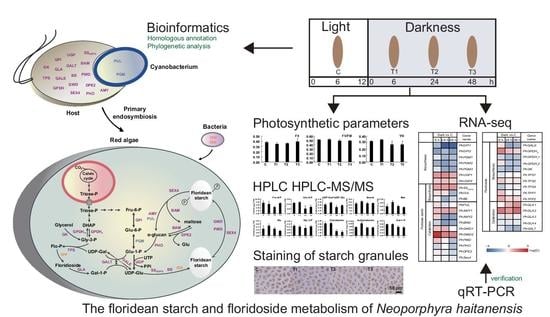
Floridean Starch and Floridoside Metabolic Pathways of Neoporphyra haitanensis and Their Regulatory Mechanism under Continuous Darkness
Abstract
:1. Introduction
2. Results
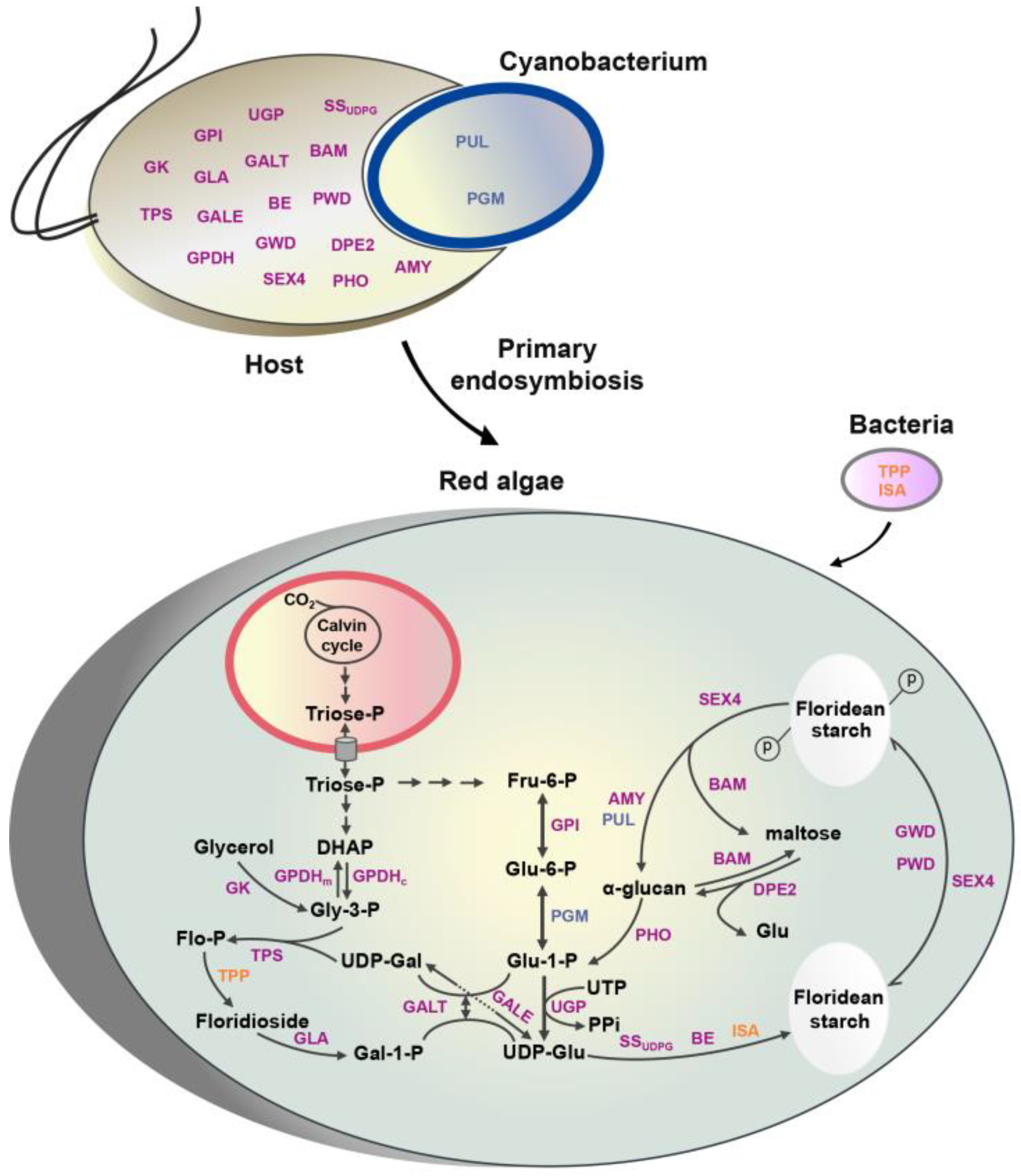
2.1. Identification of Candidate Genes
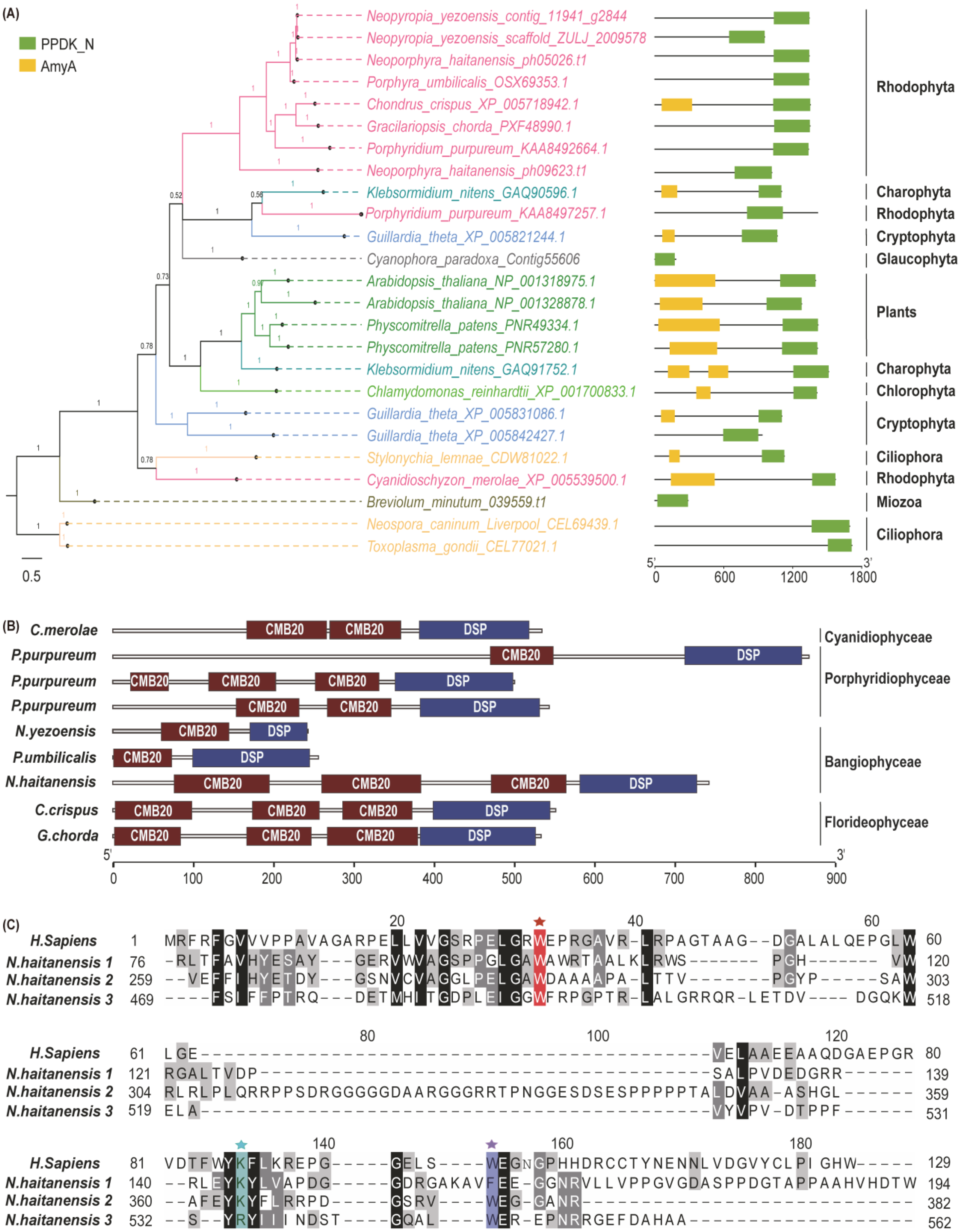
2.2. The Origin of Genes Involved in Floridean Starch and Floridoside Metabolic Pathways of N. haitanensis
2.3. Photochemical Reactions under Dark Conditions
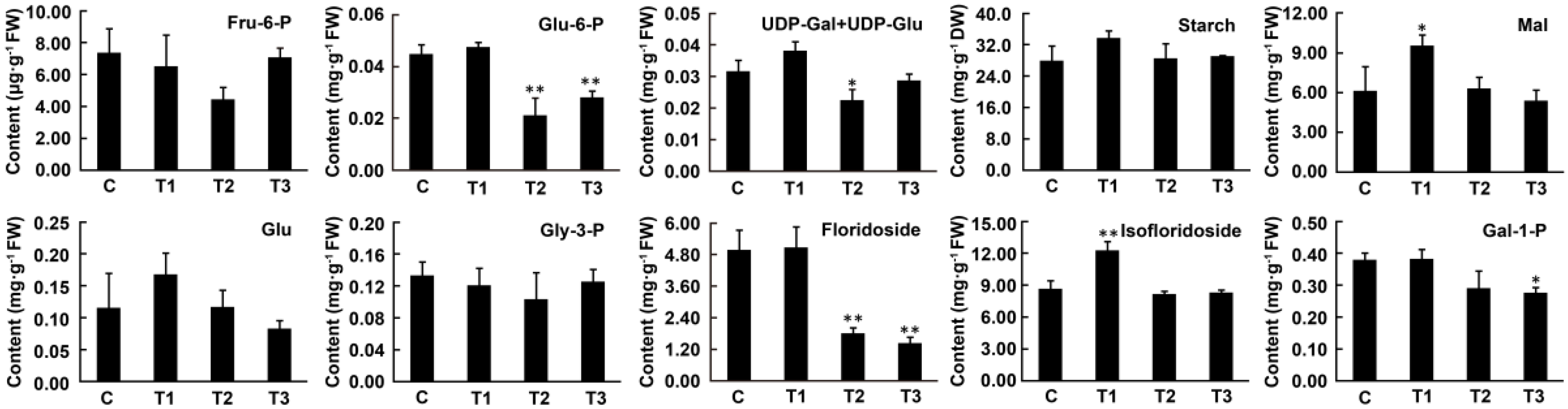
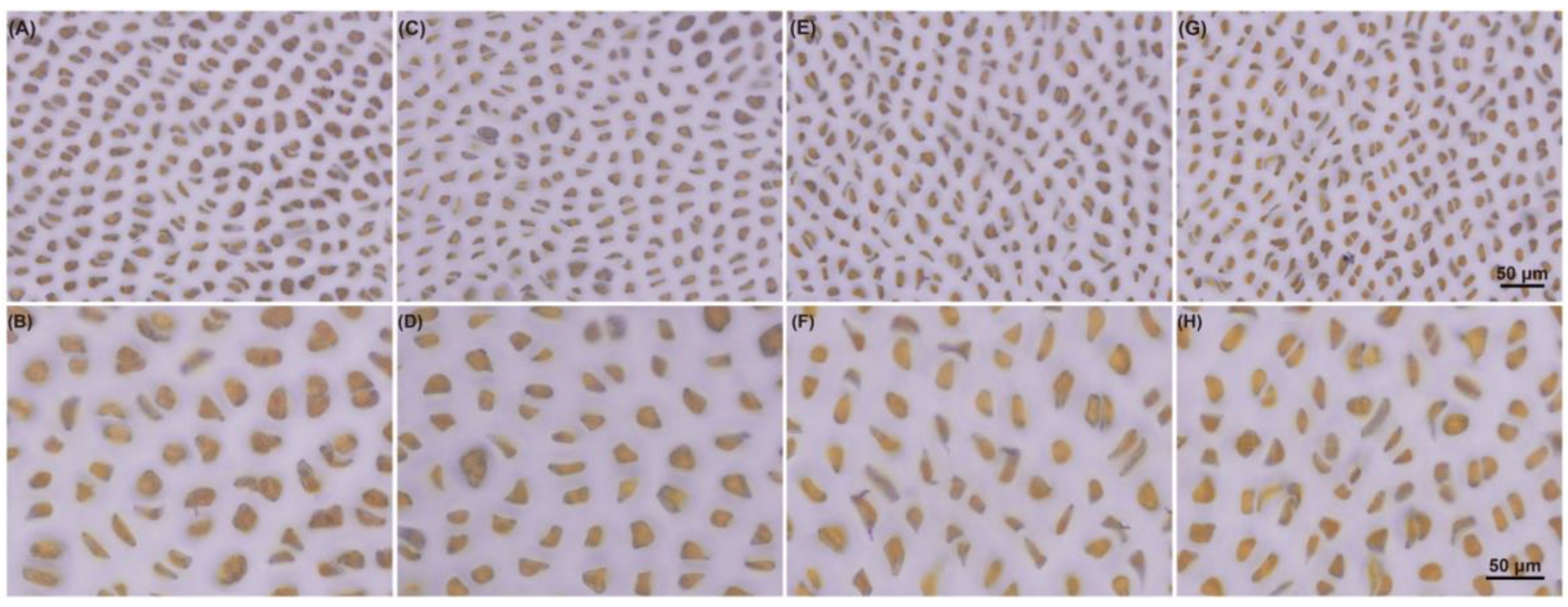
2.4. Changes in Metabolite Contents under Dark Conditions
2.5. Transcriptomic Analysis
2.6. Identification of Differentially Expressed Genes (DEGs) under Dark Conditions
3. Discussion
3.1. The “Floridean Starch–Floridoside” Metabolic Network of N. haitanensis
3.2. Integration of Genes from Different Sources Contributes to the Floridean Starch and Floridoside Metabolic Pathway Evolution in N. haitanensis
3.3. Floridean Starch and Floridoside Content in N. haitanensis Vary with Different Trends under Continuous Darkness
4. Materials and Methods
4.1. Algal Materials and Culture Conditions
4.2. Photochemical Reactions
4.3. Metabolite Detection
4.3.1. Floridean Starch
4.3.2. Floridoside and Isofloridoside
4.3.3. Soluble Sugars (Glucose and Maltose)
4.3.4. Phosphorylated Sugars (Fructose-6-Phosphate, Glucose-6-Phosphate, Glucose-1-Phosphate, and Galactose 1-Phosphate), UDP Sugars (UDP-Galactose and UDP-Glucose), and Glycerol-3-Phosphate
4.4. Staining of Starch Granules
4.5. Gene Identification
4.6. Phylogenetic Analysis
4.7. RNA Extraction, Library Construction, and Illumina Sequencing
4.8. Differential Expression Analysis and Functional Enrichment
4.9. qRT-PCR Analysis
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Blankenship, R.E. Molecular Mechanisms of Photosynthesis, 2nd ed.; Wiley/Blackwell: Chichester, UK, 2014. [Google Scholar]
- Fujiwara, T.; Hirooka, S.; Mukai, M.; Ohbayashi, R.; Kanesaki, Y.; Watanabe, S.; Miyagishima, S. Integration of a Galdieria plasma membrane sugar transporter enables heterotrophic growth of the obligate photoautotrophic red alga Cynanidioschyzon Merolae. Plant Direct 2019, 3, e00134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patron, N.J.; Keeling, P.J. Common evolutionary origin of starch biosynthetic enzymes in green and red algae 1: Evolution of starch synthesis. J. Phycol. 2005, 41, 1131–1141. [Google Scholar] [CrossRef]
- Nagao, M.; Uemura, M. Sucrose phosphate phosphatase in the green alga Klebsormidium flaccidum (Streptophyta) lacks an extensive C-terminal domain and differs from that of land plants. Planta 2012, 235, 851–861. [Google Scholar] [CrossRef]
- Rioux, L.-E.; Turgeon, S.L. Seaweed carbohydrates. In Seaweed Sustainability; Elsevier: Amsterdam, The Netherlands, 2015; pp. 141–192. [Google Scholar]
- Pfister, B.; Zeeman, S.C. Formation of starch in plant cells. Cell. Mol. Life Sci. 2016, 73, 2781–2807. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, A.M. Starch in the Arabidopsis Plant. Starch Stärke 2012, 64, 421–434. [Google Scholar] [CrossRef]
- Sheath, R.G.; Hellebust, J.A.; Sawa, T. Floridean starch metabolism of Porphyridium purpureum (Rhodophyta): III. Effects of darkness and metabolic inhibitors. Phycologia 1981, 20, 22–31. [Google Scholar] [CrossRef]
- Rincones, R.E.; Yu, S.; Pedersn, M. Effect of dark treatment on the starch degradation and the agar quality of cultivated Gracilariopsis lemaneiformis (Rhodophyta, Gracilariales) from Venezuela. Hydrobiologia 1993, 260, 633–640. [Google Scholar] [CrossRef]
- Reed, R.H.; Collins, J.C.; Russell, G. The effects of salinity upon galactosyl-glycerol content and concentration of the marine red alga Porphyra purpurea (Roth) C.Ag. J. Exp. Bot. 1980, 31, 1539–1554. [Google Scholar] [CrossRef]
- Li, S.; Lellouche, J.; Shabtai, Y.; Arad, S. Fixed carbon partitioning in the red microalga Porphyridium sp. (rhodophyta). J. Phycol. 2001, 37, 289–297. [Google Scholar] [CrossRef]
- Olatunji, O. Starch. In Aquatic Biopolymers; Springer: Berlin/Heidelberg, Germany, 2020; pp. 287–310. [Google Scholar]
- Collen, J.; Porcel, B.; Carre, W.; Ball, S.G.; Chaparro, C.; Tonon, T.; Barbeyron, T.; Michel, G.; Noel, B.; Valentin, K.; et al. Genome structure and metabolic features in the red seaweed Chondrus crispus shed light on evolution of the Archaeplastida. Proc. Natl. Acad. Sci. USA 2013, 110, 5247–5252. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharya, D.; Price, D.C.; Chan, C.X.; Qiu, H.; Rose, N.; Ball, S.; Weber, A.P.M.; Cecilia Arias, M.; Henrissat, B.; Coutinho, P.M.; et al. Genome of the red alga Porphyridium Purpureum. Nat. Commun. 2013, 4, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brawley, S.H.; Blouin, N.A.; Ficko-Blean, E.; Wheeler, G.L.; Lohr, M.; Goodson, H.V.; Jenkins, J.W.; Blaby-Haas, C.E.; Helliwell, K.E.; Chan, C.X.; et al. Insights into the red algae and eukaryotic evolution from the genome of Porphyra umbilicalis (Bangiophyceae, Rhodophyta). Proc. Natl. Acad. Sci. USA 2017, 114, E6361–E6370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ho, C.-L.; Lee, W.-K.; Lim, E.-L. Unraveling the nuclear and chloroplast genomes of an agar producing red macroalga, Gracilaria changii (Rhodophyta, Gracilariales). Genomics 2018, 110, 124–133. [Google Scholar] [CrossRef] [PubMed]
- Goulard, F.; Le Corre, G.; Diouris, M.; Deslandes, E.; Floc’h, J.-Y. NDP-sugars, floridoside and floridean starch levels in relation to activities of UDP-glucose pyrophosphorylase and UDP-glucose-4-epimerase in Solieria chordalis (Rhodophyceae) under experimental conditions. Phycological. Res. 2001, 49, 43–50. [Google Scholar] [CrossRef]
- Ishihara, K.; Oyamada, C.; Sato, Y.; Suzuki, T.; Kaneniwa, M.; Kunitake, H.; Muraoka, T. Prebiotic effect of glycerol galactoside isolated from color-faded nori in rats. Fish. Sci. 2010, 76, 1015–1021. [Google Scholar] [CrossRef]
- Bondu, S.; Kervarec, N.; Deslandes, E.; Pichon, R. Separation of floridoside and isofloridosides by HPLC and complete 1H and 13C NMR spectral assignments for D-isofloridoside. Carbohydr. Res. 2007, 342, 2470–2473. [Google Scholar] [CrossRef]
- Pade, N.; Linka, N.; Ruth, W.; Weber, A.P.M.; Hagemann, M. Floridoside and isofloridoside are synthesized by trehalose 6-phosphate synthase-like enzymes in the red alga Galdieria sulphuraria. New Phytol. 2015, 205, 1227–1238. [Google Scholar] [CrossRef]
- Sun, M.; Zhu, Z.; Chen, J.; Yang, R.; Luo, Q.; Wu, W.; Yan, X.; Chen, H. Putative trehalose biosynthesis proteins function as differential floridoside-6-phosphate synthases to participate in the abiotic stress response in the red alga Pyropia haitanensis. BMC Plant Biol. 2019, 19, 325. [Google Scholar] [CrossRef] [Green Version]
- Cao, Y.; Ashline, D.J.; Ficko-Blean, E.; Klein, A.S. Trehalose and (Iso)floridoside production under desiccation stress in red alga Porphyra Umbilicalis and the genes involved in their synthesis. J. Phycol. 2020, 56, 1468–1480. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.-E.; Zhou, W.; Hu, C.-M.; Deng, Y.-Y.; Xu, G.-P.; Zhang, T.; Russell, S.; Zhu, J.-Y.; Lu, Q.-Q.; Brodie, J. A Molecular phylogeny of the bladed bangiales (Rhodophyta) in China provides insights into biodiversity and biogeography of the genus Pyropia. Mol. Phylogenetics Evol. 2018, 120, 94–102. [Google Scholar] [CrossRef]
- Blouin, N.A.; Brodie, J.A.; Grossman, A.C.; Xu, P.; Brawley, S.H. Porphyra: A marine crop shaped by stress. Trends Plant Sci. 2011, 16, 29–37. [Google Scholar] [CrossRef]
- Chen, C.; Weng, L.; Wang, L.; Ji, D.; Xie, C.; Xu, Y. Influence of desiccation and cold preservation on the survive and growth of Porphyra haitanensis and unwanted-alg. Acta Oceanol. Sin. 2010, 29, 131–136. [Google Scholar]
- Suda, M.; Mikami, K. Reproductive responses to wounding and heat stress in gametophytic thalli of the red alga Pyropia yezoensis. Front. Mar. Sci. 2020, 7, 394. [Google Scholar] [CrossRef]
- Janeček, Š.; Mareček, F.; MacGregor, E.A.; Svensson, B. Starch-binding domains as CBM families–history, occurrence, structure, function and evolution. Biotechnol. Adv. 2019, 37, 107451. [Google Scholar] [CrossRef]
- Linka, M.; Jamai, A.; Weber, A.P.M. Functional characterization of the plastidic phosphate translocator gene family from the thermo-acidophilic red alga Galdieria sulphuraria reveals specific adaptations of primary carbon partitioning in green plants and red algae. Plant Physiol. 2008, 148, 1487–1496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deschamps, P.; Haferkamp, I.; d’Hulst, C.; Neuhaus, H.E.; Ball, S.G. The relocation of starch metabolism to chloroplasts: When, why and how. Trends Plant Sci. 2008, 13, 574–582. [Google Scholar] [CrossRef] [PubMed]
- Siow, R.-S.; Teo, S.-S.; Ho, W.-Y.; Shukor, M.Y.A.; Phang, S.-M.; Ho, C.-L. Molecular cloning and biochemical characterization of galactose-1-phosphate uridylyltransferase from Gracilaria changii (rhodophyta). J. Phycol. 2012, 48, 155–162. [Google Scholar] [CrossRef]
- Li, S.-Y.; Shabtai, Y.; Arad, S. Floridoside as a carbon precursor for the synthesis of cell-wall polysaccharide in the red microalga Porphyridium sp. (rhodophyta) 1. J. Phycol. 2002, 38, 931–938. [Google Scholar] [CrossRef]
- Chi, S.; Liu, T.; Wang, X.; Wang, R.; Wang, S.; Wang, G.; Shan, G.; Liu, C. Functional genomics analysis reveals the biosynthesis pathways of important cellular components (Alginate and Fucoidan) of Saccharina. Curr. Genet. 2018, 64, 259–273. [Google Scholar] [CrossRef]
- Jourda, C.; Cardi, C.; Gibert, O.; Giraldo Toro, A.; Ricci, J.; Mbéguié-A-Mbéguié, D.; Yahiaoui, N. Lineage-specific evolutionary histories and regulation of major starch metabolism genes during banana ripening. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [Green Version]
- Yue, C.; Cao, H.; Lin, H.; Hu, J.; Ye, Y.; Li, J.; Hao, Z.; Hao, X.; Sun, Y.; Yang, Y.; et al. Expression patterns of alpha-amylase and beta-amylase genes provide insights into the molecular mechanisms underlying the responses of tea plants (Camellia sinensis) to stress and postharvest processing treatments. Planta 2019, 250, 281–298. [Google Scholar] [CrossRef]
- Hittinger, C.T.; Carroll, S.B. Gene duplication and the adaptive evolution of a classic genetic switch. Nature 2007, 449, 677–681. [Google Scholar] [CrossRef]
- Mikkelsen, R.; Blennow, A. Functional domain organization of the potato α-glucan, water dikinase (GWD): Evidence for separate site catalysis as revealed by limited proteolysis and deletion mutants. Biochem. J. 2005, 385, 355–361. [Google Scholar] [CrossRef] [Green Version]
- Kuchtová, A.; Gentry, M.S.; Janeček, Š. The unique evolution of the carbohydrate-binding module CBM 20 in laforin. FEBS Lett. 2018, 592, 586–598. [Google Scholar] [CrossRef] [Green Version]
- Chi, S.; Wang, G.; Liu, T.; Wang, X.; Liu, C.; Jin, Y.; Yin, H.; Xu, X.; Yu, J. Transcriptomic and proteomic analysis of mannitol-metabolism-associated genes in Saccharina japonica. Genom. Proteom. Bioinform. 2020, 18, 415–429. [Google Scholar] [CrossRef] [PubMed]
- Chia, T.; Thorneycroft, D.; Chapple, A.; Messerli, G.; Chen, J.; Zeeman, S.C.; Smith, S.M.; Smith, A.M. A cytosolic glucosyltransferase is required for conversion of starch to sucrose in Arabidopsis leaves at night. Plant J. 2004, 37, 853–863. [Google Scholar] [CrossRef] [PubMed]
- Ceusters, N.; Frans, M.; Van den Ende, W.; Ceusters, J. Maltose processing and not β-amylase activity curtails hydrolytic starch degradation in the CAM orchid Phalaenopsis. Front. Plant Sci. 2019, 10, 1386. [Google Scholar] [CrossRef]
- Nagashima, H.; Ozaki, H.; Nakamura, S.; Nisizawa, K. Physiological studies on floridean starch, floridoside and trehalose in a red alga, Serraticardia maxima. Shokubutsugaku Zasshi 1969, 82, 462–473. [Google Scholar] [CrossRef] [Green Version]
- Kitajima, M.; Butler, W.L. Quenching of chlorophyll fluorescence and primary photochemistry in chloroplasts by dibromothymoquinone. Biochim. Biophys. Acta Bioenerg. 1975, 376, 105–115. [Google Scholar] [CrossRef]
- Genty, B.; Briantais, J.-M.; Baker, N.R. The relationship between the quantum yield of photosynthetic electron transport and quenching of chlorophyll fluorescence. Biochim. Biophys. Acta Gen. Subj. 1989, 990, 87–92. [Google Scholar] [CrossRef]
- McCready, R.M.; Guggolz, J.; Silviera, V.; Owens, H.S. Determination of starch and amylose in vegetables. Anal. Chem. 1950, 22, 1156–1158. [Google Scholar] [CrossRef]
- Huan, L.; Xie, X.; Zheng, Z.; Sun, F.; Wu, S.; Li, M.; Gao, S.; Gu, W.; Wang, G. Positive correlation between PSI response and oxidative pentose phosphate pathway activity during salt stress in an intertidal macroalga. Plant Cell Physiol. 2014, 55, 1395–1403. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Okonechnikov, K.; Conesa, A.; García-Alcalde, F. Qualimap 2: Advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics 2016, 32, 292–294. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Chen, C.; Xu, Y.; Ji, D.; Xie, C. Validation of housekeeping genes as internal controls for studying the gene expression in Pyropia haitanensis (Bangiales, Rhodophyta) by quantitative real-time PCR. Acta Oceanol. Sin. 2014, 33, 152–159. [Google Scholar] [CrossRef]



| Gene Name | Gene ID | Amino Acids | Open Reading Frame (bp) | Molecular Weight (kDa) | Isoelectric Point | Number of Introns | Instability Index |
|---|---|---|---|---|---|---|---|
| PhGPI1 | ph06855.t1 | 559 | 1680 | 59.63993 | 5.86 | 1 | 28.18 |
| PhGPI2 | ph10696.t1 | 638 | 1917 | 70.23072 | 5.81 | 0 | 36.16 |
| PhPGM1 | ph07345.t1 | 524 | 1575 | 53.45336 | 4.64 | 2 | 31.59 |
| PhPGM2 | ph08542.t1 | 430 | 1293 | 43.42822 | 5.43 | 1 | 31.2 |
| PhPGM3 | ph01308.t1 | 586 | 1761 | 61.88904 | 5.1 | 1 | 27.5 |
| PhUGP1 | ph10255.t1 | 504 | 1515 | 55.60583 | 6.19 | 0 | 28.03 |
| PhUGP2 | ph10249.t1 | 510 | 1533 | 56.19654 | 6.34 | 0 | 27.52 |
| PhSSUDPG | ph07441.t1 | 1767 | 5304 | 189.20083 | 6.04 | 1 | 44.02 |
| PhISA | ph06437.t1 | 786 | 2361 | 84.18759 | 5.64 | 0 | 33.81 |
| PhBE | ph11470.t1 | 755 | 2268 | 85.18558 | 5.92 | 2 | 37.93 |
| PhPUL | ph02603.t1 | 1779 | 5340 | 180.68102 | 5.5 | 1 | 41.86 |
| PhAMY1 | ph05661.t1 | 447 | 1344 | 47.39023 | 8.72 | 2 | 36.3 |
| PhAMY2 | ph09788.t1 | 505 | 1518 | 54.85313 | 5.67 | 1 | 29.27 |
| PhBAM | ph06397.t1 | 458 | 1377 | 49.31007 | 5.57 | 0 | 39.7 |
| PhGWD1 | ph05026.t1 | 1344 | 4035 | 144.2512 | 5.33 | 0 | 39.64 |
| PhGWD2 | ph09623.t1 | 1018 | 3057 | 101.82198 | 6.52 | 2 | 33.75 |
| PhPWD | ph11079.t1 | 1039 | 3120 | 103.82096 | 5.49 | 1 | 36.37 |
| PhPHO | ph09474.t1 | 883 | 2652 | 99.18552 | 6.1 | 1 | 36.27 |
| PhDPE2 | ph04961.t1 | 1014 | 3045 | 111.16318 | 5.12 | 0 | 41.86 |
| PhGALE | ph04371.t1 | 344 | 1035 | 37.82118 | 5.26 | 0 | 36.52 |
| PhSex4 | ph06018.t1 | 739 | 2220 | 75.19834 | 5.45 | 0 | 47.62 |
| PhGPDHm | ph01923.t1 | 711 | 2136 | 75.27303 | 8.58 | 1 | 33.98 |
| PhGPDHc1 | ph00687.t1 | 405 | 1218 | 40.98797 | 7.61 | 1 | 41.81 |
| PhGPDHc2 | ph05255.t1 | 386 | 1161 | 41.85018 | 5.52 | 0 | 27.84 |
| PhGK | ph03288.t1 | 576 | 1731 | 60.3038 | 4.79 | 1 | 34.82 |
| PhTPS1 | ph10158.t1 | 1153 | 3462 | 124.15916 | 6.73 | 0 | 46.18 |
| PhTPS2 | ph10797.t1 | 1505 | 4518 | 159.36145 | 5.93 | 0 | 52.21 |
| PhTPS4 | ph07423.t1 | 1007 | 3024 | 112.57108 | 5.79 | 1 | 34.78 |
| PhTPP1 | ph05156.t1 | 454 | 1365 | 46.84337 | 5.04 | 1 | 39.21 |
| PhTPP2 | ph05159.t1 | 266 | 801 | 28.07624 | 5.31 | 1 | 39.98 |
| PhGLA1 | ph02625.t1 | 1109 | 3330 | 120.26068 | 5.98 | 7 | 47.35 |
| PhGLA2 | ph00656.t1 | 1098 | 3297 | 114.41963 | 6.06 | 1 | 42.92 |
| PhGLA3 | ph08573.t1 | 1098 | 3297 | 119.0251 | 5.83 | 3 | 44.03 |
| PhGLA4 | ph05433.t1 | 542 | 1629 | 53.53138 | 9.4 | 1 | 28.14 |
| PhGALT | ph04880.t1 | 364 | 1095 | 41.01006 | 5.44 | 0 | 38.06 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, Y.; Jia, X.; Wang, W.; Jin, Y.; Liu, W.; Wang, D.; Mao, Y.; Xie, C.; Liu, T. Floridean Starch and Floridoside Metabolic Pathways of Neoporphyra haitanensis and Their Regulatory Mechanism under Continuous Darkness. Mar. Drugs 2021, 19, 664. https://doi.org/10.3390/md19120664
Yu Y, Jia X, Wang W, Jin Y, Liu W, Wang D, Mao Y, Xie C, Liu T. Floridean Starch and Floridoside Metabolic Pathways of Neoporphyra haitanensis and Their Regulatory Mechanism under Continuous Darkness. Marine Drugs. 2021; 19(12):664. https://doi.org/10.3390/md19120664
Chicago/Turabian StyleYu, Yahui, Xuli Jia, Wenlei Wang, Yuemei Jin, Weizhi Liu, Dongmei Wang, Yunxiang Mao, Chaotian Xie, and Tao Liu. 2021. "Floridean Starch and Floridoside Metabolic Pathways of Neoporphyra haitanensis and Their Regulatory Mechanism under Continuous Darkness" Marine Drugs 19, no. 12: 664. https://doi.org/10.3390/md19120664
APA StyleYu, Y., Jia, X., Wang, W., Jin, Y., Liu, W., Wang, D., Mao, Y., Xie, C., & Liu, T. (2021). Floridean Starch and Floridoside Metabolic Pathways of Neoporphyra haitanensis and Their Regulatory Mechanism under Continuous Darkness. Marine Drugs, 19(12), 664. https://doi.org/10.3390/md19120664