Structure of the Cell-Wall-Associated Polysaccharides from the Deep-Sea Marine Bacterium Devosia submarina KMM 9415T
Abstract
:1. Introduction
2. Results
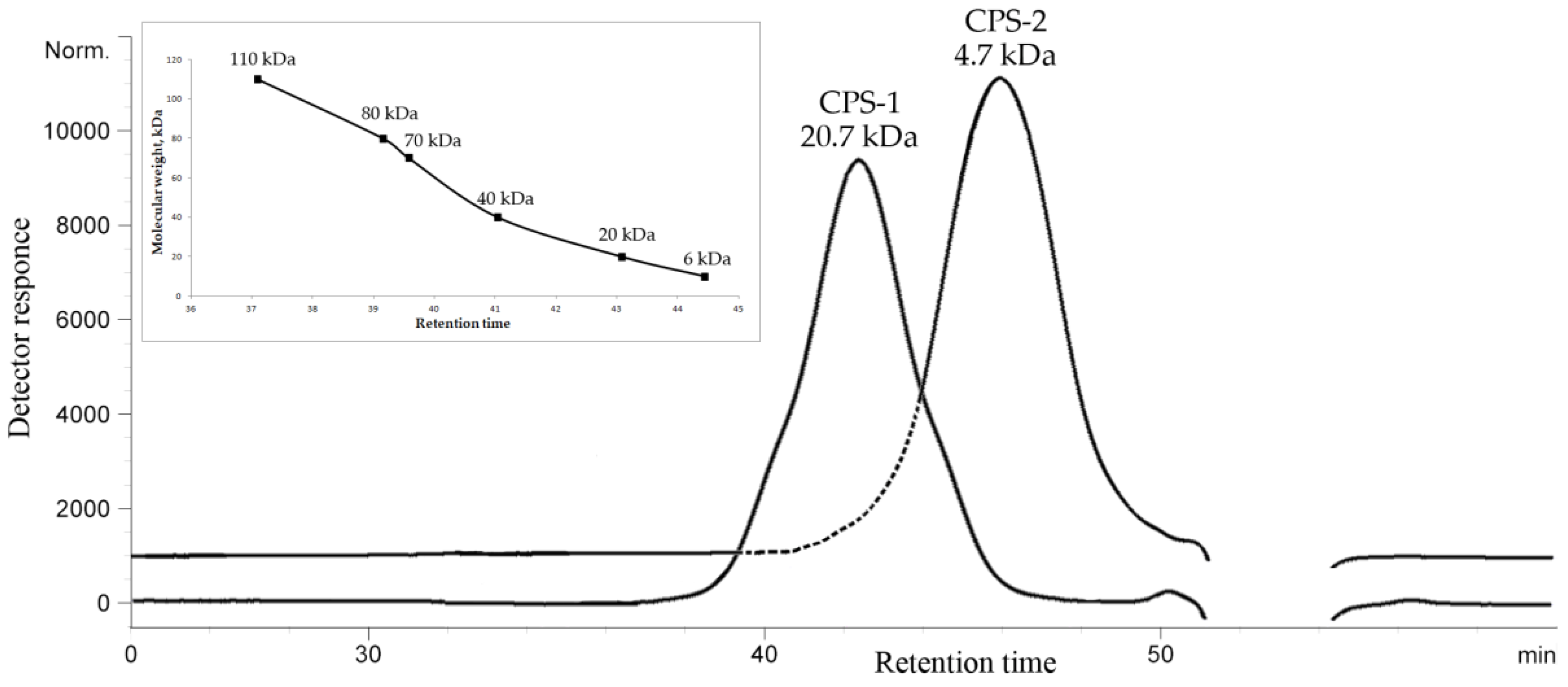
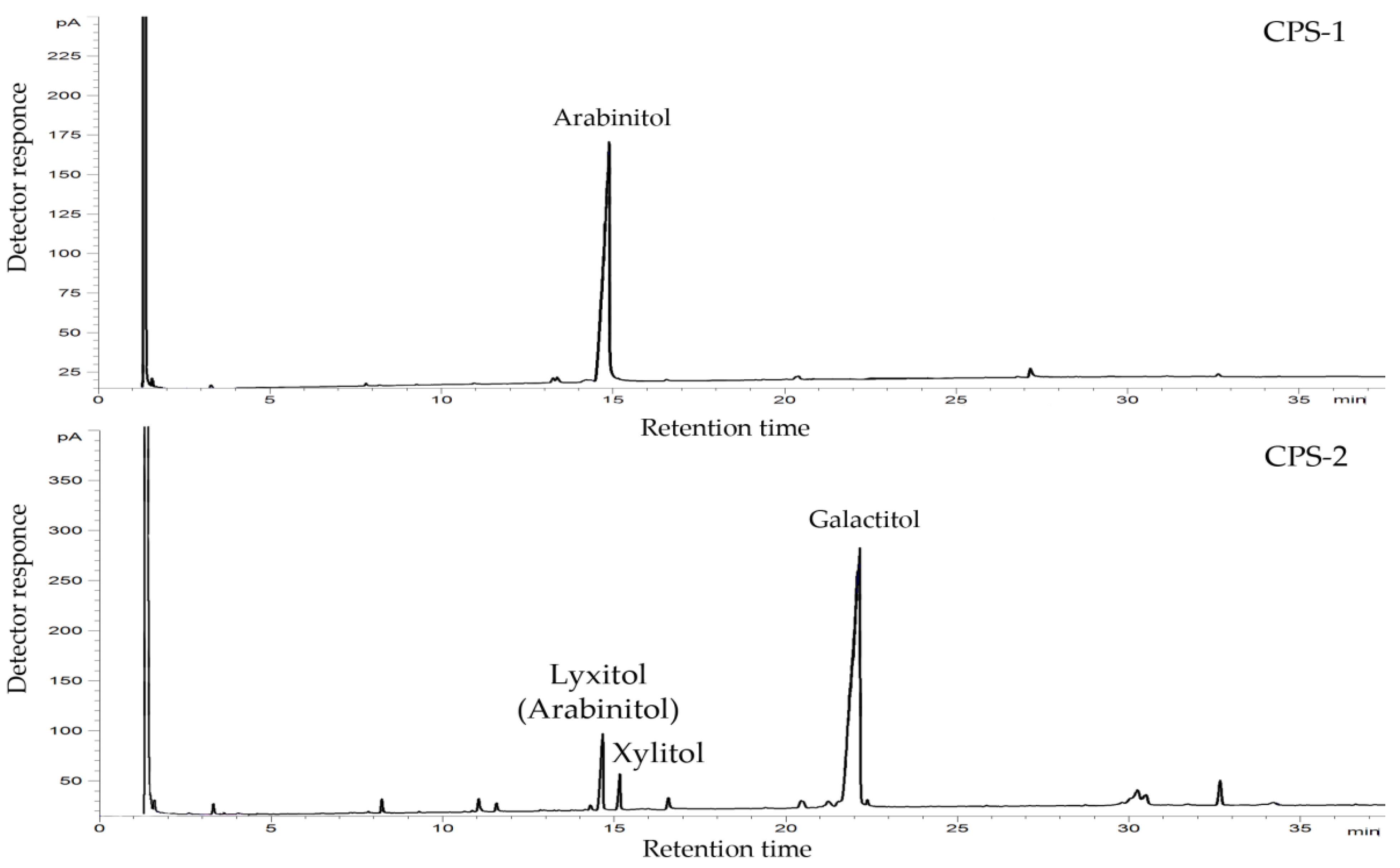
2.1. Isolation, Purification, and General Characterization of CPS
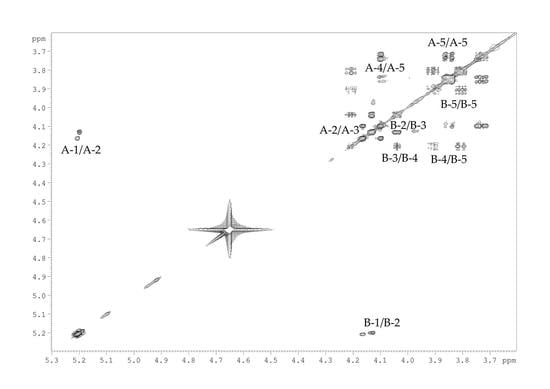
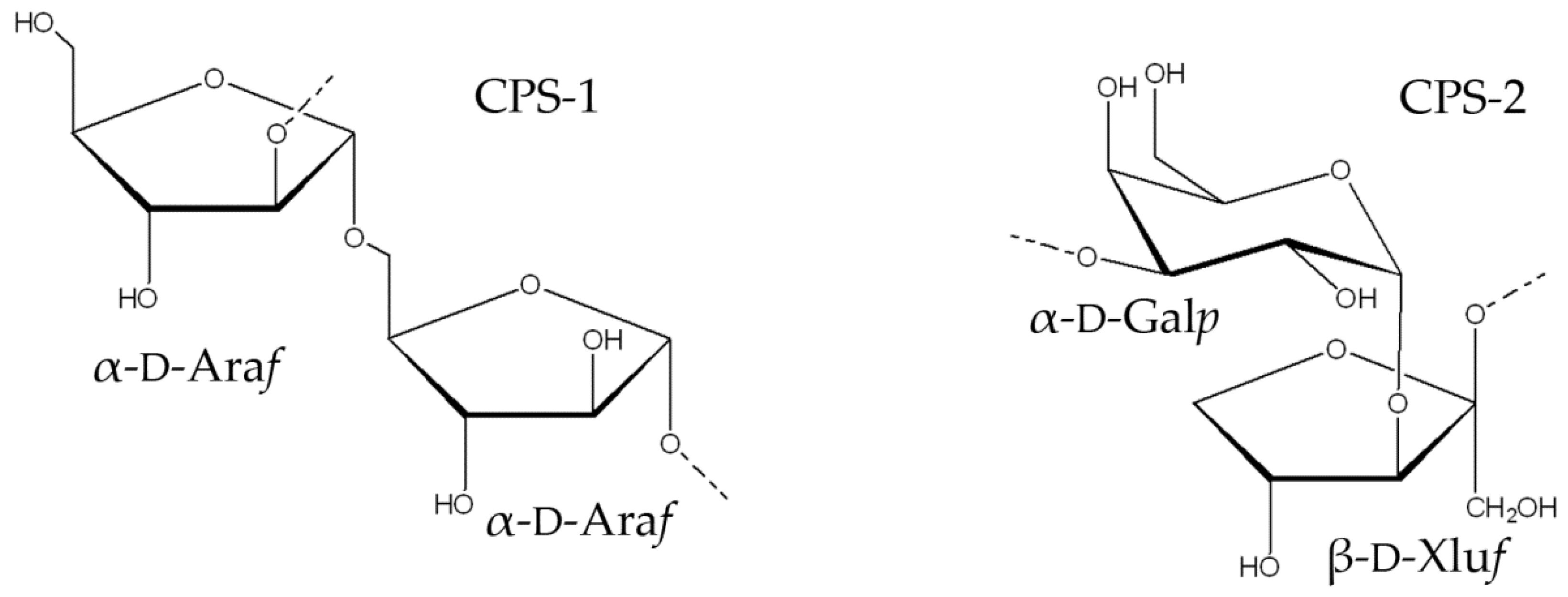
2.2. Structural Study of the CPS-1 Sample
2.3. Structural Study of the CPS-2 Sample
3. Discussion
4. Materials and Methods
4.1. Isolation and Purification of CPSs
4.2. Determination of the Molecular Weight of CPSs
4.3. Compositional Analysis of the CPS Samples
4.4. Mild Acid Hydrolysis of CPS-2
4.5. NMR Spectroscopy
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, Z.; Wu, Y.; Zhang, X.-H. Cultivation of microbes from the deep-sea environments. Deep Sea Res. Part II Top. Stud. Oceanogr. 2018, 155, 34–43. [Google Scholar] [CrossRef]
- Casillo, A.; Lanzetta, R.; Parrilli, M.; Corsaro, M.M. Exopolysaccharides from marine and marine extremophilic bacteria: Structures, properties, ecological roles and applications. Mar. Drugs 2018, 16, 69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.-N.; Meng, L.-H.; Wang, B.-G. Progress in Research on Bioactive Secondary Metabolites from Deep-Sea Derived Microorganisms. Mar. Drugs 2020, 18, 614. [Google Scholar] [CrossRef]
- Herget, S.; Toukach, P.V.; Ranzinger, R.; Hull, W.E.; Knirel, Y.A.; Von der Lieth, C.-W. Statistical analysis of the Bacterial Carbohydrate Structure Data Base (BCSDB): Characteristics and diversity of bacterial carbohydrates in comparison with mammalian glycans. BMC Struct. Biol. 2008, 8, 35. [Google Scholar] [CrossRef] [Green Version]
- Komandrova, N.A.; Kokoulin, M.S.; Kalinovsky, A.I.; Tomshich, S.V.; Romanenko, L.A.; Vaskovsky, V.E. The O-specific polysaccharide of the marine bacterium Rheinheimera pacifica KMM 1406T containing d- and L-2-acetamido-2-deoxy-galacturonic acids. Carbohydr. Res. 2014, 394, 1–6. [Google Scholar] [CrossRef]
- Kokoulin, M.S.; Komandrova, N.A.; Kalinovsky, A.I.; Tomshich, S.V.; Romanenko, L.A.; Vaskovsky, V.E. Structure of the O-specific polysaccharide from the deep-sea marine bacterium Idiomarina abyssalis KMM 227T containing a 2-O-sulfate-3-N-(4-hydroxybutanoyl)-3,6-dideoxy-d-glucose. Carbohydr. Res. 2015, 413, 100–106. [Google Scholar] [CrossRef] [PubMed]
- Kokoulin, M.S.; Sokolova, E.V.; Elkin, Y.N.; Romanenko, L.A.; Mikhailov, V.V.; Komandrova, N.A. Partial structure and immunological properties of lipopolysaccharide from marine-derived Pseudomonas stutzeri KMM 226. Antonie Van Leeuwenhoek 2017, 110, 1569–1580. [Google Scholar] [CrossRef] [PubMed]
- Kokoulin, M.S.; Kuzmich, A.S.; Romanenko, L.A.; Chikalovets, I.V. Structure and in vitro antiproliferative activity of the acidic capsular polysaccharide from the deep-sea bacterium Psychrobacter submarinus KMM 225T. Carbohydr. Polym. 2021, 262, 117941. [Google Scholar] [CrossRef]
- Romanenko, L.A.; Tanaka, N.; Svetashev, V.I. Devosia submarina sp. nov., isolated from deep-sea surface sediments. Int. J. Syst. Evol. Micr. 2013, 63, 3079–3085. [Google Scholar] [CrossRef] [Green Version]
- Bock, K.; Pedersen, C. Carbon-13 nuclear magnetic resonance spectroscopy of monosaccharides. Adv. Carbohydr. Chem. Biochem. 1983, 41, 27–66. [Google Scholar]
- Gilbert, M.; Mandrell, R.E.; Parker, C.T.; Li, J.; Vinogradov, E. Structural analysis of the capsular polysaccharide from Campylobacter jejuni RM1221. ChemBioChem 2007, 8, 625–631. [Google Scholar] [CrossRef]
- Klyne, J. The Configuration of the Anomeric Carbon Atoms in some Cardiac Glycosides. Biochem. J. 1950, 47, xli. [Google Scholar] [PubMed]
- Poli, A.; Anzelmo, G.; Nicolaus, B. Bacterial exopolysaccharides from extreme marine habitats: Production, characterization and biological activities. Mar. Drugs 2010, 8, 1779–1802. [Google Scholar] [CrossRef]
- Toukach, P.V.; Egorova, K.S. Carbohydrate structure database merged from bacterial, archaeal, plant and fungal parts. Nucleic Acids Res. 2016, 44, D1229–D1236. [Google Scholar] [CrossRef] [PubMed]
- Leelayuwapan, H.; Ruchirawat, S.; Boonyarattanakalin, S. Rapid synthesis and immunogenicity of mycobacterial (1→5)-α-d-arabinofuranan. Carbohydr. Polym. 2019, 206, 262–267. [Google Scholar] [CrossRef]
- Zhang, S.; Li, Z.; Wang, X.; An, L.; Bao, J.; Zhang, J.; Cui, J.; Li, Y.; Jin, D.-Q.; Tuerhong, M.; et al. Isolation, structural elucidation, and immunoregulation properties of an arabinofuranan from the rinds of Garcinia mangostana. Carbohydr. Polym. 2020, 246, 116567. [Google Scholar] [CrossRef]
- Wang, H.; Wang, X.; Li, Y.; Zhang, S.; Li, Z.; Li, Y.; Cui, J.; Lan, X.; Zhang, E.; Yuan, L.; et al. Structural properties and in vitro and in vivo immunomodulatory activity of an arabinofuranan from the fruits of Akebia quinata. Carbohydr. Polym. 2021, 256, 117521. [Google Scholar] [CrossRef] [PubMed]
- Perepelov, A.V.; Senchenkova, S.N.; Kalinchuk, N.A.; Shashkov, A.S.; Knirel, Y.A. Structure of O-polysaccharide of Escherichia coli O95: A disaccharide repeating unit containing d-fucose and d-threo-pent-2-ulose (xylulose). Russ. Chem. Bull. 2018, 67, 1931–1933. [Google Scholar] [CrossRef]
- Gorshkova, R.P.; Isakov, V.V.; Kalmykova, E.N.; Ovodov, Y.S. Structural studies of O-specific polysaccharide chains of the lipopolysaccharide from Yersinia enterocolitica serovar O:10. Carbohydr. Res. 1995, 268, 249–255. [Google Scholar] [CrossRef]
- Sato, N.; Nakazawa, F.; Ito, T.; Hoshino, T.; Hoshino, E. The structure of the antigenic polysaccharide produced by Eubacterium saburreum T15. Carbohydr. Res. 2003, 338, 923–930. [Google Scholar] [CrossRef]
- Arena, A.; Gugliandolo, C.; Stassi, G.; Pavone, B.; Iannello, D.; Bisignano, G.; Maugeri, T.L. An exopolysaccharide produced by Geobacillus thermodenitrificans strain B3-72: Antiviral activity on immunocompetent cells. Immunol. Lett. 2009, 123, 132–137. [Google Scholar] [CrossRef] [PubMed]
- Arena, A.; Maugeri, T.L.; Pavone, B.; Iannello, D.; Gugliandolo, C.; Bisignano, G. Antiviral and immunoregulatory effect of a novel exopolysaccharide from a marine thermotolerant Bacillus licheniformis. Int. Immunopharmacol. 2006, 6, 8–13. [Google Scholar] [CrossRef]
- Kambourova, M.; Mandeva, R.; Dimova, D.; Poli, A.; Nicolaus, B.; Tommonaro, G. Production and characterization of a microbial glucan, synthesized by Geobacillus tepidamans V264 isolated from Bulgarian hot spring. Carbohydr. Polym. 2009, 77, 338–343. [Google Scholar] [CrossRef]
- Cox, A.D.; Taylor, C.J.; Anderson, A.J.; Perry, M.B.; Wilkinson, S.G. Structures of the two Polymers Present in the Lipopolysaccharide of Burkholderia (Pseudomonas) Cepacia Serogroup O4. Eur. J. Biochem. 1995, 231, 784–789. [Google Scholar] [CrossRef]
- Cerantola, S.; Montrozier, H. Structural elucidation of two polysaccharides present in the lipopolysaccharide of a clinical isolate of Burkholderia cepacia. Eur. J. Biochem. 1997, 246, 360–366. [Google Scholar] [CrossRef] [Green Version]
- Lipinski, T.; Zatonsky, G.V.; Kocharova, N.A.; Jaquinod, M.; Forest, E.; Shashkov, A.M.; Gamian, A.; Knirel, Y.A. Structures of two O-chain polysaccharides of Citrobacter gillenii O9a,9b lipopolysaccharide: A new homopolymer of 4-amino-4,6-dideoxy-d-mannose (perosamine) Eur. J. Biochem. 2002, 269, 93–99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Castro, C.; Molinaro, A.; Wallace, A.; Grant, W.D.; Forest, E.; Parrilli, M. The O-specific chain structure of the major component from the lipopolysaccharide fraction of Halomonas magadii strain 21 MI (NCIMB 13595). Eur. J. Org. Chem. 2003, 6, 1029–1034. [Google Scholar] [CrossRef]
- De Castro, C.; Carannante, A.; Lanzetta, R.; Nunziata, R.; Piscopo, V.; Parrilli, M. Elucidation of two O-chain structures from the lipopolysaccharide fraction of Agrobacterium tumefaciens F/1. Carbohydr. Res. 2004, 339, 2451–2455. [Google Scholar] [CrossRef] [PubMed]
- Ovchinnikova, O.G.; Kocharova, N.A.; Katzenellenbogen, E.; Zatonsky, G.V.; Shashkov, A.M.; Knirel, Y.A.; Lipinski, T.; Gamian, A. Structures of two O-polysaccharides of the lipopolysaccharide of Citrobacter youngae PCM 1538 (serogroup O9). Carbohydr. Res. 2004, 339, 881–884. [Google Scholar] [CrossRef]
- Zdorovenko, E.L.; Varbanets, L.D.; Zatonsky, G.V.; Ostapchuk, A.N. Structures of two putative O-specific polysaccharides from the Rahnella aquatilis 3-95 lipopolysaccharide. Carbohydr. Res. 2006, 341, 164–168. [Google Scholar] [CrossRef] [PubMed]
- Fernandez de Cordoba, F.J.; Rodriguez-Carvajal, M.A.; Tejero-Mateo, P.; Corzo, J.; Gil-Serrano, A.M. Structure of the O-Antigen of the Main Lipopolysaccharide Isolated from Sinorhizobium fredii SMH12. Biomacromolecules 2008, 9, 678–685. [Google Scholar] [CrossRef] [PubMed]
- Zdorovenko, E.L.; Varbanets, L.D.; Zatonsky, G.V.; Zdorovenko, G.M.; Shashkov, A.M.; Knirel, Y.A. Isolation and structure elucidation of two different polysaccharides from the lipopolysaccharide of Rahnella aquatilis 33071T. Carbohydr. Res. 2009, 344, 1259–1262. [Google Scholar] [CrossRef] [PubMed]
- Ierano, T.; Silipo, A.; Cescutti, P.; Leone, M.R.; Rizzo, R.; Lanzetta, R.; Parrilli, M.; Molinaro, A. Structural Study and Conformational Behavior of the Two Different Lipopolysaccharide O-Antigens Produced by the Cystic Fibrosis Pathogen Burkholderia multivorans. Chem. Eur. J. 2009, 15, 7156–7166. [Google Scholar] [CrossRef] [PubMed]
- Komandrova, N.A.; Isakov, V.V.; Tomshich, S.V.; Romanenko, L.A.; Perepelov, A.V.; Shashkov, A.S. Structure of an acidic O-specific polysaccharide of the marine bacterium Pseudoalteromonas agarivorans KMM 232 (R-form). Biochem. Mosc. 2010, 75, 623–628. [Google Scholar] [CrossRef] [PubMed]
- Silipo, A.; Leone, M.R.; Lanzetta, R.; Parrilli, M.; Lackner, G.; Busch, B.; Hertweck, C.; Molinaro, A. Structural characterization of two lipopolysaccharide O-antigens produced by the endofungal bacterium Burkholderia sp. HKI-402 (B4). Carbohydr. Res. 2012, 347, 95–98. [Google Scholar] [CrossRef]
- Sigida, E.N.; Fedonenko, Y.P.; Shashkov, A.S.; Zdorovenko, E.L.; Konnova, S.A.; Ignatov, V.V.; Knirel, Y.A. Structure of the polysaccharides from the lipopolysaccharide of Azospirillum brasilense Jm125A2. Carbohydr. Res. 2015, 416, 37–40. [Google Scholar] [CrossRef]
- Kokoulin, M.S.; Tomshich, S.V.; Kalinovsky, A.I.; Romanenko, L.A.; Komandrova, N.A. Structure of polysaccharide moiety of Pseudomonas xanthomarina KMM 1447T lipopolysaccharide. Carbohydr. Res. 2016, 434, 6–11. [Google Scholar] [CrossRef]
- Kokoulin, M.S.; Lizanov, I.N.; Romanenko, L.A.; Chikalovets, I.V. Structure of phosphorylated and sulfated polysaccharides from lipopolysaccharide of marine bacterium Marinicella litoralis KMM 3900T. Carbohydr. Res. 2020, 490, 107961. [Google Scholar] [CrossRef]
- Ren, K.; Li, Y.; Shi, F.; Wang, X. Separation of lipopolysaccharides containing different fatty acid chains using hydrophobic interaction chromatography. Anal. Methods 2012, 4, 838. [Google Scholar] [CrossRef]
- Leontein, K.; Lindberg, B.; Lönngren, J. Assignment of absolute configuration of sugars by g.l.c. of their acetylated glycosides formed from chiral alcohols. Carb. Res. 1978, 62, 359–362. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
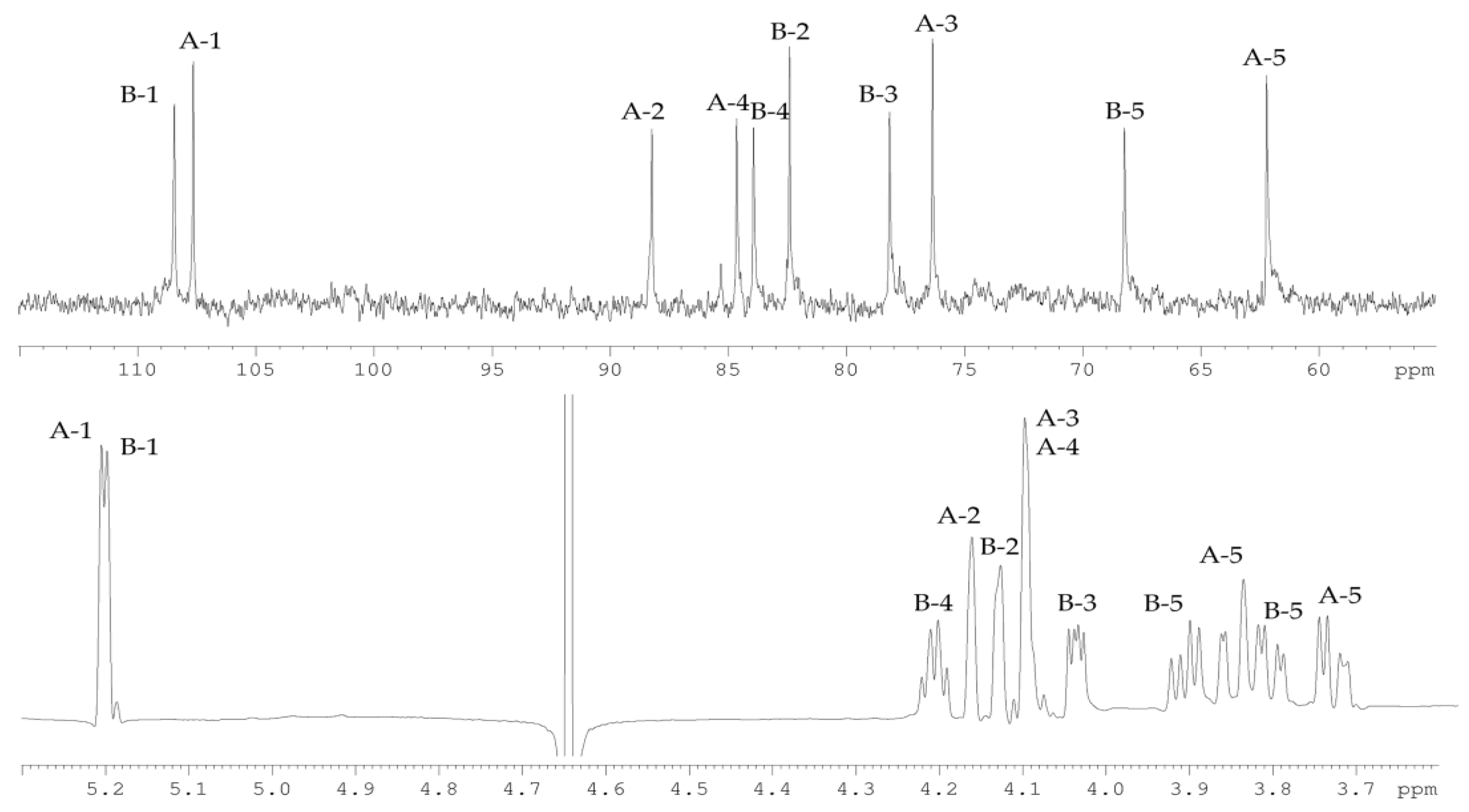

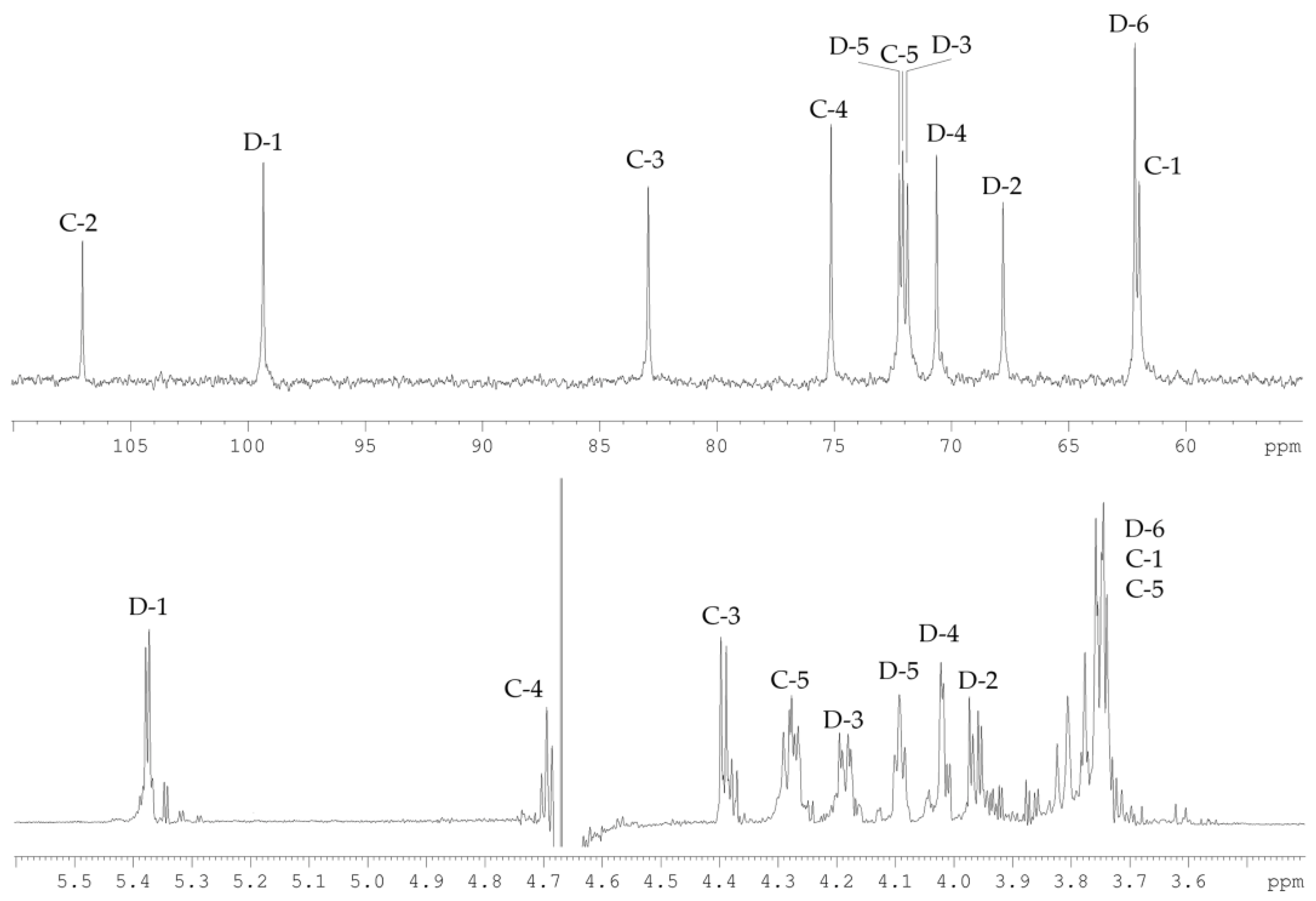
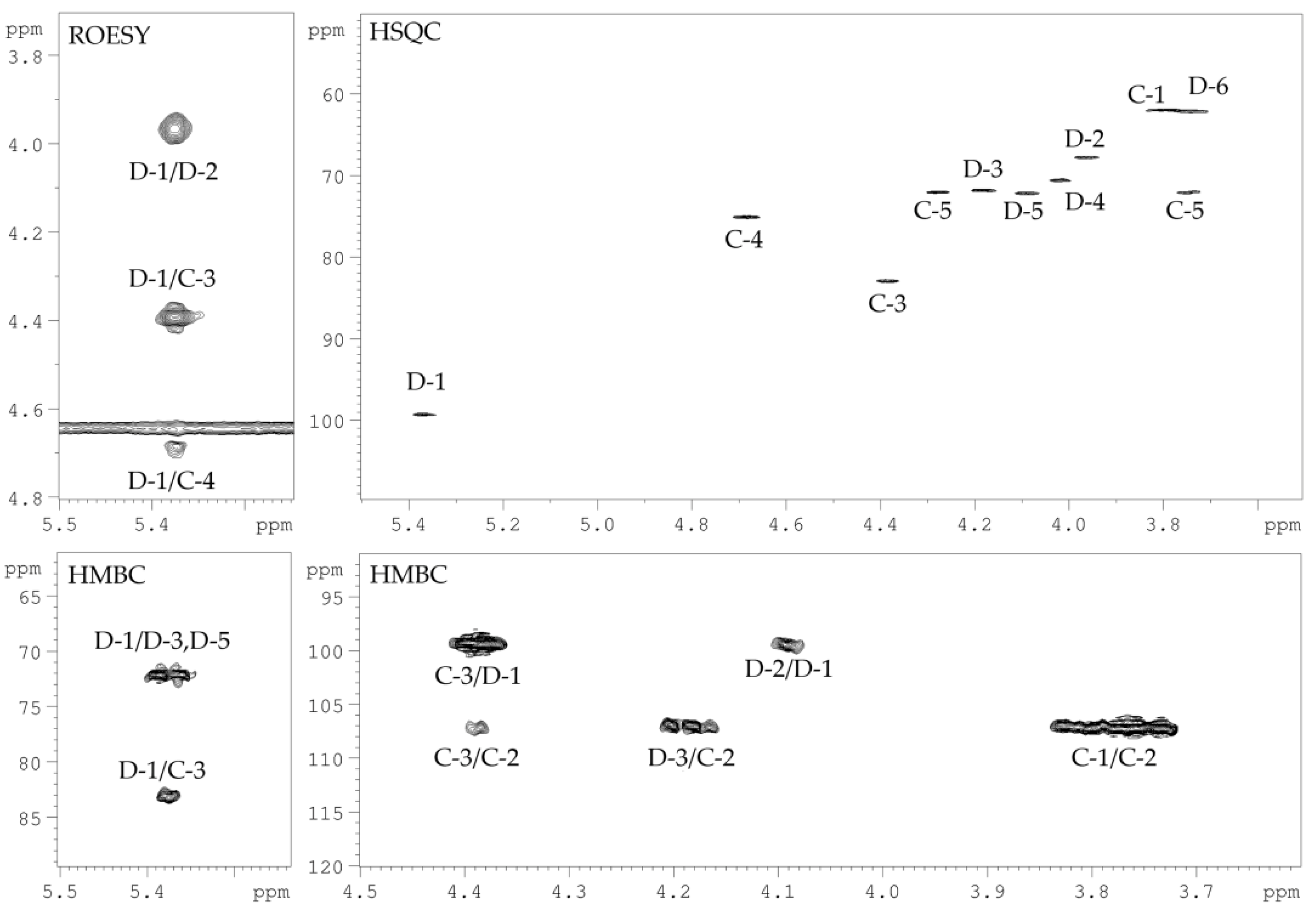
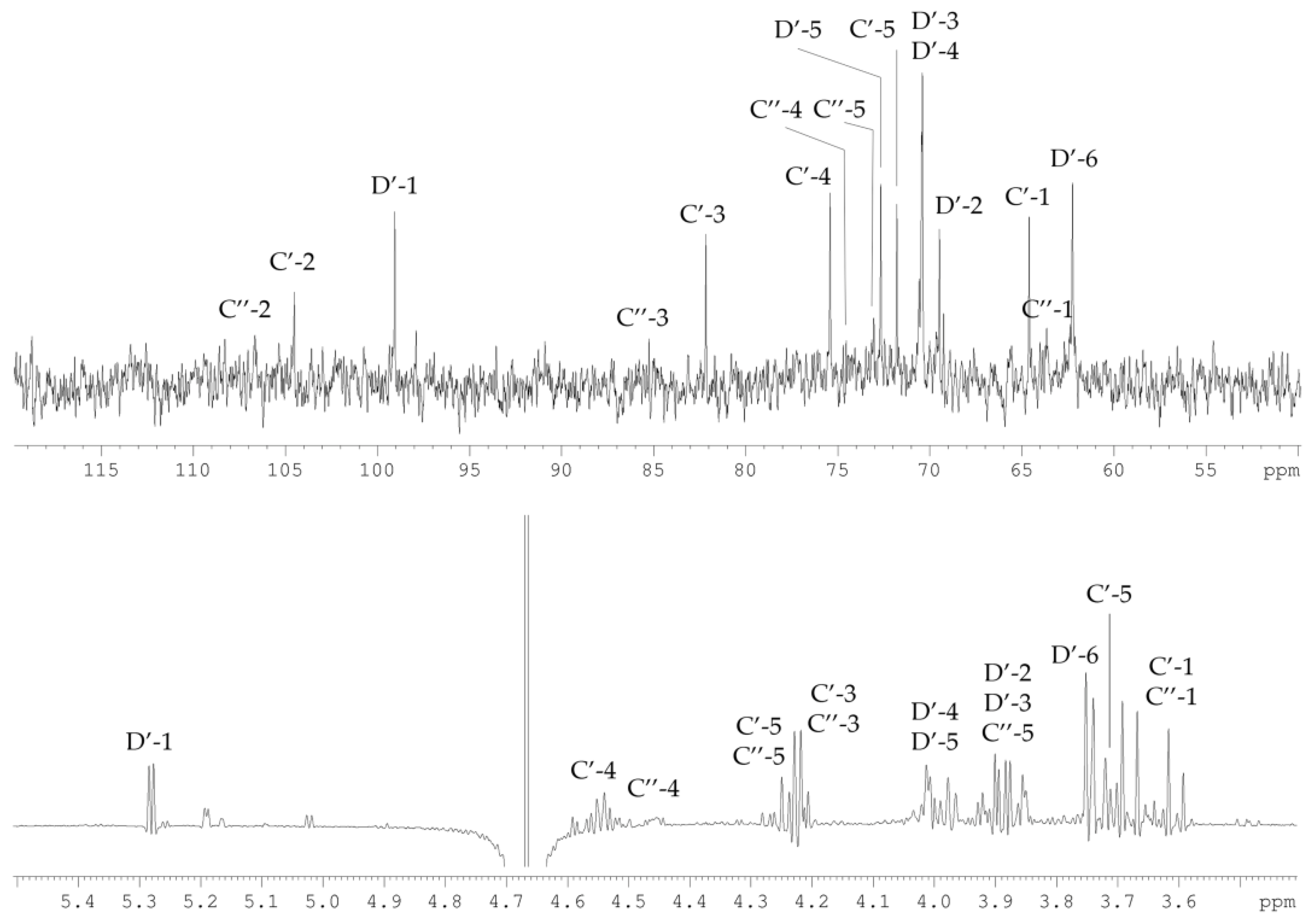
| Sugar Residue | H-1 | H-2 | H-3 | H-4 | H-5 |
|---|---|---|---|---|---|
| C-1 | C-2 | C-3 | C-4 | C-5 | |
| →2)-α-d-Araf-(1→ | 5.20 | 4.16 | 4.10 | 4.10 | 3.85, 3.73 |
| A | 107.6 | 88.2 | 76.3 | 84.6 | 62.2 |
| →5)-α-d-Araf-(1→ | 5.19 | 4.13 | 4.04 | 4.21 | 3.91, 3.80 |
| B | 108.4 | 82.4 | 78.2 | 83.9 | 68.2 |
| Sugar Residue | H-1 | H-2 | H-3 | H-4 | H-5 | H-6 |
|---|---|---|---|---|---|---|
| C-1 | C-2 | C-3 | C-4 | C-5 | C-6 | |
| →3)-β-d-Xluf-(2→ | 3.71, 3.76 | 4.39 | 4.69 | 3.75, 4.28 | ||
| C | 62.0 | 107.0 | 83.0 | 75.2 | 72.1 | |
| →3)-α-d-Galp-(1→ | 5.38 | 3.96 | 4.19 | 4.02 | 4.09 | 3.75, 3.75 |
| D | 99.3 | 67.8 | 71.9 | 70.6 | 72.2 | 62.2 |
| Sugar Residue | H-1 | H-2 | H-3 | H-4 | H-5 | H-6 |
|---|---|---|---|---|---|---|
| C-1 | C-2 | C-3 | C-4 | C-5 | C-6 | |
| α-d-Galp-(1→ | 5.28 | 3.89 | 3.92 | 4.01 | 3.98 | 3.75, 3.75 |
| D’ | 99.1 | 69.5 | 70.4 | 70.4 | 72.6 | 62.2 |
| →3)-β-d-Xluf | 3.60, 3.68 | 4.22 | 4.55 | 3.71, 4.23 | ||
| C’ | 64.6 | 104.5 | 82.1 | 75.4 | 71.8 | |
| →3)-α-d-Xluf | 3.71, 3.71 | 4.21 | 4.46 | 3.91, 4.26 | ||
| C’’ | 63.7 | 106.6 | 85.3 | 74.6 | 73.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kokoulin, M.S.; Romanenko, L.A.; Kuzmich, A.S.; Chernikov, O. Structure of the Cell-Wall-Associated Polysaccharides from the Deep-Sea Marine Bacterium Devosia submarina KMM 9415T. Mar. Drugs 2021, 19, 665. https://doi.org/10.3390/md19120665
Kokoulin MS, Romanenko LA, Kuzmich AS, Chernikov O. Structure of the Cell-Wall-Associated Polysaccharides from the Deep-Sea Marine Bacterium Devosia submarina KMM 9415T. Marine Drugs. 2021; 19(12):665. https://doi.org/10.3390/md19120665
Chicago/Turabian StyleKokoulin, Maxim S., Lyudmila A. Romanenko, Aleksandra S. Kuzmich, and Oleg Chernikov. 2021. "Structure of the Cell-Wall-Associated Polysaccharides from the Deep-Sea Marine Bacterium Devosia submarina KMM 9415T" Marine Drugs 19, no. 12: 665. https://doi.org/10.3390/md19120665
APA StyleKokoulin, M. S., Romanenko, L. A., Kuzmich, A. S., & Chernikov, O. (2021). Structure of the Cell-Wall-Associated Polysaccharides from the Deep-Sea Marine Bacterium Devosia submarina KMM 9415T. Marine Drugs, 19(12), 665. https://doi.org/10.3390/md19120665