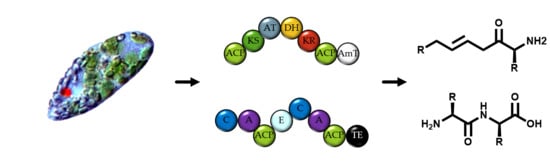
Mining Natural Product Biosynthesis in Eukaryotic Algae
Abstract
1. Introduction
2. Results and Discussion
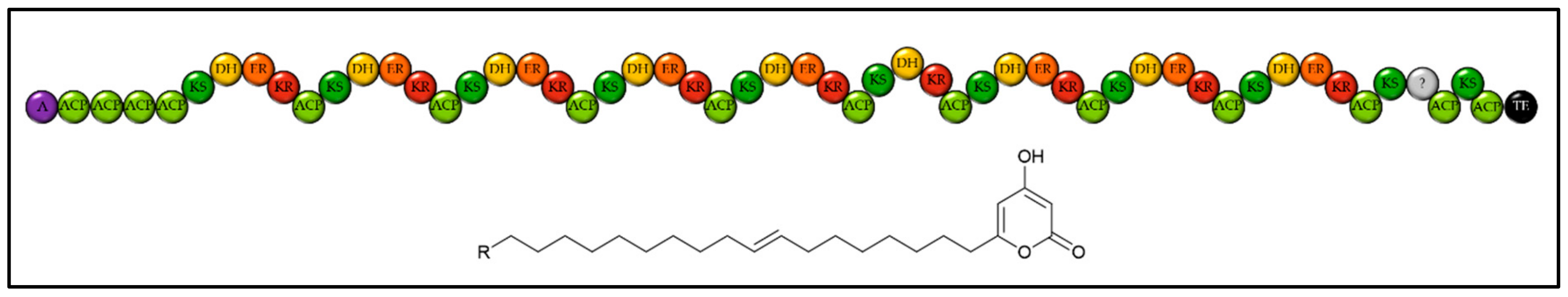
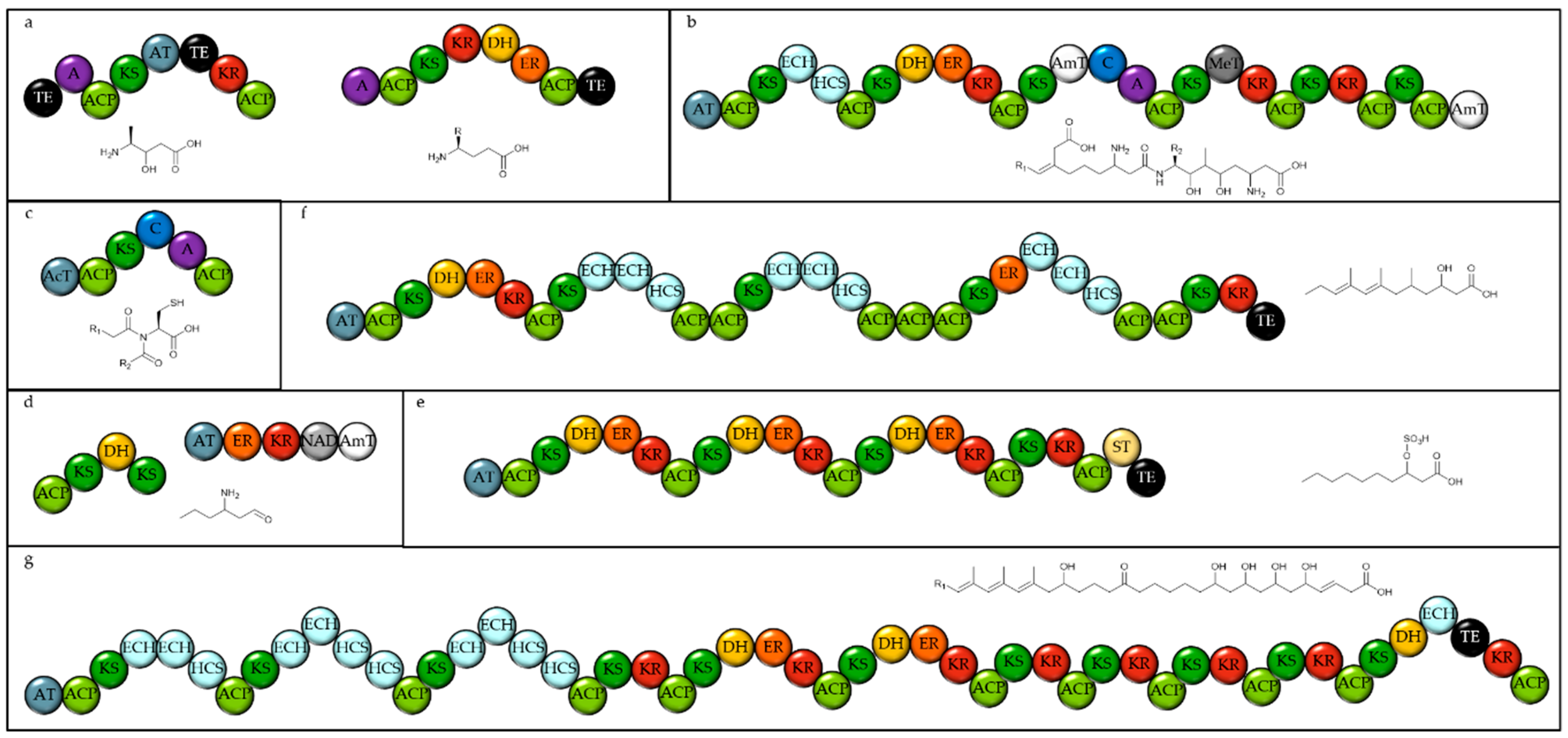
2.1. Identifying Natural Product Biosynthetic Genes in Algal Genomes
2.2. Distribution of Biosynthetic Gene Clusters Amongst Algal Classes
2.2.1. Green Algae (Chlorophytes)
2.2.2. Red Algae (Rhodophytes)
2.2.3. Glaucophytes
2.2.4. Brown Algae (Phaeophytes)
2.2.5. Haptophytes
2.2.6. Diatoms (Bacillariophyta)
2.2.7. Dinoflagellates
2.2.8. Cryptophytes
2.2.9. Chromerids
2.2.10. Chlorarachniophytes
2.2.11. Euglenophytes
3. Conclusions
4. Materials and Methods
Gene Cluster Identification
Funding
Acknowledgments
Conflicts of Interest
References
- Tyrrell, T.; Merico, A. Emiliania huxleyi: Bloom observations and the conditions that induce them. In Coccolithophores: From Molecular Processes to Global Impact; Thierstein, H.R., Young, J.R., Eds.; Springer: Berlin/Heidelberg, Germany, 2004; pp. 75–97. [Google Scholar]
- Brown, E.R.; Cepeda, M.R.; Mascuch, S.J.; Poulson-Ellestad, K.L.; Kubanek, J. Chemical ecology of the marine plankton. Nat. Prod. Res. 2019, 36, 1093–1116. [Google Scholar] [CrossRef] [PubMed]
- Wagstaff, B.A.; Hems, E.S.; Rejzek, M.; Pratscher, J.; Brooks, E.; Kuhaudomlarp, S.; O’Neill, E.C.; Donaldson, M.I.; Lane, S.; Currie, J.; et al. Insights into toxic Prymnesium parvum blooms: The role of sugars and algal viruses. Biochem. Soc. Trans. 2018, 46, 413–421. [Google Scholar] [CrossRef] [PubMed]
- De Clerck, O.; Bogaert, K.A.; Leliaert, F. Chapter Two-Diversity and Evolution of Algae: Primary Endosymbiosis. In Advances in Botanical Research; Piganeau, G., Ed.; Academic Press: Cambridge, MA, USA, 2012; Volume 64, pp. 55–86. [Google Scholar]
- Oborník, M. Endosymbiotic Evolution of Algae, Secondary Heterotrophy and Parasitism. Biomolecules 2019, 9, 266. [Google Scholar]
- Archibald, J.M. Chapter Three-The Evolution of Algae by Secondary and Tertiary Endosymbiosis. In Advances in Botanical Research; Piganeau, G., Ed.; Academic Press: Cambridge, MA, USA, 2012; Volume 64, pp. 87–118. [Google Scholar]
- Larkum, A.W.D.; Lockhart, P.J.; Howe, C.J. Shopping for plastids. Trends Plant Sci. 2007, 12, 189–195. [Google Scholar] [CrossRef]
- Grossman, A.R. In the Grip of Algal Genomics. In Transgenic Microalgae as Green Cell Factories; León, R., Galván, A., Fernández, E., Eds.; Springer: New York, NY, USA, 2007; pp. 54–76. [Google Scholar]
- Blaby-Haas, C.E.; Merchant, S.S. Comparative and functional algal genomics. Annu. Rev. Plant Biol. 2019, 70, 605–638. [Google Scholar] [CrossRef]
- O’Neill, E.C.; Saalbach, G.; Field, R.A. Gene discovery for synthetic biology. Method. Enzymol. 2016, 576, 99–120. [Google Scholar]
- Shelest, E.; Heimerl, N.; Fichtner, M.; Sasso, S. Multimodular type I polyketide synthases in algae evolve by module duplications and displacement of AT domains in trans. BMC Genomics 2015, 16, 1015. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef]
- Kautsar, S.A.; Suarez Duran, H.G.; Blin, K.; Osbourn, A.; Medema, M.H. plantiSMASH: Automated identification, annotation and expression analysis of plant biosynthetic gene clusters. Nucleic Acids Res. 2017, 45, W55–W63. [Google Scholar] [CrossRef]
- Ebenezer, T.E.; Zoltner, M.; Burrell, A.; Nenarokova, A.; Novák Vanclová, A.M.G.; Prasad, B.; Soukal, P.; Santana-Molina, C.; O’Neill, E.; Nankissoor, N.N.; et al. Transcriptome, proteome and draft genome of Euglena gracilis. BMC Biol. 2019, 17, 11. [Google Scholar] [CrossRef]
- O’Neill, E.C.; Trick, M.; Henrissat, B.; Field, R.A. Euglena in time: Evolution, control of central metabolic processes and multi-domain proteins in carbohydrate and natural product biochemistry. Perspect. Sci. 2015, 6, 84–93. [Google Scholar] [CrossRef]
- Geib, E.; Baldeweg, F.; Doerfer, M.; Nett, M.; Brock, M. Cross-Chemistry Leads to Product Diversity from Atromentin Synthetases in Aspergilli from Section Nigri. Cell Chem. Biol. 2019, 26, 223–234e226. [Google Scholar] [CrossRef] [PubMed]
- Heimerl, N.; Hommel, E.; Westermann, M.; Meichsner, D.; Lohr, M.; Hertweck, C.; Grossman, A.R.; Mittag, M.; Sasso, S. A giant type I polyketide synthase participates in zygospore maturation in Chlamydomonas reinhardtii. Plant J. 2018, 95, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Meslet-Cladière, L.; Delage, L.; Leroux, C.J.-J.; Goulitquer, S.; Leblanc, C.; Creis, E.; Gall, E.A.; Stiger-Pouvreau, V.; Czjzek, M.; Potin, P. Structure/Function Analysis of a Type III Polyketide Synthase in the Brown Alga Ectocarpus siliculosus Reveals a Biochemical Pathway in Phlorotannin Monomer Biosynthesis. Plant Cell 2013, 25, 3089–3103. [Google Scholar]
- Campbell, E.L.; Cohen, M.F.; Meeks, J.C. A polyketide-synthase-like gene is involved in the synthesis of heterocyst glycolipids in Nostoc punctiforme strain ATCC 29133. Arch. Microbiol. 1997, 167, 251–258. [Google Scholar] [CrossRef]
- John, U.; Beszteri, B.; Derelle, E.; Van de Peer, Y.; Read, B.; Moreau, H.; Cembella, A. Novel Insights into Evolution of Protistan Polyketide Synthases through Phylogenomic Analysis. Protist 2008, 159, 21–30. [Google Scholar] [CrossRef]
- John, U.; Beszteri, S.; Glöckner, G.; Singh, R.; Medlin, L.; Cembella, A.D. Genomic characterisation of the ichthyotoxic prymnesiophyte Chrysochromulina polylepis, and the expression of polyketide synthase genes in synchronized cultures. Eur. J. Phycol. 2010, 45, 215–229. [Google Scholar] [CrossRef]
- Ziemert, N.; Podell, S.; Penn, K.; Badger, J.H.; Allen, E.; Jensen, P.R. The natural product domain seeker NaPDoS: A phylogeny nased bioinformatic tool to classify secondary metabolite gene diversity. PLoS ONE 2012, 7, e34064. [Google Scholar] [CrossRef]
- Hovde, B.T.; Deodato, C.R.; Hunsperger, H.M.; Ryken, S.A.; Yost, W.; Jha, R.K.; Patterson, J.; Monnat, R.J., Jr.; Barlow, S.B.; Starkenburg, S.R.; et al. Genome sequence and transcriptome analyses of Chrysochromulina tobin: Metabolic tools for enhanced algal fitness in the prominent order Prymnesiales (Haptophyceae). PLoS Genet. 2015, 11, e1005469. [Google Scholar] [CrossRef]
- Butcher, R.A.; Schroeder, F.C.; Fischbach, M.A.; Straight, P.D.; Kolter, R.; Walsh, C.T.; Clardy, J. The identification of bacillaene, the product of the PksX megacomplex in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 2007, 104, 1506–1509. [Google Scholar] [CrossRef]
- Bowler, C.; Allen, A.E.; Badger, J.H.; Grimwood, J.; Jabbari, K.; Kuo, A.; Maheswari, U.; Martens, C.; Maumus, F.; Otillar, R.P.; et al. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 2008, 456, 239–244. [Google Scholar] [CrossRef] [PubMed]
- Kellmann, R.; Stüken, A.; Orr, R.J.; Svendsen, H.M.; Jakobsen, K.S. Biosynthesis and molecular genetics of polyketides in marine dinoflagellates. Mar. Drugs 2010, 8, 1011–1048. [Google Scholar] [CrossRef] [PubMed]
- Wisecaver, J.H.; Hackett, J.D. Dinoflagellate Genome Evolution. Annu. Rev. Microbiol. 2011, 65, 369–387. [Google Scholar] [CrossRef] [PubMed]
- Kohli, G.S.; John, U.; Figueroa, R.I.; Rhodes, L.L.; Harwood, D.T.; Groth, M.; Bolch, C.J.S.; Murray, S.A. Polyketide synthesis genes associated with toxin production in two species of Gambierdiscus (Dinophyceae). BMC Genomics 2015, 16, 410. [Google Scholar] [CrossRef]
- Stüken, A.; Orr, R.J.S.; Kellmann, R.; Murray, S.A.; Neilan, B.A.; Jakobsen, K.S. Discovery of nuclear-encoded genes for the neurotoxin saxitoxin in dinoflagellates. PLoS ONE 2011, 6, e20096. [Google Scholar] [CrossRef]
- Woo, Y.H.; Ansari, H.; Otto, T.D.; Klinger, C.M.; Kolisko, M.; Michálek, J.; Saxena, A.; Shanmugam, D.; Tayyrov, A.; Veluchamy, A.; et al. Chromerid genomes reveal the evolutionary path from photosynthetic algae to obligate intracellular parasites. eLife 2015, 4, e06974. [Google Scholar] [CrossRef]
- O’Neill, E.C.; Trick, M.; Hill, L.; Rejzek, M.; Dusi, R.G.; Hamilton, C.J.; Zimba, P.V.; Henrissat, B.; Field, R.A. The transcriptome of Euglena gracilis reveals unexpected metabolic capabilities for carbohydrate and natural product biochemistry. Mol. Biosyst. 2015, 11, 2808–2820. [Google Scholar] [CrossRef]
- Zimba, P.V.; Rowan, M.; Triemer, R. Identification of Euglenoid algae that produce ichthyotoxin(s). J. Fish Dis. 2004, 27, 115–117. [Google Scholar] [CrossRef]
- O’Neill, E.C.; Schorn, M.; Larson, C.B.; Millán-Aguiñaga, N. Targeted antibiotic discovery through biosynthesis-associated resistance determinants: Target directed genome mining. Crit. Rev. Microbiol. 2019, 45, 255–277. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017, 45, D200–D203. [Google Scholar] [CrossRef]
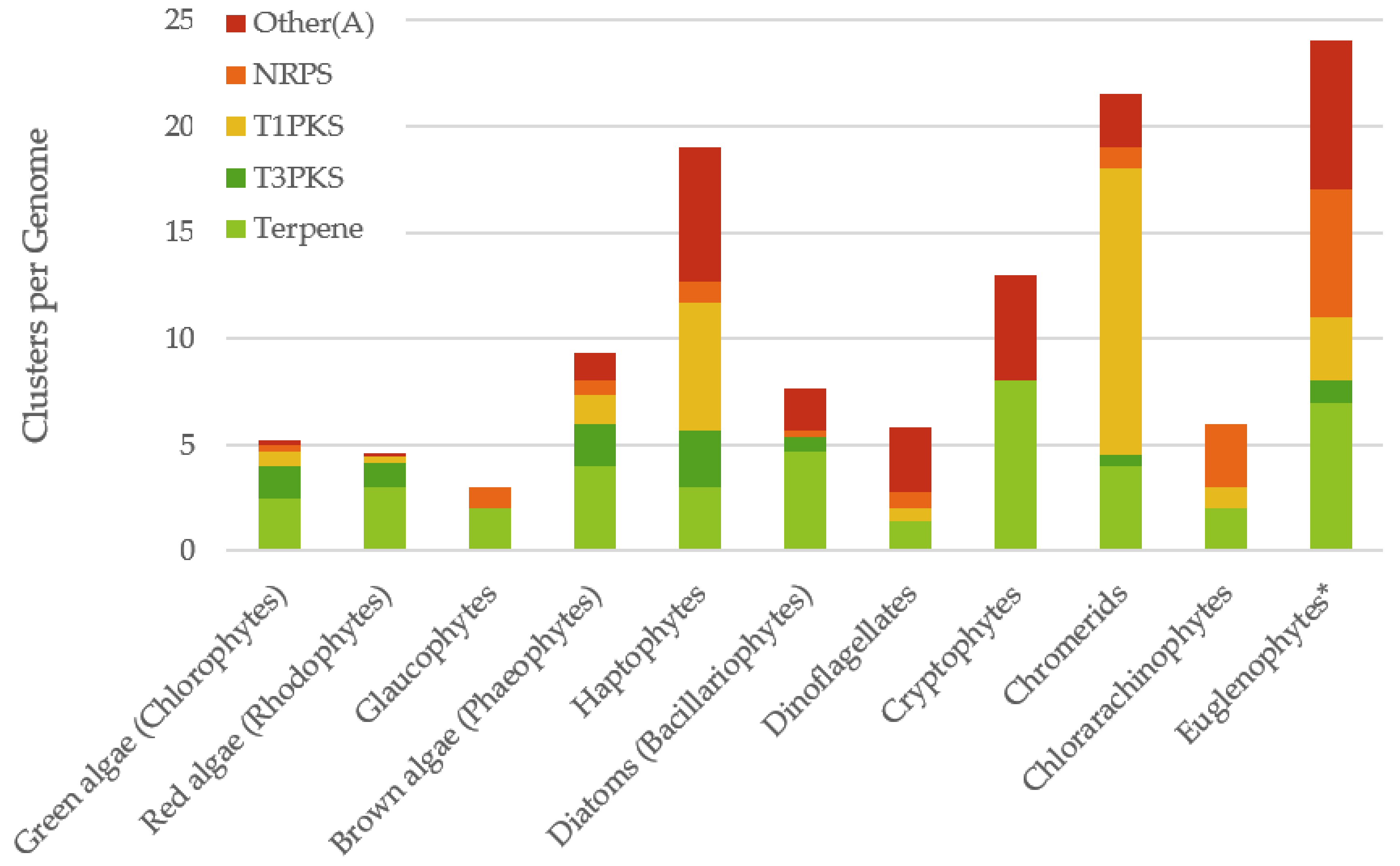

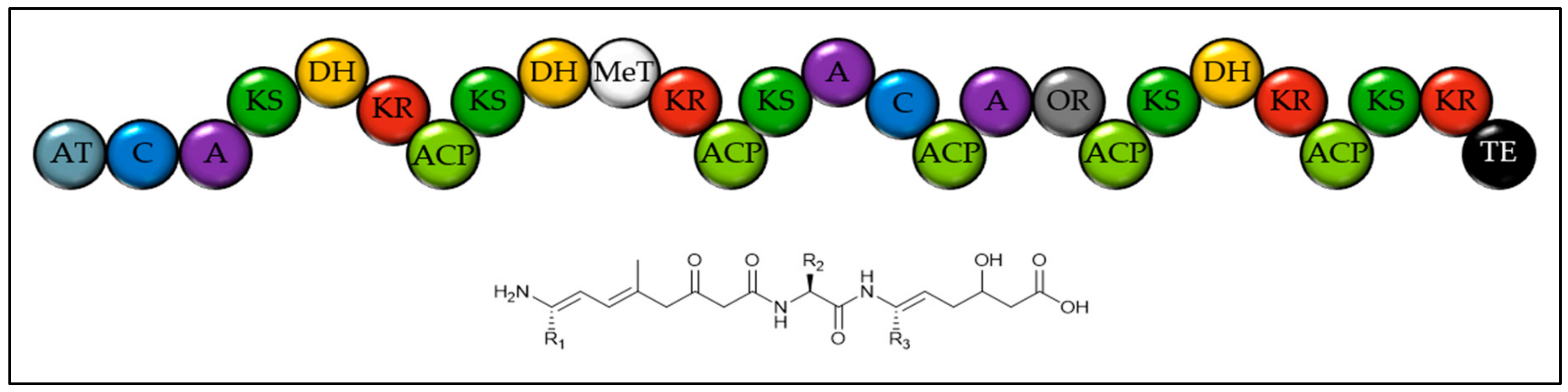
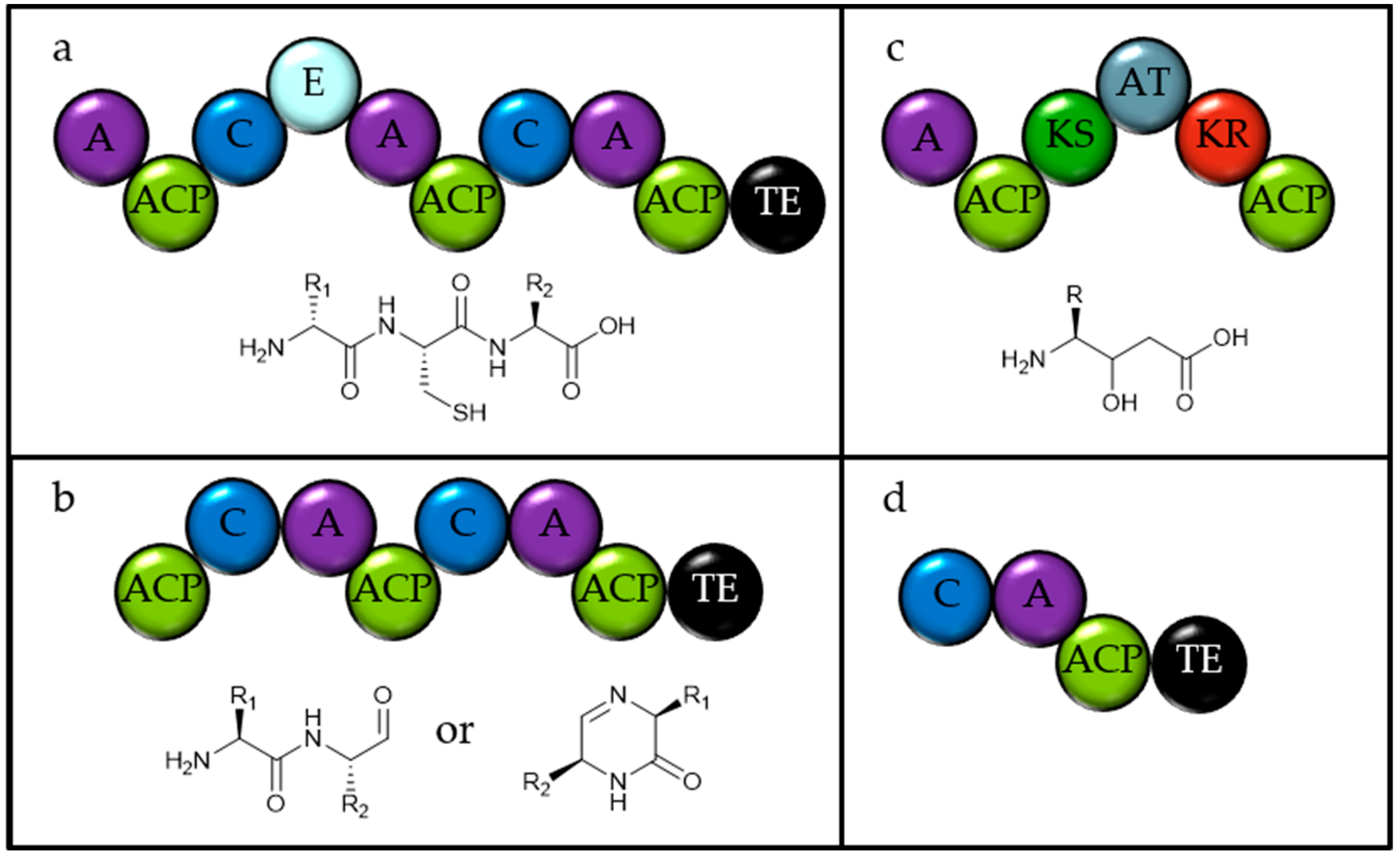

| Class | Species | Ungapped Sequence Length | Accession | Number of Gene Clusters | ||||
|---|---|---|---|---|---|---|---|---|
| Green Algae (Chlorophytes) | Terpene | T3PKS | T1PKS | NRPS | Other(A) | |||
| Chlamydomonas reinhardtii | 107,048,224 | GCA_000002595 | 1 | 3 | 1 | 0 | 0 | |
| Micromonas pusilla | 21,706,984 | GCA_000151265 | 5 | 0 | 2 | 0 | 0 | |
| Volvox carteri f. nagariensis | 125,467,762 | GCA_000143455 | 3 | 2 | 0 | 0 | 0 | |
| Chlorella variabilis | 42,214,557 | GCA_000147415 | 5 | 1 | 0 | 1 | 0 | |
| Coccomyxa subellipsoidea C-169 | 48,826,616 | GCA_000258705 | 1 | 5 | 1 | 0 | 0 | |
| Auxenochlorella pyrenoidosa | 48,566,231 | GCA_001430745 | 2 | 0 | 0 | 0 | 0 | |
| Helicosporidium sp. | 12,373,820 | GCA_000690575 | 1 | 2 | 0 | 0 | 0 | |
| Parachlorella kessleri | 59,187,803 | GCA_001598975 | 1 | 0 | 1 | 2 | 0 | |
| Prototheca cutis | 19,644,471 | GCA_002897115 | 2 | 1 | 1 | 0 | 0 | |
| Eudorina sp. | 182,993,185 | GCA_003117195 | 2 | 3 | 0 | 0 | 0 | |
| Yamagishiella unicocca | 134,234,618 | GCA_003116995 | 1 | 1 | 0 | 0 | 1 | |
| Trebouxia gelatinosa | 60,898,934 | GCA_000818905 | 1 | 0 | 0 | 0 | 0 | |
| Micractinium conductrix | 61,018,900 | GCA_002245815 | 2 | 0 | 0 | 0 | 0 | |
| Dunaliella salina | 280,838,039 | GCA_002284615 | 2 | 1 | 0 | 0 | 0 | |
| Botryococcus braunii | 179,769,887 | GCA_002005505 | 2 | 4 | 0 | 0 | 0 | |
| Tetrabaena socialis | 97,974,014 | GCA_002891735 | 2 | 0 | 0 | 0 | 0 | |
| Picocystis sp. ML | 29,646,247 | GCA_003665715 | 7 | 3 | 3 | 0 | 0 | |
| Ostreococcus tauri | 14,758,467 | GCA_002158475 | 5 | 0 | 1 | 0 | 2 | |
| Gonium pectorale | 117,596,311 | GCA_001584585 | 2 | 3 | 1 | 0 | 0 | |
| Cymbomonas tetramitiformis | 262,008,979 | GCA_001247695 | 1 | 3 | 4 | 3 | 2 | |
| Klebsormidium nitens | 103,146,182 | GCA_000708835.1 | 2 | 0 | 0 | 0 | 1 | |
| Chara braunii | 1,429,941,810 | GCA_003427395 | 5 | 1 | 0 | 0 | 0 | |
| Average | 2.5 | 1.5 | 0.7 | 0.3 | 0.3 | |||
| Red Algae (Rhodophytes) | ||||||||
| Gracilariopsis chorda | 92,180,038 | GCA_003194525 | 3 | 0 | 0 | 0 | 0 | |
| Porphyridium purpureum | 19,451,899 | GCA_000397085 | 1 | 1 | 0 | 0 | 0 | |
| Galdieria sulphuraria | 13,419,354 | GCA_000341285 | 3 | 1 | 0 | 0 | 0 | |
| Chondrus crispus | 104,085,276 | GCA_000350225 | 3 | 1 | 0 | 0 | 0 | |
| Porphyra umbilicalis | 87,766,581 | GCA_002049455 | 4 | 1 | 0 | 0 | 0 | |
| Gracilariopsis lemaneiformis | 86,759,375 | GCA_003346895 | 4 | 1 | 1 | 0 | 1 | |
| Kappaphycus alvarezii | 336,721,358 | GCA_002205965 | 3 | 3 | 1 | 0 | 0 | |
| Average | 3 | 1.1 | 0.3 | 0 | 0.1 | |||
| Glaucophytes | ||||||||
| Cyanophora paradoxa | 99,940,401 | GCA_004431415 | 2 | 0 | 0 | 1 | 0 | |
| Brown Algae (Phaeophytes) | ||||||||
| Ectocarpus siliculosus | 191,106,465 | GCA_000310025 | 1 | 3 | 1 | 0 | 1 | |
| Saccharina japonica | 537,522,535 | GCA_000978595 | 1 | 1 | 0 | 0 | 1 | |
| Cladosiphon okamuranus | 166,898,169 | GCA_001742925 | 10 | 2 | 3 | 1 | 3 | |
| Average | 4 | 2 | 1.3 | 0.3 | 1.7 | |||
| Haptophytes | ||||||||
| Emiliania huxleyi | 155,930,723 | GCA_000372725 | 2 | 2 | 8 | 0 | 8 | |
| Chrysochromulina parva | 65,764,750 | GCA_002887195 | 3 | 3 | 5 | 2 | 5 | |
| Chrysochromulina tobinii | 59,073,094 | GCA_001275005 | 4 | 3 | 5 | 1 | 6 | |
| Average | 3 | 2.7 | 6 | 1 | 6.3 | |||
| Diatoms (Bacillariophytes) | ||||||||
| Thalassiosira pseudonana | 32,272,629 | GCA_000149405 | 5 | 1 | 0 | 0 | 1 | |
| Thalassiosira oceanica | 92,185,637 | GCA_000296195 | 4 | 0 | 0 | 1 | 1 | |
| Phaeodactylum tricornutum | 27,017,695 | GCA_000150955 | 5 | 1 | 0 | 0 | 4 | |
| Average | 4.7 | 0. 7 | 0 | 0.3 | 2 | |||
| Dinoflagellates | ||||||||
| Symbiodinium microadriaticum | 745,992,902 | GCA_001939145 | 1 | 0 | 2 | 1 | 5 | |
| Symbiodinium sp. clade A Y106 | 756,831,958 | GCA_003297005 | 2 | 0 | 0 | 0 | 1 | |
| Symbiodinium sp. clade C Y103 | 674,313,450 | CA_003297045 | 1 | 0 | 0 | 0 | 3 | |
| Breviolum minutum | 603,733,232 | GCA_000507305 | 1 | 0 | 1 | 2 | 5 | |
| Prorocentrum minimum | 29,349,011 | GCA_001652855 | 2 | 0 | 0 | 1 | 1 | |
| Average | 1.4 | 0 | 0.6 | 0.8 | 3 | |||
| Cryptophyte | ||||||||
| Guillardia theta | 83,457,412 | GCA_000315625 | 8 | 0 | 0 | 0 | 3 | |
| Chromerids | ||||||||
| Vitrella brassicaformis | 71,768,979 | GCA_001179505 | 3 | 0 | 18 | 1 | 2 | |
| Chromera velia | 187,454,854 | GCA_000585135 | 5 | 1 | 9 | 1 | 3 | |
| Average | 4 | 0.5 | 13.5 | 1 | 2.5 | |||
| Chlorarachinophytes | ||||||||
| Bigelowiella natans | 91,405,885 | GCA_000320545 | 2 | 0 | 1 | 3 | 0 | |
| Euglenophytes | ||||||||
| Euglena gracilis | 1,435,499,417 | GCA_900893395 | 7 | 1 | 3 | 6 | 7 | |
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
O’Neill, E. Mining Natural Product Biosynthesis in Eukaryotic Algae. Mar. Drugs 2020, 18, 90. https://doi.org/10.3390/md18020090
O’Neill E. Mining Natural Product Biosynthesis in Eukaryotic Algae. Marine Drugs. 2020; 18(2):90. https://doi.org/10.3390/md18020090
Chicago/Turabian StyleO’Neill, Ellis. 2020. "Mining Natural Product Biosynthesis in Eukaryotic Algae" Marine Drugs 18, no. 2: 90. https://doi.org/10.3390/md18020090
APA StyleO’Neill, E. (2020). Mining Natural Product Biosynthesis in Eukaryotic Algae. Marine Drugs, 18(2), 90. https://doi.org/10.3390/md18020090