Sciuto, K., da Silva, J. P., & Moro, I. (2022). Total Phenolic Levels, In Vitro Antioxidant Properties, and Fatty Acid Profile of Two Microalgae, Tetraselmis marina Strain IMA043 and Naviculoid Diatom Strain IMA053, Isolated from the North Adriatic Sea. Marine Drugs, 20(3), 207. https://doi.org/10.3390/md20030207







