Deep Hypersaline Anoxic Basins as Untapped Reservoir of Polyextremophilic Prokaryotes of Biotechnological Interest
Abstract
1. Introduction
2. Prokaryotic Assemblages of DHABs
3. Biotechnological Potential of Prokaryotes Inhabiting DHABs
3.1. DHABs as a Hidden Treasure for Biodiscovery of Pharmaceuticals
3.2. DHABs as a Reservoir of Polyextreme Enzymes
3.3. DHAB-Derived Prokaryotes: Promising Candidates for Enhanced Bioremediation of Oil Hydrocarbons
4. Conclusions and Future Directions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Danovaro, R.; Corinaldesi, C.; Dell’Anno, A.; Snelgrove, P.V.R. The deep-sea under global change. Curr. Biol. 2017, 27, R461–R465. [Google Scholar] [CrossRef] [PubMed]
- Danovaro, R.; Snelgrove, P.V.R.; Tyler, P. Challenging the paradigms of deep-sea ecology. Trends Ecol. Evol. 2014, 29, 465–475. [Google Scholar] [CrossRef] [PubMed]
- Herring, P.J. The biology of the deep ocean; Oxford University Press: New York, NY, USA, 2001; pp. 1–25. [Google Scholar]
- Bartlett, D.H. Microbial Life in the Trenches. Mar. Technol. Soc. J. 2009, 43, 128–131. [Google Scholar] [CrossRef]
- Jørgensen, B.B.; Boetius, A. Feast and famine - Microbial life in the deep-sea bed. Nat. Rev. Microbiol. 2007, 5, 770–781. [Google Scholar] [CrossRef]
- Ramirez-Llodra, E.; Brandt, A.; Danovaro, R.; De Mol, B.; Escobar, E.; German, C.R.; Levin, L.A.; Martinez Arbizu, P.; Menot, L.; Buhl-Mortensen, P.; et al. Deep, diverse and definitely different: Unique attributes of the world’s largest ecosystem. Biogeosciences 2010, 7, 2851–2899. [Google Scholar] [CrossRef]
- Merino, N.; Aronson, H.S.; Bojanova, D.P.; Feyhl-Buska, J.; Wong, M.L.; Zhang, S.; Giovannelli, D. Living at the extremes: Extremophiles and the limits of life in a planetary context. Front. Microbiol. 2019, 10, 1–25. [Google Scholar] [CrossRef]
- Merlino, G.; Barozzi, A.; Michoud, G.; Ngugi, D.K.; Daffonchio, D. Microbial ecology of deep-sea hypersaline anoxic basins. FEMS Microbiol. Ecol. 2018, 94, 1–15. [Google Scholar] [CrossRef]
- Barone, G.; Varrella, S.; Tangherlini, M.; Rastelli, E.; Dell’Anno, A.; Danovaro, R.; Corinaldesi, C. Marine Fungi: Biotechnological Perspectives from Deep-Hypersaline Anoxic Basins. Diversity 2019, 11, 113. [Google Scholar] [CrossRef]
- Jongsma, D.; Fortuin, A.R.; Huson, W.; Troelstra, S.R.; Klaver, G.T.; Peters, J.M.; Van Harten, D.; De Lange, G.J.; Ten Haven, L. Discovery of an anoxic basin within the strabo trench, eastern mediterranean. Nature 1983, 305, 795–797. [Google Scholar] [CrossRef]
- Camerlenghi, A. Anoxic basins of the eastern Mediterranean: Geological framework. Mar. Chem. 1990, 31, 1–19. [Google Scholar] [CrossRef]
- Hartmann, M.; Scholten, J.C.; Stoffers, P.; Wehner, F. Hydrographic structure of brine-filled deeps in the Red Sea - new results from the Shaban, Kebrit, Atlantis II, and Discovery Deep. Mar. Geol. 1998, 144, 311–330. [Google Scholar] [CrossRef]
- Pautot, G.; Guennoc, P.; Coutelle, A.; Lyberis, N. Discovery of a large brine deep in the northern Red Sea. Nature 1984, 310, 133–136. [Google Scholar] [CrossRef]
- Backer, H.; Schoell, M. New Deeps with Brines and Metalliferous Sediments in the Red Sea. Nat. Phys. Sci. 1972, 240, 153–158. [Google Scholar] [CrossRef]
- Shokes, R.F.; Trabant, P.K.; Presley, B.J.; Reid, D.F. Anoxic, hypersaline basin in the northern Gulf of Mexico. Science 1977, 4297, 1443–1446. [Google Scholar] [CrossRef] [PubMed]
- La Cono, V.; Smedile, F.; Bortoluzzi, G.; Arcadi, E.; Maimone, G.; Messina, E.; Borghini, M.; Oliveri, E.; Mazzola, S.; L’Haridon, S.; et al. Unveiling microbial life in new deep-sea hypersaline Lake Thetis. Part I: Prokaryotes and environmental settings. Environ. Microbiol. 2011, 13, 2250–2268. [Google Scholar] [CrossRef] [PubMed]
- Yakimov, M.M.; La Cono, V.; Spada, G.L.; Bortoluzzi, G.; Messina, E.; Smedile, F.; Arcadi, E.; Borghini, M.; Ferrer, M.; Schmitt-Kopplin, P.; et al. Microbial community of the deep-sea brine Lake Kryos seawater-brine interface is active below the chaotropicity limit of life as revealed by recovery of mRNA. Environ. Microbiol. 2015, 17, 364–382. [Google Scholar] [CrossRef]
- La Cono, V.; Bortoluzzi, G.; Messina, E.; La Spada, G.; Smedile, F.; Giuliano, L.; Borghini, M.; Stumpp, C.; Schmitt-Kopplin, P.; Harir, M.; et al. The discovery of Lake Hephaestus, the youngest athalassohaline deep-sea formation on Earth. Sci. Rep. 2019, 9, 1679. [Google Scholar] [CrossRef]
- Mapelli, F.; Barozzi, A.; Michoud, G.; Merlino, G.; Crotti, E.; Borin, S.; Daffonchio, D. An updated view of the microbial diversity in deep hypersaline anoxic basins. In Adaption of Microbial Life to Environmental Extremes; Stan-Lotter, H., Fendrihan, S., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 23–40. ISBN 9783319483276. [Google Scholar]
- Eder, W.; Ludwig, W.; Huber, R. Novel 16S rRNA gene sequences retrieved from highly saline brine sediments of Kebrit Deep, Red Sea. Arch. Microbiol. 1999, 172, 213–218. [Google Scholar] [CrossRef]
- Daffonchio, D.; Borin, S.; Brusa, T.; Brusetti, L.; van der Wielen, P.W.J.J.; Bolhuis, H.; Yakimov, M.M.; D’Auria, G.; Giuliano, L.; Marty, D.; et al. Stratified prokaryote network in the oxic–anoxic transition of a deep-sea halocline. Nature 2006, 440, 203–207. [Google Scholar] [CrossRef]
- Yakimov, M.M.; La Cono, V.; Denaro, R.; D’Auria, G.; Decembrini, F.; Timmis, K.N.; Golyshin, P.N.; Giuliano, L. Primary producing prokaryotic communities of brine, interface and seawater above the halocline of deep anoxic lake L’Atalante, Eastern Mediterranean Sea. ISME J. 2007, 1, 743–755. [Google Scholar] [CrossRef]
- Borin, S.; Brusetti, L.; Mapelli, F.; D’Auria, G.; Brusa, T.; Marzorati, M.; Rizzi, A.; Yakimov, M.; Marty, D.; De Lange, G.J.; et al. Sulfur cycling and methanogenesis primarily drive microbial colonization of the highly sulfidic Urania deep hypersaline basin. Proc. Natl. Acad. Sci. USA 2009, 106, 9151–9156. [Google Scholar] [CrossRef] [PubMed]
- Joye, S.B.; MacDonald, I.R.; Montoya, J.P.; Peccini, M. Geophysical and geochemical signatures of Gulf of Mexico seafloor brines. Biogeosciences 2005, 2, 295–309. [Google Scholar] [CrossRef]
- Cita, M.B. Exhumation of Messinian evaporites in the deep-sea and creation of deep anoxic brine-filled collapsed basins. Sediment. Geol. 2006, 188–189, 357–378. [Google Scholar] [CrossRef]
- Hsü, K.J.; Cita, M.B.; Ryan, W.B.F. The origin of the Mediterranean evaporites. Initial Reports Deep Sea Drill. Proj. 1973, 13, 1203–1231. [Google Scholar]
- van der Wielen, P.W.J.J.; Bolhuis, H.; Borin, S.; Daffonchio, D.; Corselli, C.; Giuliano, L.; D’Auria, G.; de Lange, G.J.; Huebner, A.; Varnavas, S.P.; et al. The enigma of prokaryotic life in deep hypersaline anoxic basins. Science 2005, 307, 121–123. [Google Scholar] [CrossRef]
- Barozzi, A.; Mapelli, F.; Michoud, G.; Crotti, E.; Merlino, G.; Molinari, F.; Borin, S.; Daffonchio, D. Microbial Diversity and Biotechnological Potential of Microorganisms Thriving in the Deep-Sea Brine Pools. In Extremophiles From Biology to Biotechnology; Durvasula, R., Rao, D.S., Eds.; CRC Press: Boca Raton, FL, USA; Taylor & Francis, a CRC title, part of the Taylor & Francis imprint, a member of the Taylor & Francis Group, the academic division of T&F Informa plc: Boca Raton, FL, USA, 2018; pp. 19–32. ISBN 9781498774925. [Google Scholar]
- Swift, S.A.; Bower, A.S.; Schmitt, R.W. Vertical, horizontal, and temporal changes in temperature in the Atlantis II and Discovery hot brine pools, Red Sea. Deep. Res. Part I Oceanogr. Res. Pap. 2012, 64. [Google Scholar] [CrossRef]
- Ngugi, D.K.; Blom, J.; Alam, I.; Rashid, M.; Ba-Alawi, W.; Zhang, G.; Hikmawan, T.; Guan, Y.; Antunes, A.; Siam, R.; et al. Comparative genomics reveals adaptations of a halotolerant thaumarchaeon in the interfaces of brine pools in the Red Sea. ISME J. 2015, 9, 396–411. [Google Scholar] [CrossRef]
- Yakimov, M.M.; La Cono, V.; Slepak, V.Z.; La Spada, G.; Arcadi, E.; Messina, E.; Borghini, M.; Monticelli, L.S.; Rojo, D.; Barbas, C.; et al. Microbial life in the Lake Medee, the largest deep-sea salt-saturated formation. Sci. Rep. 2013, 3, 1–9. [Google Scholar] [CrossRef]
- Karbe, L. Hot Brines and the Deep Sea Environment. In Red Sea; Edwards, A.J., Head, S.J., Eds.; Elsevier: Oxford, UK, 1987; pp. 70–89. [Google Scholar]
- MacDonald, I.R.; Guinasso, N.L.; Reilly, J.F.; Brooks, J.M.; Callender, W.R.; Gabrielle, S.G. Gulf of Mexico hydrocarbon seep communities: VI. Patterns in community structure and habitat. Geo-Marine Lett. 1990, 10, 244–252. [Google Scholar] [CrossRef]
- De Lange, G.J.; Middelburg, J.J.; Van der Weijden, C.H.; Catalano, G.; Luther, G.W.; Hydes, D.J.; Woittiez, J.R.W.; Klinkhammer, G.P. Composition of anoxic hypersaline brines in the Tyro and Bannock Basins, eastern Mediterranean. Mar. Chem. 1990, 31, 63–88. [Google Scholar] [CrossRef]
- Van Cappellen, P.; Viollier, E.; Roychoudhury, A.; Clark, L.; Ingall, E.; Lowe, K.; Dichristina, T. Biogeochemical cycles of manganese and iron at the oxic-anoxic transition of a stratified marine basin (Orca Basin, Gulf of Mexico). Environ. Sci. Technol. 1998, 32, 2931–2939. [Google Scholar] [CrossRef]
- Schmidt, M.; Al-Farawati, R.; Botz, R. Geochemical Classification of Brine-Filled Red Sea Deeps. In The Red Sea; RasulIan, N.M.A., Stewart, C.F., Eds.; Springer Berlin Heidelberg: Cham, Switzerland, 2015; pp. 219–233. [Google Scholar]
- Stock, A.; Filker, S.; Yakimov, M.; Stoeck, T. Deep Hypersaline Anoxic Basins as Model Systems for Environmental Selection of Microbial Plankton. In Polyextremophiles Life Under Multiple Forms of Stress Life Under Multiple Forms of Stress; Springer Netherlands: Dordrecht, The Netherlands, 2013; pp. 499–515. [Google Scholar]
- Eder, W.; Schmidt, M.; Koch, M.; Garbe-Schönberg, D.; Huber, R. Prokaryotic phylogenetic diversity and corresponding geochemical data of the brine-seawater interface of the Shaban Deep, Red Sea. Environ. Microbiol. 2002, 4, 758–763. [Google Scholar] [CrossRef] [PubMed]
- Sass, A.M.; Sass, H.; Coolen, M.J.L.; Cypionka, H.; Overmann, J. Microbial Communities in the Chemocline of a Hypersaline Deep-Sea Basin (Urania Basin, Mediterranean Sea). Appl. Environ. Microbiol. 2001, 67, 5392–5402. [Google Scholar] [CrossRef] [PubMed]
- van der Wielen, P.W.J.J.; Heijs, S.K. Sulfate-reducing prokaryotic communities in two deep hypersaline anoxic basins in the Eastern Mediterranean deep sea. Environ. Microbiol. 2007, 9, 1335–1340. [Google Scholar] [CrossRef]
- Danovaro, R.; Corinaldesi, C.; Dell’Anno, A.; Fabiano, M.; Corselli, C. Viruses, prokaryotes and DNA in the sediments of a deep-hypersaline anoxic basin (DHAB) of the Mediterranean Sea. Environ. Microbiol. 2005, 7, 586–592. [Google Scholar] [CrossRef]
- Corinaldesi, C.; Tangherlini, M.; Luna, G.M.; Dell’Anno, A. Extracellular DNA can preserve the genetic signatures of present and past viral infection events in deep hypersaline anoxic basins. Proc. R. Soc. B Biol. Sci. 2014, 281, 1–10. [Google Scholar] [CrossRef]
- Danovaro, R.; Gambi, C.; Dell’Anno, A.; Corinaldesi, C.; Pusceddu, A.; Neves, R.C.; Kristensen, R.M. The challenge of proving the existence of metazoan life in permanently anoxic deep-sea sediments. BMC Biol. 2016, 14, 1–7. [Google Scholar] [CrossRef]
- Pachiadaki, M.G.; Yakimov, M.M.; Lacono, V.; Leadbetter, E.; Edgcomb, V. Unveiling microbial activities along the halocline of Thetis, a deep-sea hypersaline anoxic basin. ISME J. 2014, 8, 2478–2489. [Google Scholar] [CrossRef]
- Edgcomb, V.P.; Orsi, W.; Breiner, H.W.; Stock, A.; Filker, S.; Yakimov, M.M.; Stoeck, T. Novel active kinetoplastids associated with hypersaline anoxic basins in the Eastern Mediterranean deep-sea. Deep. Res. Part I Oceanogr. Res. Pap. 2011, 58, 1040–1048. [Google Scholar] [CrossRef]
- Alexander, E.; Stock, A.; Breiner, H.W.; Behnke, A.; Bunge, J.; Yakimov, M.M.; Stoeck, T. Microbial eukaryotes in the hypersaline anoxic L’Atalante deep-sea basin. Environ. Microbiol. 2009, 11, 360–381. [Google Scholar] [CrossRef]
- Danovaro, R.; Dell’Anno, A.; Pusceddu, A.; Gambi, C.; Heiner, I.; Kristensen, R.M. The first metazoa living in permanently anoxic conditions. BMC Biol. 2010, 8, 30. [Google Scholar] [CrossRef] [PubMed]
- Eder, W.; Jahnke, L.L.; Schmidt, M.; Huber, R. Microbial Diversity of the Brine-Seawater Interface of the Kebrit Deep, Red Sea, Studied via 16S rRNA Gene Sequences and Cultivation Methods. Appl. Environ. Microbiol. 2001, 67, 3077–3085. [Google Scholar] [CrossRef] [PubMed]
- Cavalazzi, B.; Barbieri, R.; Gómez, F.; Capaccioni, B.; Olsson-Francis, K.; Pondrelli, M.; Rossi, A.P.; Hickman-Lewis, K.; Agangi, A.; Gasparotto, G.; et al. The Dallol Geothermal Area, Northern Afar (Ethiopia)—An Exceptional Planetary Field Analog on Earth. Astrobiology 2019, 19, 553–578. [Google Scholar] [CrossRef] [PubMed]
- Poli, A.; Finore, I.; Romano, I.; Gioiello, A.; Lama, L.; Nicolaus, B. Microbial Diversity in Extreme Marine Habitats and Their Biomolecules. Microorganisms 2017, 5, 25. [Google Scholar] [CrossRef]
- Gunde-Cimerman, N.; Plemenitaš, A.; Oren, A. Strategies of adaptation of microorganisms of the three domains of life to high salt concentrations. FEMS Microbiol. Rev. 2018, 42, 353–375. [Google Scholar] [CrossRef]
- Giddings, L.-A.; Newman, D.J. Bioactive Compounds from Extremophiles. In Bioactive Compounds from Extremophiles; Springer: Cham, Switzerland, 2015; pp. 1–44. [Google Scholar]
- Coker, J.A. Extremophiles and biotechnology: Current uses and prospects. F1000Research 2016, 5, 396–403. [Google Scholar] [CrossRef]
- Sorokin, D.Y.; Kublanov, I.V.; Gavrilov, S.N.; Rojo, D.; Roman, P.; Golyshin, P.N.; Slepak, V.Z.; Smedile, F.; Ferrer, M.; Messina, E.; et al. Elemental sulfur and acetate can support life of a novel strictly anaerobic haloarchaeon. ISME J. 2016, 10, 240–252. [Google Scholar] [CrossRef]
- Kormas, K.A.; Pachiadaki, M.G.; Karayanni, H.; Leadbetter, E.R.; Bernhard, J.M.; Edgcomb, V.P. Inter-comparison of the potentially active prokaryotic communities in the halocline sediments of Mediterranean deep-sea hypersaline basins. Extremophiles 2015, 19, 949–960. [Google Scholar] [CrossRef]
- Mwirichia, R.; Alam, I.; Rashid, M.; Vinu, M.; Ba-Alawi, W.; Anthony Kamau, A.; Kamanda Ngugi, D.; Goker, M.; Klenk, H.P.; Bajic, V.; et al. Metabolic traits of an uncultured archaeal lineage-MSBL1-from brine pools of the Red Sea. Sci. Rep. 2016, 6, 1–14. [Google Scholar] [CrossRef]
- La Cono, V.; Arcadi, E.; Spada, G.; Barreca, D.; Laganà, G.; Bellocco, E.; Catalfamo, M.; Smedile, F.; Messina, E.; Giuliano, L.; et al. A Three-Component Microbial Consortium from Deep-Sea Salt-Saturated Anoxic Lake Thetis Links Anaerobic Glycine Betaine Degradation with Methanogenesis. Microorganisms 2015, 3, 500–517. [Google Scholar] [CrossRef]
- Guan, Y.; Hikmawan, T.; Antunes, A.; Ngugi, D.; Stingl, U. Diversity of methanogens and sulfate-reducing bacteria in the interfaces of five deep-sea anoxic brines of the Red Sea. Res. Microbiol. 2015, 166, 688–699. [Google Scholar] [CrossRef] [PubMed]
- Nigro, L.M.; Hyde, A.S.; MacGregor, B.J.; Teske, A. Phylogeography, Salinity Adaptations and Metabolic Potential of the Candidate Division KB1 Bacteria Based on a Partial Single Cell Genome. Front. Microbiol. 2016, 7, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Joye, S.B.; Samarkin, V.A.; Orcutt, B.N.; MacDonald, I.R.; Hinrichs, K.U.; Elvert, M.; Teske, A.P.; Lloyd, K.G.; Lever, M.A.; Montoya, J.P.; et al. Metabolic variability in seafloor brines revealed by carbon and sulphur dynamics. Nat. Geosci. 2009, 2, 349–354. [Google Scholar] [CrossRef]
- Stahl, D.A.; de la Torre, J.R. Physiology and Diversity of Ammonia-Oxidizing Archaea. Annu. Rev. Microbiol. 2012, 66, 83–101. [Google Scholar] [CrossRef]
- Könneke, M.; Bernhard, A.E.; de la Torre, J.R.; Walker, C.B.; Waterbury, J.B.; Stahl, D.A. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 2005, 437, 543–546. [Google Scholar] [CrossRef]
- Oren, A. Molecular ecology of extremely halophilic Archaea and Bacteria. FEMS Microbiol. Ecol. 2002, 39, 1–7. [Google Scholar] [CrossRef]
- Oren, A. Life at High Salt Concentrations. In The Prokaryotes; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer Berlin Heidelberg: Berlin, Germany, 2013; pp. 421–440. [Google Scholar]
- Guan, Y.; Ngugi, D.K.; Vinu, M.; Blom, J.; Alam, I.; Guillot, S.; Ferry, J.G.; Stingl, U. Comparative Genomics of the Genus Methanohalophilus, Including a Newly Isolated Strain From Kebrit Deep in the Red Sea. Front. Microbiol. 2019, 10, 1–11. [Google Scholar] [CrossRef]
- De Vitis, V.; Guidi, B.; Contente, M.L.; Granato, T.; Conti, P.; Molinari, F.; Crotti, E.; Mapelli, F.; Borin, S.; Daffonchio, D.; et al. Marine Microorganisms as Source of Stereoselective Esterases and Ketoreductases: Kinetic Resolution of a Prostaglandin Intermediate. Mar. Biotechnol. 2015, 17, 144–152. [Google Scholar] [CrossRef]
- Messina, E.; Sorokin, D.Y.; Kublanov, I.V.; Toshchakov, S.; Lopatina, A.; Arcadi, E.; Smedile, F.; La Spada, G.; La Cono, V.; Yakimov, M.M. Complete genome sequence of “Halanaeroarchaeum sulfurireducens” M27-SA2, a sulfur-reducing and acetate-oxidizing haloarchaeon from the deep-sea hypersaline anoxic lake Medee. Stand. Genomic Sci. 2016, 11, 1–15. [Google Scholar] [CrossRef]
- Antunes, A.; Taborda, M.; Huber, R.; Moissl, C.; Nobre, M.F.; da Costa, M.S. Halorhabdus tiamatea sp. nov., a non-pigmented extremely halophilic archaeon from a deep-sea hypersaline anoxic basin of the Red Sea, and emended description of the genus Halorhabdus. Int. J. Syst. Evol. Microbiol. 2008, 58, 215–220. [Google Scholar] [CrossRef]
- Antunes, A.; Rainey, F.A.; Wanner, G.; Taborda, M.; Pätzold, J.; Nobre, M.F.; da Costa, M.S.; Huber, R. A New Lineage of Halophilic, Wall-Less, Contractile Bacteria from a Brine-Filled Deep of the Red Sea. J. Bacteriol. 2008, 190, 3580–3587. [Google Scholar] [CrossRef] [PubMed]
- Antunes, A.; Francça, L.; Rainey, F.A.; Huber, R.; Fernanda Nobre, M.; Edwards, K.J.; da Costa, M.S. Marinobacter salsuginis sp. nov., isolated from the brine-seawater interface of the Shaban Deep, Red Sea. Int. J. Syst. Evol. Microbiol. 2007, 57, 1035–1040. [Google Scholar] [CrossRef] [PubMed]
- Edgcomb, V.P.; Pachiadaki, M.G.; Mara, P.; Kormas, K.A.; Leadbetter, E.R.; Bernhard, J.M. Gene expression profiling of microbial activities and interactions in sediments under haloclines of E. Mediterranean deep hypersaline anoxic basins. ISME J. 2016, 10, 2643–2657. [Google Scholar] [CrossRef] [PubMed]
- Siam, R.; Mustafa, G.A.; Sharaf, H.; Moustafa, A.; Ramadan, A.R.; Antunes, A.; Bajic, V.B.; Stingl, U.; Marsis, N.G.R.; Coolen, M.J.L.; et al. Unique Prokaryotic Consortia in Geochemically Distinct Sediments from Red Sea Atlantis II and Discovery Deep Brine Pools. PLoS ONE 2012, 7, e42872. [Google Scholar] [CrossRef] [PubMed]
- Tortorella, E.; Tedesco, P.; Esposito, F.P.; January, G.G.; Fani, R.; Jaspars, M.; De Pascale, D.; Palma Esposito, F.; January, G.G.; Fani, R.; et al. Antibiotics from deep-sea microorganisms: Current discoveries and perspectives. Mar. Drugs 2018, 16, 355. [Google Scholar] [CrossRef]
- Corinaldesi, C.; Barone, G.; Marcellini, F.; Dell’Anno, A.; Danovaro, R. Marine microbial-derived molecules and their potential use in cosmeceutical and cosmetic products. Mar. Drugs 2017, 15, 118. [Google Scholar] [CrossRef]
- Lo Giudice, A.; Rizzo, C. Bacteria Associated with Marine Benthic Invertebrates from Polar Environments: Unexplored Frontiers for Biodiscovery? Diversity 2018, 10, 80. [Google Scholar] [CrossRef]
- Andryukov, B.; Mikhailov, V.; Besednova, N. The Biotechnological Potential of Secondary Metabolites from Marine Bacteria. J. Mar. Sci. Eng. 2019, 7, 176. [Google Scholar] [CrossRef]
- Núñez-Montero, K.; Barrientos, L. Advances in Antarctic Research for Antimicrobial Discovery: A Comprehensive Narrative Review of Bacteria from Antarctic Environments as Potential Sources of Novel Antibiotic Compounds Against Human Pathogens and Microorganisms of Industrial Importance. Antibiotics 2018, 7, 90. [Google Scholar] [CrossRef]
- Corinaldesi, C. New perspectives in benthic deep-sea microbial ecology. Front. Mar. Sci. 2015, 2, 1–12. [Google Scholar] [CrossRef]
- Kim, S.K. Handbook of Marine biotechnology; Springer-Verlag Berlin Heidelberg: Berlin, Germany, 2015; pp. 307–326. ISBN 9783642539718. [Google Scholar]
- Ferrer, M.; Martínez-Martínez, M.; Bargiela, R.; Streit, W.R.; Golyshina, O.V.; Golyshin, P.N. Estimating the success of enzyme bioprospecting through metagenomics: Current status and future trends. Microb. Biotechnol. 2016, 9, 22–34. [Google Scholar] [CrossRef] [PubMed]
- Ziko, L.; Adel, M.; Malash, M.N.; Siam, R. Insights into red sea brine pool specialized metabolism gene clusters encoding potential metabolites for biotechnological applications and extremophile survival. Mar. Drugs 2019, 17, 273. [Google Scholar] [CrossRef] [PubMed]
- Ziko, L.; Saqr, A.-H.A.; Ouf, A.; Gimpel, M.; Aziz, R.K.; Neubauer, P.; Siam, R. Antibacterial and anticancer activities of orphan biosynthetic gene clusters from Atlantis II Red Sea brine pool. Microb. Cell Fact. 2019, 18, 56. [Google Scholar] [CrossRef] [PubMed]
- Sagar, S.; Esau, L.; Holtermann, K.; Hikmawan, T.; Zhang, G.; Stingl, U.; Bajic, V.B.; Kaur, M. Induction of apoptosis in cancer cell lines by the Red Sea brine pool bacterial extracts. BMC Complement. Altern. Med. 2013, 13, 344. [Google Scholar] [CrossRef]
- Esau, L.; Zhang, G.; Sagar, S.; Stingl, U.; Bajic, V.B.; Kaur, M. Mining the deep Red-Sea brine pool microbial community for anticancer therapeutics. BMC Complement. Altern. Med. 2019, 19. [Google Scholar] [CrossRef]
- Sagar, S.; Esau, L.; Hikmawan, T.; Antunes, A.; Holtermann, K.; Stingl, U.; Bajic, V.B.; Kaur, M. Cytotoxic and apoptotic evaluations of marine bacteria isolated from brine-seawater interface of the Red Sea. BMC Complement. Altern. Med. 2013, 13, 1–8. [Google Scholar] [CrossRef]
- Pruesse, E.; Peplies, J.; Glöckner, F.O. SINA: Accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 2012, 28, 1823–1829. [Google Scholar] [CrossRef]
- Lewis, K.; Epstein, S.; D’Onofrio, A.; Ling, L.L. Uncultured microorganisms as a source of secondary metabolites. J. Antibiot. 2010, 63, 468–476. [Google Scholar] [CrossRef]
- Ksouri, R.; Ksouri, W.M.; Jallali, I.; Debez, A.; Magné, C.; Hiroko, I.; Abdelly, C. Medicinal halophytes: Potent source of health promoting biomolecules with medical, nutraceutical and food applications. Crit. Rev. Biotechnol. 2012, 32, 289–326. [Google Scholar] [CrossRef]
- Raddadi, N.; Cherif, A.; Daffonchio, D.; Neifar, M.; Fava, F. Biotechnological applications of extremophiles, extremozymes and extremolytes. Appl. Microbiol. Biotechnol. 2015, 99, 7907–7913. [Google Scholar] [CrossRef]
- Singh, O.V. Extremophiles; Singh, O.V., Ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2012; pp. 1–429. ISBN 9781118394144. [Google Scholar]
- Rampelotto, P.H.; Trincone, A. Grand Challenges in Marine Biotechnology; Springer International Publishing: Cham, Switzerland, 2016; pp. 1–260. ISBN 978-3-319-69075-9. [Google Scholar]
- Babu, P.; Chandel, A.K.; Singh, O.V. Extremophiles and Their Applications in Medical Processes; SpringerBriefs in Microbiology: Cham, Switzerland, 2015; pp. 1–54. ISBN 978-3-319-12807-8. [Google Scholar]
- Shin, D.S.; Pratt, A.J.; Tainer, J.A. Archaeal genome guardians give insights into eukaryotic DNA replication and damage response proteins. Archaea 2014, 2014, 206735. [Google Scholar] [CrossRef] [PubMed]
- Majhi, M.C.; Behera, A.K.; Kulshreshtha, N.M.; Mahmooduzafar, D.; Kumar, R.; Kumar, A. ExtremeDB: A Unified Web Repository of Extremophilic Archaea and Bacteria. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Cárdenas, J.P.; Valdés, J.; Quatrini, R.; Duarte, F.; Holmes, D.S. Lessons from the genomes of extremely acidophilic bacteria and archaea with special emphasis on bioleaching microorganisms. Appl. Microbiol. Biotechnol. 2010, 88, 605–620. [Google Scholar] [CrossRef] [PubMed]
- Land, M.; Hauser, L.; Jun, S.R.; Nookaew, I.; Leuze, M.R.; Ahn, T.H.; Karpinets, T.; Lund, O.; Kora, G.; Wassenaar, T.; et al. Insights from 20 years of bacterial genome sequencing. Funct. Integr. Genomics 2015, 15, 141–161. [Google Scholar] [CrossRef] [PubMed]
- Baltz, R.H. Molecular beacons to identify gifted microbes for genome mining. J. Antibiot. 2017, 70, 639–646. [Google Scholar] [CrossRef] [PubMed]
- Brown, E.D.; Wright, G.D. Antibacterial drug discovery in the resistance era. Nature 2016, 529, 336–343. [Google Scholar] [CrossRef]
- Holohan, C.; Van Schaeybroeck, S.; Longley, D.B.; Johnston, P.G. Cancer drug resistance: An evolving paradigm. Nat. Rev. Cancer 2013, 13, 714–726. [Google Scholar] [CrossRef]
- Ventola, C.L. The antibiotic resistance crisis: Part 1: Causes and threats. P T 2015, 40, 277–283. [Google Scholar]
- Housman, G.; Byler, S.; Heerboth, S.; Lapinska, K.; Longacre, M.; Snyder, N.; Sarkar, S. Drug resistance in cancer: An overview. Cancers 2014, 6, 1769–1792. [Google Scholar] [CrossRef]
- Trindade, M.; van Zyl, L.J.; Navarro-Fernández, J.; Elrazak, A.A. Targeted metagenomics as a tool to tap into marine natural product diversity for the discovery and production of drug candidates. Front. Microbiol. 2015, 6, 1–14. [Google Scholar] [CrossRef]
- Malve, H. Exploring the ocean for new drug developments: Marine pharmacology. J. Pharm. Bioallied Sci. 2016, 8, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Wever, W.J.; Walsh, C.T.; Bowers, A.A. Dithiolopyrrolones: Biosynthesis, synthesis, and activity of a unique class of disulfide-containing antibiotics. Nat. Prod. Rep. 2014, 31, 905–923. [Google Scholar] [CrossRef] [PubMed]
- Imada, C.; Maeda, M.; Hara, S.; Taga, N.; Simidu, U. Purification and characterization of subtilisin inhibitors ‘Marinostatin’ produced by marine Alteromonas sp. J. Appl. Bacteriol. 1986, 60, 469–476. [Google Scholar] [CrossRef]
- Gustafson, K.; Roman, M.; Fenical, W. The Macrolactins, a Novel Class of Antiviral and Cytotoxic Macrolides from a Deep-Sea Marine Bacterium. J. Am. Chem. Soc. 1989, 111, 7519–7524. [Google Scholar] [CrossRef]
- Nagao, T.; Adachi, K.; Sakai, M.; Nishijima, M.; Sano, H. Novel macrolactins as antibiotic lactones from a marine bacterium. J. Antibiot. 2001, 54, 333–339. [Google Scholar] [CrossRef]
- Jaruchoktaweechai, C.; Suwanborirux, K.; Tanasupawatt, S.; Kittakoop, P.; Menasveta, P. New macrolactins from a marine Bacillus sp. Sc026. J. Nat. Prod. 2000, 63, 984–986. [Google Scholar] [CrossRef]
- Xue, C.; Tian, L.; Xu, M.; Deng, Z.; Lin, W. A new 24-membered lactone and a new polyene δ-lactone from the marine bacterium Bacillus marinus. J. Antibiot. 2008, 61, 668–674. [Google Scholar] [CrossRef]
- Berger, E.; Crampton, M.C.; Nxumalo, N.P.; Louw, M.E. Extracellular secretion of a recombinant therapeutic peptide by Bacillus halodurans utilizing a modified flagellin type III secretion system. Microb. Cell Fact. 2011, 10. [Google Scholar] [CrossRef]
- Gerard, J.M.; Haden, P.; Kelly, M.T.; Andersen, R.J. Loloatins A-D, cyclic decapeptide antibiotics produced in culture by a tropical marine bacterium. J. Nat. Prod. 1999, 62, 80–85. [Google Scholar] [CrossRef]
- Rahman, H.; Austin, B.; Mitchell, W.J.; Morris, P.C.; Jamieson, D.J.; Adams, D.R.; Spragg, A.M.; Schweizer, M. Novel anti-infective compounds from marine bacteria. Mar. Drugs 2010, 8, 498–518. [Google Scholar] [CrossRef]
- Mondol, M.A.M.; Shin, H.J.; Islam, M.T. Diversity of secondary metabolites from marine Bacillus species: Chemistry and biological activity. Mar. Drugs 2013, 11, 2846–2872. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.L.; Hua, H.M.; Pei, Y.H.; Yao, X.S. Three New Cytotoxic Cyclic Acylpeptides from Marine Bacillus sp. Chem. Pharm. Bull. 2004, 52, 1029–1030. [Google Scholar] [CrossRef] [PubMed]
- Oku, N.; Adachi, K.; Matsuda, S.; Kasai, H.; Takatsuki, A.; Shizuri, Y. Ariakemicins A and B, novel polyketide-peptide antibiotics from a marine gliding bacterium of the genus Rapidithrix. Org. Lett. 2008, 10, 2481–2484. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; MacMillan, J.B. Erythrazoles A-B, cytotoxic benzothiazoles from a marine-derived Erythrobacter sp. Org. Lett. 2011, 13, 6580–6583. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Legako, A.G.; Espindola, A.P.D.M.; MacMillan, J.B. Erythrolic acids A-E, meroterpenoids from a marine-derived Erythrobacter sp. J. Org. Chem. 2012, 77, 3401–3407. [Google Scholar] [CrossRef] [PubMed]
- Di Lorenzo, F.; Palmigiano, A.; Paciello, I.; Pallach, M.; Garozzo, D.; Bernardini, M.-L.L.; Cono, V.L.; Yakimov, M.M.; Molinaro, A.; Silipo, A.; et al. The deep-sea polyextremophile Halobacteroides lacunaris TB21 rough-type LPS: Structure and inhibitory activity towards toxic LPS. Mar. Drugs 2017, 15, 201. [Google Scholar] [CrossRef]
- Homann, V.V.; Sandy, M.; Tincu, J.A.; Templeton, A.S.; Tebo, B.M.; Butler, A. Loihichelins A-F, a suite of amphiphilic siderophores produced by the marine bacterium Halomonas LOB-5. J. Nat. Prod. 2009, 72, 884–888. [Google Scholar] [CrossRef]
- Wang, L.; Große, T.; Stevens, H.; Brinkhoff, T.; Simon, M.; Liang, L.; Bitzer, J.; Bach, G.; Zeeck, A.; Tokuda, H.; et al. Bioactive hydroxyphenylpyrrole-dicarboxylic acids from a new marine Halomonas sp.: Production and structure elucidation. Appl. Microbiol. Biotechnol. 2006, 72, 816–822. [Google Scholar] [CrossRef]
- Bitzer, J.; Große, T.; Wang, L.; Lang, S.; Beil, W.; Zeeck, A. New aminophenoxazinones from a marine Halomonas sp.: Fermentation, structure elucidation, and biological activity. J. Antibiot. 2006, 59, 86–92. [Google Scholar] [CrossRef]
- Silipo, A.; Lanzetta, R.; Parrilli, M.; Sturiale, L.; Garozzo, D.; Nazarenko, E.L.; Gorshkova, R.P.; Ivanova, E.P.; Molinaro, A. The complete structure of the core carbohydrate backbone from the LPS of marine halophilic bacterium Pseudoalteromonas carrageenovora type strain IAM 12662T. Carbohydr. Res. 2005, 340, 1475–1482. [Google Scholar] [CrossRef]
- Mitova, M.; Tutino, M.L.; Infusini, G.; Marino, G.; De Rosa, S. Exocellular peptides from antarctic psychrophile Pseudoalteromonas haloplanktis. Mar. Biotechnol. 2005, 7, 523–531. [Google Scholar] [CrossRef] [PubMed]
- Shiozawa, H.; Shimada, A.; Takahashi, S. Thiomarinols D, E, F and G, new hybrid antimicrobial antibiotics produced by a marine bacterium. Isolated, structure, and antimicrobial activity. J. Antibiot. 1997, 50, 449–452. [Google Scholar] [CrossRef] [PubMed]
- Shiozawa, H.; Kagasaki, T.; Takahashi, S.; Kinoshita, T.; Haruyama, H.; Domon, H.; Utsuib, Y.; Kodama, K. Thiomarinol, a new hybrid antimicrobial antibiotic produced by a marine bacterium fermentation, isolation, structure, and antimicrobial activity. J. Antibiot. 1993, 46, 1834–1842. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, S.S.; Nicholson, B.; Teisan, S.; Lam, K.S.; Potts, B.C.M. Aureoverticillactam, a novel 22-atom macrocyclic lactam from the marine actinomycete Streptomyces aureoverticillatus. J. Nat. Prod. 2004, 67, 1400–1402. [Google Scholar] [CrossRef]
- Pesic, A.; Baumann, H.I.; Kleinschmidt, K.; Ensle, P.; Wiese, J.; Süssmuth, R.D.; Imhoff, J.F. Champacyclin, a new cyclic octapeptide from Streptomyces strain C42 isolated from the Baltic Sea. Mar. Drugs 2013, 11, 4834–4857. [Google Scholar] [CrossRef]
- Kasanah, N.; Triyanto, T. Bioactivities of Halometabolites from Marine Actinobacteria. Biomolecules 2019, 9, 225. [Google Scholar] [CrossRef]
- Martin, G.D.A.; Tan, L.T.; Jensen, P.R.; Dimayuga, R.E.; Fairchild, C.R.; Raventos-Suarez, C.; Fenical, W. Marmycins A and B, cytotoxic pentacyclic C-glycosides from a marine sediment-derived actinomycete related to the genus Streptomyces. J. Nat. Prod. 2007, 70, 1406–1409. [Google Scholar] [CrossRef]
- Zhou, X.; Huang, H.; Li, J.; Song, Y.; Jiang, R.; Liu, J.; Zhang, S.; Hua, Y.; Ju, J. New anti-infective cycloheptadepsipeptide congeners and absolute stereochemistry from the deep sea-derived Streptomyces drozdowiczii SCSIO 10141. Tetrahedron 2014, 70, 7795–7801. [Google Scholar] [CrossRef]
- Bruntner, C.; Binder, T.; Pathom-Aree, W.; Goodfellow, M.; Bull, A.T.; Potterat, O.; Puder, C.; Hörer, S.; Schmid, A.; Bolek, W.; et al. Frigocyclinone, a novel angucyclinone antibiotic produced by a Streptomyces griseus strain from Antarctica. J. Antibiot. 2005, 58, 346–349. [Google Scholar] [CrossRef]
- El-Gendy, M.M.A.; Shaaban, M.; Shaaban, K.A.; El-Bondkly, A.M.; Laatsch, H. Essramycin: A first triazolopyrimidine antibiotic isolated from nature. J. Antibiot. 2008, 61, 149–157. [Google Scholar] [CrossRef]
- Song, Y.; Huang, H.; Chen, Y.; Ding, J.; Zhang, Y.; Sun, A.; Zhang, W.; Ju, J. Cytotoxic and antibacterial marfuraquinocins from the deep south china sea-derived Streptomyces niveus scsio 3406. J. Nat. Prod. 2013, 76, 2263–2268. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Li, Q.; Liu, X.; Chen, Y.; Zhang, Y.; Sun, A.; Zhang, W.; Zhang, J.; Ju, J. Cyclic hexapeptides from the deep South China sea-derived Streptomyces scopuliridis SCSIO ZJ46 active against pathogenic gram-positive bacteria. J. Nat. Prod. 2014, 77, 1937–1941. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, A.; Kurasawa, S.; Ikeda, D.; Okami, Y.; Takeuchi, T. Altemicidin, a new acaricidal and antitumor substance. I. Taxonomy, fermentation, isolation and physico-chemical and biological properties. J. Antibiot. 1989, 42, 1556–1561. [Google Scholar] [CrossRef] [PubMed]
- Pan, H.Q.; Zhang, S.Y.; Wang, N.; Li, Z.L.; Hua, H.M.; Hu, J.C.; Wang, S.J. New spirotetronate antibiotics, lobophorins H and I, from a South China Sea-derived Streptomyces sp. 12A35. Mar. Drugs 2013, 11, 3891–3901. [Google Scholar] [CrossRef]
- Moon, K.; Ahn, C.H.; Shin, Y.; Won, T.H.; Ko, K.; Lee, S.K.; Oh, K.B.; Shin, J.; Nam, S.I.; Oh, D.C. New Benzoxazine Secondary Metabolites from an Arctic Actinomycete. Mar. Drugs 2014, 12, 2526–5238. [Google Scholar] [CrossRef]
- Schultz, A.W.; Oh, D.C.; Carney, J.R.; Williamson, R.T.; Udwary, D.W.; Jensen, P.R.; Gould, S.J.; Fenical, W.; Moore, B.S. Biosynthesis and structures of cyclomarins and cyclomarazines, prenylated cyclic peptides of marine actinobacterial origin. J. Am. Chem. Soc. 2008, 130, 4507–4516. [Google Scholar] [CrossRef]
- Renner, M.K.; Shen, Y.C.; Cheng, X.C.; Jensen, P.R.; Frankmoelle, W.; Kauffman, C.A.; Fenical, W.; Lobkovsky, E.; Clardy, J. Cyclomarins A-C, new antiinflammatory cyclic peptides produced by a marine bacterium (Streptomyces sp.). J. Am. Chem. Soc. 1999, 121, 11273–11276. [Google Scholar] [CrossRef]
- Hughes, C.C.; Prieto-Davo, A.; Jensen, P.R.; Fenical, W. The marinopyrroles, antibiotics of an unprecedented structure class from a marine Streptomyces sp. Org. Lett. 2008, 10, 629–631. [Google Scholar] [CrossRef]
- Hughes, C.C.; Kauffman, C.A.; Jensen, P.R.; Fenical, W. Structures, Reactivities, and Antibiotic Properties of the Marinopyrroles A–F. J. Org. Chem. 2010, 75, 3240–3250. [Google Scholar] [CrossRef]
- Asolkar, R.N.; Jensen, P.R.; Kauffman, C.A.; Fenical, W. Daryamides A-C, weakly cytotoxic polyketides from a marine-derived actinomycete of the genus Streptomyces strain CNQ-085. J. Nat. Prod. 2006, 69, 1756–1759. [Google Scholar] [CrossRef]
- Hughes, C.C.; MacMillan, J.B.; Gaudêncio, S.P.; Jensen, P.R.; Fenical, W. The ammosamides: Structures of cell cycle modulators from a marine-derived Streptomyces species. Angew. Chemi. Int. Ed. 2009, 48, 725–727. [Google Scholar] [CrossRef]
- Hughes, C.C.; Fenical, W. Total synthesis of the ammosamides. J. Am. Chem. Soc. 2010, 132, 2528–2529. [Google Scholar] [CrossRef] [PubMed]
- Pan, E.; Jamison, M.; Yousufuddin, M.; MacMillan, J.B. Ammosamide D, an oxidatively ring opened ammosamide analog from a marine-derived Streptomyces variabilis. Org. Lett. 2012, 14, 2390–2393. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Maskey, R.P.; Qin, S.; Sattler, I.; Fiebig, H.H.; Maier, A.; Zeeck, A.; Laatsch, H. Chinikomycins A and B: Isolation, structure elucidation, and biological activity of novel antibiotics from a marine Streptomyces sp. isolate M045. J. Nat. Prod. 2005, 68, 349–353. [Google Scholar] [CrossRef] [PubMed]
- Pérez, M.; Crespo, C.; Schleissner, C.; Rodríguez, P.; Zúñiga, P.; Reyes, F. Tartrolon D, a cytotoxic macrodiolide from the marine-derived actinomycete Streptomyces sp. MDG-04-17-069. J. Nat. Prod. 2009, 72, 2192–2194. [Google Scholar] [CrossRef] [PubMed]
- Hawas, U.W.; Shaaban, M.; Shaaban, K.A.; Speitling, M.; Maier, A.; Kelter, G.; Fiebig, H.H.; Meiners, M.; Helmke, E.; Laatsch, H. Mansouramycins A-D, cytotoxic isoquinolinequinones from a marine Streptomycete. J. Nat. Prod. 2009, 72, 2120–2124. [Google Scholar] [CrossRef] [PubMed]
- Nachtigall, J.; Schneider, K.; Bruntner, C.; Bull, A.T.; Goodfellow, M.; Zinecker, H.; Imhoff, J.F.; Nicholson, G.; Irran, E.; Süssmuth, R.D.; et al. Benzoxacystol, a benzoxazine-type enzyme inhibitor from the deep-sea strain Streptomyces sp. NTK 935. J. Antibiot. 2011, 64, 453–457. [Google Scholar] [CrossRef]
- Zhang, W.; Liu, Z.; Li, S.; Yang, T.; Zhang, Q.; Ma, L.; Tian, X.; Zhang, H.; Huang, C.; Zhang, S.; et al. Spiroindimicins A-D: New bisindole alkaloids from a deep-sea-derived actinomycete. Org. Lett. 2012, 14, 3364–3367. [Google Scholar] [CrossRef]
- Song, Y.; Liu, G.; Li, J.; Huang, H.; Zhang, X.; Zhang, H.; Ju, J. Cytotoxic and antibacterial angucycline- and prodigiosin-analogues from the deep-sea derived Streptomyces sp. SCSIO 11594. Mar. Drugs 2015, 13, 1304–1316. [Google Scholar] [CrossRef]
- You, Z.Y.; Wang, Y.H.; Zhang, Z.G.; Xu, M.J.; Xie, S.J.; Han, T.S.; Feng, L.; Li, X.G.; Xu, J. Identification of two novel anti-fibrotic benzopyran compounds produced by engineered strains derived from Streptomyces xiamenensis M1-94P that originated from deep-sea sediments. Mar. Drugs 2013, 11, 4035–4049. [Google Scholar] [CrossRef]
- Miller, E.D.; Kauffman, C.A.; Jensen, P.R.; Fenical, W. Piperazimycins: Cytotoxic hexadepsipeptides from a marine-derived bacterium of the genus Streptomyces. J. Org. Chem. 2007, 72, 323–330. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.-L.; Liu, S.-B.; Qiao, L.-P.; Chen, X.-L.; Pang, X.; Shi, M.; Zhang, X.-Y.; Qin, Q.-L.; Zhou, B.-C.; Zhang, Y.-Z.; et al. A novel exopolysaccharide from deep-sea bacterium Zunongwangia profunda SM-A87: Low-cost fermentation, moisture retention, and antioxidant activities. Appl. Microbiol. Biotechnol. 2014, 98, 7437–7445. [Google Scholar] [CrossRef] [PubMed]
- Chater, K.F. Recent advances in understanding Streptomyces. F1000Research 2016, 5, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Hettiarachchi, S.A.; Lee, S.-J.; Lee, Y.; Kwon, Y.-K.; De Zoysa, M.; Moon, S.; Jo, E.; Kim, T.; Kang, D.-H.; Heo, S.-J.; et al. A rapid and efficient screening method for antibacterial compound-producing bacteria. J. Microbiol. Biotechnol. 2017, 27, 1441–1448. [Google Scholar] [PubMed]
- Andreo-Vidal, A.; Sanchez-Amat, A.; Campillo-Brocal, J. The Pseudoalteromonas luteoviolacea L-amino Acid Oxidase with Antimicrobial Activity Is a Flavoenzyme. Mar. Drugs 2018, 16, 499. [Google Scholar] [CrossRef] [PubMed]
- Kalitnik, A.A.; Byankina Barabanova, A.O.; Nagorskaya, V.P.; Reunov, A.V.; Glazunov, V.P.; Solov’eva, T.F.; Yermak, I.M. Low molecular weight derivatives of different carrageenan types and their antiviral activity. J. Appl. Phycol. 2013, 25, 65–72. [Google Scholar] [CrossRef]
- Queiroz, E.A.I.F.; Fortes, Z.B.; da Cunha, M.A.A.; Sarilmiser, H.K.; Barbosa Dekker, A.M.; Öner, E.T.; Dekker, R.F.H.; Khaper, N. Levan promotes antiproliferative and pro-apoptotic effects in MCF-7 breast cancer cells mediated by oxidative stress. Int. J. Biol. Macromol. 2017, 102, 565–570. [Google Scholar] [CrossRef]
- Ruiz-Ruiz, C.; Srivastava, G.K.; Carranza, D.; Mata, J.A.; Llamas, I.; Santamaría, M.; Quesada, E.; Molina, I.J. An exopolysaccharide produced by the novel halophilic bacterium Halomonas stenophila strain B100 selectively induces apoptosis in human T leukaemia cells. Appl. Microbiol. Biotechnol. 2011, 89, 345–355. [Google Scholar] [CrossRef]
- Lorenz, P.; Eck, J. Metagenomics and industrial applications. Nat. Rev. Microbiol. 2005, 3, 510–516. [Google Scholar] [CrossRef]
- Prasad, S.; Roy, I. Converting Enzymes into Tools of Industrial Importance. Recent Pat. Biotechnol. 2018, 12, 33–56. [Google Scholar] [CrossRef]
- Bommarius, A.S.; Paye, M.F. Stabilizing biocatalysts. Chem. Soc. Rev. 2013, 42, 6534–6565. [Google Scholar] [CrossRef] [PubMed]
- Madhavan, A.; Sindhu, R.; Binod, P.; Sukumaran, R.K.; Pandey, A. Strategies for design of improved biocatalysts for industrial applications. Bioresour. Technol. 2017, 245, 1304–1313. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Martínez, M.; Bargiela, R.; Ferrer, M. Metagenomics and the Search for Industrial Enzymes. In Biotechnology of Microbial Enzymes: Production, Biocatalysis and Industrial Applications; Brahmachari, G., Demain, A.L., Adrio, J.L., Eds.; Academic Press: Cambridge, MA, USA, 2016; pp. 1–608. [Google Scholar]
- Chapman, J.; Ismail, A.; Dinu, C. Industrial Applications of Enzymes: Recent Advances, Techniques, and Outlooks. Catalysts 2018, 8, 238. [Google Scholar] [CrossRef]
- Dumorné, K.; Córdova, D.C.; Astorga-Eló, M.; Renganathan, P. Extremozymes: A potential source for industrial applications. J. Microbiol. Biotechnol. 2017, 27, 649–659. [Google Scholar] [CrossRef] [PubMed]
- Dalmaso, G.; Ferreira, D.; Vermelho, A. Marine Extremophiles: A Source of Hydrolases for Biotechnological Applications. Mar. Drugs 2015, 13, 1925–1965. [Google Scholar] [CrossRef] [PubMed]
- Sarmiento, F.; Peralta, R.; Blamey, J.M. Cold and Hot Extremozymes: Industrial Relevance and Current Trends. Front. Bioeng. Biotechnol. 2015, 3, 1–15. [Google Scholar] [CrossRef]
- Di Donato, P.; Buono, A.; Poli, A.; Finore, I.; Abbamondi, G.R.; Nicolaus, B.; Lama, L. Exploring marine environments for the identification of extremophiles and their enzymes for sustainable and green bioprocesses. Sustainability 2019, 11, 149. [Google Scholar] [CrossRef]
- Bruno, S.; Coppola, D.; di Prisco, G.; Giordano, D.; Verde, C. Enzymes from Marine Polar Regions and Their Biotechnological Applications. Mar. Drugs 2019, 17, 544. [Google Scholar] [CrossRef]
- Yamanaka, Y.; Kazuoka, T.; Yoshida, M.; Yamanaka, K.; Oikawa, T.; Soda, K. Thermostable aldehyde dehydrogenase from psychrophile, Cytophaga sp. KUC-1: Enzymological characteristics and functional properties. Biochem. Biophys. Res. Commun. 2002, 298, 632–637. [Google Scholar] [CrossRef]
- Anburajan, L.; Meena, B.; Narendar Sivvaswamy, S. First report on molecular characterization of novel betaine aldehyde dehydrogenase from the halotolerant eubacteria, Bacillus halodurans. Gene Reports 2017, 9, 131–135. [Google Scholar] [CrossRef]
- Kim, H.J.; Joo, W.A.; Cho, C.W.; Kim, C.W. Halophile aldehyde dehydrogenase from Halobacterium salinarum. J. Proteome Res. 2006, 5, 192–195. [Google Scholar] [CrossRef] [PubMed]
- Akal, A.L.; Karan, R.; Hohl, A.; Alam, I.; Vogler, M.; Grötzinger, S.W.; Eppinger, J.; Rueping, M. A polyextremophilic alcohol dehydrogenase from the Atlantis II Deep Red Sea brine pool. FEBS Open Bio 2019, 9, 194–205. [Google Scholar] [CrossRef] [PubMed]
- Borchert, E.; Knobloch, S.; Dwyer, E.; Flynn, S.; Jackson, S.A.; Jóhannsson, R.; Marteinsson, V.T.; O’Gara, F.; Dobson, A.D.W.W.; O’Gara, F.; et al. Biotechnological Potential of Cold Adapted Pseudoalteromonas spp. Isolated from ‘Deep Sea’ Sponges. Mar. Drugs 2017, 15, 184. [Google Scholar] [CrossRef] [PubMed]
- Izotova, L.S.; Strongin, A.Y.; Chekulaeva, L.N.; Sterkin, V.E.; Ostoslavskaya, V.I.; Lyublinskaya, L.A.; Timokhina, E.A.; Stepanov, V.M. Purification and properties of serine protease from Halobacterium halobium. J. Bacteriol. 1983, 155, 826–830. [Google Scholar] [CrossRef] [PubMed]
- Venugopal, M.; Saramma, A.V. An alkaline protease from Bacillus circulans BM15, newly isolated from a mangrove station: Characterization and application in laundry detergent formulations. Indian J. Microbiol. 2007, 47, 298–303. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Liu, G.; Zhang, D.; Li, C.; Sun, C. Purification and biochemical characterization of an alkaline protease from marine bacteria Pseudoalteromonas sp. 129-1. J. Basic Microbiol. 2015, 55, 1427–1434. [Google Scholar] [CrossRef]
- Ibrahim, A.S.S.S.; Al-Salamah, A.A.; El-Badawi, Y.B.; El-Tayeb, M.A.; Antranikian, G. Detergent-, solvent- and salt-compatible thermoactive alkaline serine protease from halotolerant alkaliphilic Bacillus sp. NPST-AK15: Purification and characterization. Extremophiles 2015, 19, 961–971. [Google Scholar] [CrossRef]
- Raval, V.H.; Pillai, S.; Rawal, C.M.; Singh, S.P. Biochemical and structural characterization of a detergent-stable serine alkaline protease from seawater haloalkaliphilic bacteria. Process Biochem. 2014, 49, 955–962. [Google Scholar] [CrossRef]
- Mothe, T.; Sultanpuram, V.R. Production, purification and characterization of a thermotolerant alkaline serine protease from a novel species Bacillus caseinilyticus. 3 Biotech 2016, 6, 1–10. [Google Scholar] [CrossRef]
- Sanchez-Porro, C.; Martin, S.; Mellado, E.; Ventosa, A. Diversity of moderately halophilic bacteria producing extracellular hydrolytic enzymes. J. Appl. Microbiol. 2003, 94, 295–300. [Google Scholar] [CrossRef]
- Sánchez-Porro, C.; Mellado, E.; Pugsley, A.P.; Francetic, O.; Ventosa, A. The haloprotease CPI produced by the moderately halophilic bacterium Pseudoalteromonas ruthenica is secreted by the type II secretion pathway. Appl. Environ. Microbiol. 2009, 75, 4197–4201. [Google Scholar] [CrossRef] [PubMed]
- Lama, L.; Romano, I.; Calandrelli, V.; Nicolaus, B.; Gambacorta, A. Purification and characterization of a protease produced by an aerobic haloalkaliphilic species belonging to the Salinivibrio genus. Res. Microbiol. 2005, 156, 478–484. [Google Scholar] [CrossRef] [PubMed]
- Behera, B.C.; Sethi, B.K.; Mishra, R.R.; Dutta, S.K.; Thatoi, H.N. Microbial cellulases—Diversity & biotechnology with reference to mangrove environment: A review. J. Genet. Eng. Biotechnol. 2017, 15, 197–210. [Google Scholar] [PubMed]
- Maki, M.; Leung, K.T.; Qin, W. The prospects of cellulase-producing bacteria for the bioconversion of lignocellulosic biomass. Int. J. Biol. Sci. 2009, 5, 500–516. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, Y.Q.; de Veras, B.O.; de França, A.F.J.; Gorlach-Lira, K.; Velasques, J.; Migliolo, L.; Dos Santos, E.A. A new salt-tolerant thermostable cellulase from a marine bacillus sp. Strain. J. Microbiol. Biotechnol. 2018, 28, 1078–1085. [Google Scholar] [CrossRef]
- Werner, J.; Ferrer, M.; Michel, G.; Mann, A.J.; Huang, S.; Juarez, S.; Ciordia, S.; Albar, J.P.; Alcaide, M.; La Cono, V.; et al. Halorhabdus tiamatea: Proteogenomics and glycosidase activity measurements identify the first cultivated euryarchaeon from a deep-sea anoxic brine lake as potential polysaccharide degrader. Environ. Microbiol. 2014, 16, 2525–2537. [Google Scholar] [CrossRef]
- Zhang, T.; Datta, S.; Eichler, J.; Ivanova, N.; Axen, S.D.; Kerfeld, C.A.; Chen, F.; Kyrpides, N.; Hugenholtz, P.; Cheng, J.F.; et al. Identification of a haloalkaliphilic and thermostable cellulase with improved ionic liquid tolerance. Green Chem. 2011, 13, 2083–2090. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, X.; Zhang, W.; Zhao, Y.; Lu, X. Expression and characterization of a glucose-tolerant β-1,4-glucosidase with wide substrate specificity from Cytophaga hutchinsonii. Appl. Microbiol. Biotechnol. 2017, 101, 1919–1926. [Google Scholar] [CrossRef]
- Makhdoumi, A.; Dehghani-Joybari, Z.; Mashreghi, M.; Jamialahmadi, K.; Asoodeh, A. A novel halo-alkali-tolerant and thermo-tolerant chitinase from Pseudoalteromonas sp. DC14 isolated from the Caspian Sea. Int. J. Environ. Sci. Technol. 2015, 12, 3895–3904. [Google Scholar] [CrossRef]
- Roman, D.L.; Roman, M.; Sletta, H.; Ostafe, V.; Isvoran, A. Assessment of the properties of chitin deacetylases showing different enzymatic action patterns. J. Mol. Graph. Model. 2019, 88, 41–48. [Google Scholar] [CrossRef]
- Zhao, Y.; Park, R.D.; Muzzarelli, R.A.A. Chitin deacetylases: Properties and applications. Mar. Drugs 2010, 8, 24–46. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.B.; Lee, W.; Ryu, Y.W. Cloning and characterization of thermostable esterase from Archaeoglobus fulgidus. J. Microbiol. 2008, 46, 100–107. [Google Scholar] [CrossRef] [PubMed]
- Ghati, A.; Paul, G. Purification and characterization of a thermo-halophilic, alkali-stable and extremely benzene tolerant esterase from a thermo-halo tolerant Bacillus cereus strain AGP-03, isolated from ‘Bakreshwar’ hot spring, India. Process Biochem. 2015, 50, 771–781. [Google Scholar] [CrossRef]
- Mohamed, Y.M.; Ghazy, M.A.; Sayed, A.; Ouf, A.; El-Dorry, H.; Siam, R. Isolation and characterization of a heavy metal-resistant, thermophilic esterase from a Red Sea Brine Pool. Sci. Rep. 2013, 3, 3358. [Google Scholar] [CrossRef]
- Ferrer, M.; Golyshina, O.V.; Chernikova, T.N.; Khachane, A.N.; Martins, V.A.P.; Santos, D.; Yakimov, M.M.; Timmis, K.N.; Golyshin, P.N. Microbial Enzymes Mined from the Urania Deep-Sea Hypersaline Anoxic Basin. Chem. Biol. 2005, 12, 895–904. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Wu, G.; Liu, Z.; Shao, Z.; Liu, Z. Characterization of EstB, a novel cold-active and organic solvent-tolerant esterase from marine microorganism Alcanivorax dieselolei B-5(T). Extremophiles 2014, 18, 251–259. [Google Scholar] [CrossRef]
- Rahman, M.A.; Culsum, U.; Tang, W.; Zhang, S.W.; Wu, G.; Liu, Z. Characterization of a novel cold active and salt tolerant esterase from Zunongwangia profunda. Enzyme Microb. Technol. 2016, 85, 1–11. [Google Scholar] [CrossRef]
- Garczarek, F.; Dong, M.; Typke, D.; Witkowska, H.E.; Hazen, T.C.; Nogales, E.; Biggin, M.D.; Glaeser, R.M. Octomeric pyruvate-ferredoxin oxidoreductase from Desulfovibrio vulgaris. J. Struct. Biol. 2007, 159, 9–18. [Google Scholar] [CrossRef]
- Bock, A.K.; Schönheit, P.; Teixeira, M. The iron-sulfur centers of the pyruvate:ferredoxin oxidoreductase from Methanosarcina barkeri (Fusaro). FEBS Lett. 1997, 414, 209–212. [Google Scholar]
- Chen, C.K.M.; Lee, G.C.; Ko, T.P.; Guo, R.T.; Huang, L.M.; Liu, H.J.; Ho, Y.F.; Shaw, J.F.; Wang, A.H.J. Structure of the Alkalohyperthermophilic Archaeoglobus fulgidus Lipase Contains a Unique C-Terminal Domain Essential for Long-Chain Substrate Binding. J. Mol. Biol. 2009, 390, 672–685. [Google Scholar] [CrossRef]
- Akbari, E.; Beheshti-Maal, K.; Nayeri, H. A novel halo-alkalo-tolerant bacterium, Marinobacter alkaliphilus ABN-IAUF-1, isolated from Persian Gulf suitable for alkaline lipase production. Int. J. Environ. Sci. Technol. 2018, 15, 1767–1776. [Google Scholar] [CrossRef]
- Jeon, J.H.; Kim, J.-T.; Kim, Y.J.; Kim, H.-K.; Lee, H.S.; Kang, S.G.; Kim, S.-J.; Lee, J.-H. Cloning and characterization of a new cold-active lipase from a deep-sea sediment metagenome. Appl. Microbiol. Biotechnol. 2009, 81, 865–874. [Google Scholar] [CrossRef] [PubMed]
- Loperena, L.; Soria, V.; Varela, H.; Lupo, S.; Bergalli, A.; Guigou, M.; Pellegrino, A.; Bernardo, A.; Calviño, A.; Rivas, F.; et al. Extracellular enzymes produced by microorganisms isolated from maritime Antarctica. World J. Microbiol. Biotechnol. 2012, 28, 2249–2256. [Google Scholar] [CrossRef] [PubMed]
- Kiran, G.S.; Lipton, A.N.; Kennedy, J.; Dobson, A.D.W.; Selvin, J. A halotolerant thermostable lipase from the marine bacterium Oceanobacillus sp. PUMB02 with an ability to disrupt bacterial biofilms. Bioeng. Bugs 2014, 5, 305–318. [Google Scholar]
- Ciok, A.; Dziewit, L. Exploring the genome of Arctic Psychrobacter sp. DAB_AL32B and construction of novel Psychrobacter-specific cloning vectors of an increased carrying capacity. Arch. Microbiol. 2019, 201, 559–569. [Google Scholar] [CrossRef] [PubMed]
- Sayed, A.; Ghazy, M.A.; Ferreira, A.J.S.; Setubal, J.C.; Chambergo, F.S.; Ouf, A.; Adel, M.; Dawe, A.S.; Archer, J.A.C.; Bajic, V.B.; et al. A novel mercuric reductase from the unique deep brine environment of Atlantis II in the Red sea. J. Biol. Chem. 2014, 289, 1675–1687. [Google Scholar] [CrossRef]
- Zhou, P.; Huo, Y.Y.; Xu, L.; Wu, Y.H.; Meng, F.X.; Wang, C.S.; Xu, X.W. Investigation of mercury tolerance in Chromohalobacter israelensis DSM 6768T and Halomonas zincidurans B6T by comparative genomics with Halomonas xinjiangensis TRM 0175T. Mar. Genomics 2014, 19, 15–16. [Google Scholar] [CrossRef]
- Sonbol, S.A.; Ferreira, A.J.S.; Siam, R. Red Sea Atlantis II brine pool nitrilase with unique thermostability profile and heavy metal tolerance. BMC Biotechnol. 2016, 16, 1–13. [Google Scholar] [CrossRef]
- Hii, S.L.; Tan, J.S.; Ling, T.C.; Ariff, A. Bin Pullulanase: Role in Starch Hydrolysis and Potential Industrial Applications. Enzyme Res. 2012, 2012, 1–14. [Google Scholar] [CrossRef]
- Chakdar, H.; Kumar, M.; Pandiyan, K.; Singh, A.; Nanjappan, K.; Kashyap, P.L.; Srivastava, A.K. Bacterial xylanases: Biology to biotechnology. 3 Biotech 2016, 6, 150. [Google Scholar] [CrossRef]
- Araki, T.; Tani, S.; Maeda, K.; Hashikawa, S.; Nakagawa, H.; Morishita, T. Purification and Characterization of β-1,3-Xylanase from a Marine Bacterium, Vibrio sp. XY-214. Biosci. Biotechnol. Biochem. 1999, 63, 2017–2019. [Google Scholar] [CrossRef] [PubMed]
- Wejse, P.L.; Ingvorsen, K.; Mortensen, K.K. Purification and characterisation of two extremely halotolerant xylanases from a novel halophilic bacterium. Extremophiles 2003, 7, 423–431. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.J.; Kwon, Y.K.; Kim, J.H.; Heo, S.J.; Lee, Y.; Lee, S.J.; Shim, W.B.; Jung, W.K.; Hyun, J.H.; Kwon, K.K.; et al. Effective microwell plate-based screening method for microbes producing cellulase and xylanase and its application. J. Microbiol. Biotechnol. 2014, 24, 1559–1565. [Google Scholar] [CrossRef] [PubMed]
- Møller, M.F.; Kjeldsen, K.U.; Ingvorsen, K. Marinimicrobium haloxylanilyticum sp. nov., a new moderately halophilic, polysaccharide-degrading bacterium isolated from Great Salt Lake, Utah. Antonie van Leeuwenhoek, Int. J. Gen. Mol. Microbiol. 2010, 98, 553–565. [Google Scholar] [CrossRef]
- Rattu, G.; Joshi, S.; Satyanarayana, T. Bifunctional recombinant cellulase–xylanase (rBhcell-xyl) from the polyextremophilic bacterium Bacillus halodurans TSLV1 and its utility in valorization of renewable agro-residues. Extremophiles 2016, 20, 831–842. [Google Scholar] [CrossRef]
- Kim, J.; Hong, S.-K. Isolation and Characterization of an Agarase-Producing Bacterial Strain, Alteromonas sp. GNUM-1, from the West Sea, Korea. J. Microbiol. Biotechnol. 2012, 22, 1621–1628. [Google Scholar] [CrossRef]
- Fu, X.T.; Kim, S.M. Agarase: Review of major sources, categories, purification method, enzyme characteristics and applications. Mar. Drugs 2010, 8, 200–218. [Google Scholar] [CrossRef]
- Han, X.; Lin, B.; Ru, G.; Zhang, Z.; Liu, Y.; Hu, Z. Gene Cloning and Characterization of an α-Amylase from Alteromonas macleodii B7 for Enteromorpha Polysaccharide Degradation. J. Microbiol. Biotechnol. 2014, 24, 254–263. [Google Scholar] [CrossRef]
- Sewalt, V.J.; Reyes, T.F.; Bui, Q. Safety evaluation of two α-amylase enzyme preparations derived from Bacillus licheniformis expressing an α-amylase gene from Cytophaga species. Regul. Toxicol. Pharmacol. 2018, 98, 140–150. [Google Scholar] [CrossRef]
- Amoozegar, M.A.; Siroosi, M.; Atashgahi, S.; Smidt, H.; Ventosa, A. Systematics of haloarchaea and biotechnological potential of their hydrolytic enzymes. Microbiology 2017, 163, 623–645. [Google Scholar] [CrossRef]
- Qin, Y.; Huang, Z.; Liu, Z. A novel cold-active and salt-tolerant α-amylase from marine bacterium Zunongwangia profunda: Molecular cloning, heterologous expression and biochemical characterization. Extremophiles 2014, 18, 271–281. [Google Scholar] [CrossRef] [PubMed]
- Amoozegar, M.A.; Malekzadeh, F.; Malik, K.A. Production of amylase by newly isolated moderate halophile, Halobacillus sp. strain MA-2. J. Microbiol. Methods 2003, 52, 353–359. [Google Scholar] [CrossRef]
- de Lourdes Moreno, M.; Pérez, D.; García, M.; Mellado, E.; De Lourdes Moreno, M.; Pérez, D.; García, M.T.; Mellado, E. Halophilic Bacteria as a Source of Novel Hydrolytic Enzymes. Life 2013, 3, 38–51. [Google Scholar] [CrossRef] [PubMed]
- Kiran, K.; Koteswaraiah, P.; Chandra, T.S. Production of Halophilic α-Amylase by Immobilized Cells of Moderately Halophilic Bacillus sp. Strain TSCVKK. Br. Microbiol. Res. J. 2012, 2, 146–157. [Google Scholar] [CrossRef]
- Uotsu-Tomita, R.; Tonozuka, T.; Sakai, H.; Sakano, Y. Novel glucoamylase-type enzymes from Thermoactinomyces vulgaris and Methanococcus jannaschii whose genes are found in the flanking region of the α-amylase genes. Appl. Microbiol. Biotechnol. 2001, 56, 465–473. [Google Scholar] [CrossRef]
- Kim, J.W.; Flowers, L.O.; Whiteley, M.; Peeples, T.L. Biochemical confirmation and characterization of the family-57-like α-amylase of Methanococcus jannaschii. Folia Microbiol. 2001, 46, 467–473. [Google Scholar] [CrossRef]
- Aghajari, N.; Feller, G.; Gerday, C.; Haser, R. Structures of the psychrophilic Alteromonas haloplanctis α-amylase give insights into cold adaptation at a molecular level. Structure 1998, 6, 1503–1516. [Google Scholar] [CrossRef]
- Mageswari, A.; Subramanian, P.; Chandrasekaran, S.; Sivashanmugam, K.; Babu, S.; Gothandam, K.M. Optimization and immobilization of amylase obtained from halotolerant bacteria isolated from solar salterns. J. Genet. Eng. Biotechnol. 2012, 10, 201–208. [Google Scholar] [CrossRef]
- Coronado, M.J.; Vargas, C.; Mellado, E.; Tegos, G.; Drainas, C.; Nieto, J.J.; Ventosa, A. The α-amylase gene amyH of the moderate halophile Halomonas meridiana: Cloning and molecular characterization. Microbiology 2000, 146, 861–868. [Google Scholar] [CrossRef]
- Singh, G.; Verma, A.K.; Kumar, V. Catalytic properties, functional attributes and industrial applications of β-glucosidases. 3 Biotech 2016, 6, 1–14. [Google Scholar] [CrossRef]
- Sun, J.; Wang, W.; Yao, C.; Dai, F.; Zhu, X.; Liu, J.; Hao, J. Overexpression and characterization of a novel cold-adapted and salt-tolerant GH1 β-glucosidase from the marine bacterium Alteromonas sp. L82. J. Microbiol. 2018, 56, 656–664. [Google Scholar] [CrossRef] [PubMed]
- Hebbale, D.; Bhargavi, R.; Ramachandra, T.V. Saccharification of macroalgal polysaccharides through prioritized cellulase producing bacteria. Heliyon 2019, 5, e01372. [Google Scholar] [CrossRef]
- Zhu, B.; Ning, L. Purification and Characterization of a New k-Carrageenase from the Marine Bacterium Vibrio sp. NJ-2. J. Microbiol. Biotechnol. 2016, 26, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Xiao, A.; Zeng, J.; Li, J.; Zhu, Y.; Xiao, Q.; Ni, H. Molecular cloning, characterization, and heterologous expression of a new κ-carrageenase gene from Pseudoalteromonas carrageenovora ASY5. J. Food Biochem. 2018, 42. [Google Scholar] [CrossRef]
- Blanco, K.C.; De Lima, C.J.B.; Monti, R.; Martins, J.; Bernardi, N.S.; Contiero, J. Bacillus lehensis—An alkali-tolerant bacterium isolated from cassava starch wastewater: Optimization of parameters for cyclodextrin glycosyltransferase production. Ann. Microbiol. 2012, 62, 329–337. [Google Scholar] [CrossRef]
- Suriya, J.; Bharathiraja, S.; Krishnan, M.; Manivasagan, P.; Kim, S.-K. Marine Microbial Amylases. In Advances in food and nutrition research; Elsevier Inc. Academic Press: Cambridge, MA, USA, 2016; Volume 79, pp. 161–177. ISBN 978-0-12-804714-9. [Google Scholar]
- Mehta, D.; Satyanarayana, T. Bacterial and archaeal α-amylases: Diversity and amelioration of the desirable characteristics for industrial applications. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef]
- Jabbour, D.; Sorger, A.; Sahm, K.; Antranikian, G. A highly thermoactive and salt-tolerant α-amylase isolated from a pilot-plant biogas reactor. Appl. Microbiol. Biotechnol. 2013, 97, 2971–2978. [Google Scholar] [CrossRef]
- Cowan, D.; Cramp, R.; Pereira, R.; Graham, D.; Almatawah, Q. Biochemistry and biotechnology of mesophilic and thermophilic nitrile metabolizing enzymes. Extremophiles 1998, 2, 207–216. [Google Scholar] [CrossRef]
- Gupta, N.; Balomajumder, C.; Agarwal, V.K. Enzymatic mechanism and biochemistry for cyanide degradation: A review. J. Hazard. Mater. 2010, 176, 1–13. [Google Scholar] [CrossRef]
- Nigam, V.K.; Arfi, T.; Kumar, V.; Shukla, P. Bioengineering of Nitrilases Towards Its Use as Green Catalyst: Applications and Perspectives. Indian J. Microbiol. 2017, 57, 131–138. [Google Scholar] [CrossRef]
- Kuddus, M. Cold-active enzymes in food biotechnology: An updated mini review. J. Appl. Biol. Biotechnol. 2018, 6, 58–63. [Google Scholar]
- Hasan, F.; Shah, A.A.; Hameed, A. Industrial applications of microbial lipases. Enzyme Microb. Technol. 2006, 39, 235–251. [Google Scholar] [CrossRef]
- Mahjoubi, M.; Cappello, S.; Souissi, Y.; Jaouani, A.; Cherif, A. Microbial Bioremediation of Petroleum Hydrocarbon–Contaminated Marine Environments. In Recent Insights in Petroleum Science and Engineering; Zoveidavianpoor, M., Ed.; InTechOpen: London, UK, 2018; pp. 325–350. [Google Scholar]
- Hassanshahian, M.; Cappello, S. Crude Oil Biodegradation in the Marine Environments. In Biodegradation—Engineering and Technology; Rolando Chamy, Ed.; InTechOpen: London, UK, 2013; pp. 101–135. [Google Scholar]
- Floodgate, G.D. Biodegradation of hydrocarbons in the sea. In Water Pollution Microbiology; Mitchell, R., Ed.; Wiley-Interscience: New York, NY, USA, 1972; pp. 153–171. [Google Scholar]
- Jarvis, I.W.H.; Dreij, K.; Mattsson, Å.; Jernström, B.; Stenius, U. Interactions between polycyclic aromatic hydrocarbons in complex mixtures and implications for cancer risk assessment. Toxicology 2014, 321, 27–39. [Google Scholar] [CrossRef] [PubMed]
- Moreno, R.; Jover, L.; Diez, C.; Sardà, F.; Sanpera, C. Ten Years after the Prestige Oil Spill: Seabird Trophic Ecology as Indicator of Long-Term Effects on the Coastal Marine Ecosystem. PLoS ONE 2013, 8. [Google Scholar] [CrossRef]
- Sikkema, J.; de Bont, J.A.; Poolman, B. Mechanisms of Membrane Toxicity of Hydrocarbons. Microbiol. Rev. 1995, 59, 201–222. [Google Scholar] [CrossRef]
- Catania, V.; Santisi, S.; Signa, G.; Vizzini, S.; Mazzola, A.; Cappello, S.; Yakimov, M.M.; Quatrini, P. Intrinsic bioremediation potential of a chronically polluted marine coastal area. Mar. Pollut. Bull. 2015, 99, 138–149. [Google Scholar] [CrossRef]
- Durval, I.J.B.; Resende, A.H.M.; Figueiredo, M.A.; Luna, J.M.; Rufino, R.D.; Sarubbo, L.A. Studies on Biosurfactants Produced using Bacillus cereus Isolated from Seawater with Biotechnological Potential for Marine Oil-Spill Bioremediation. J. Surfactants Deterg. 2018, 22, 349–363. [Google Scholar] [CrossRef]
- Hazen, T.C.; Dubinsky, E.A.; DeSantis, T.Z.; Andersen, G.L.; Piceno, Y.M.; Singh, N.; Jansson, J.K.; Probst, A.; Borglin, S.E.; Fortney, J.L.; et al. Deep-sea oil plume enriches indigenous oil-degrading bacteria. Science 2010, 330, 204–208. [Google Scholar] [CrossRef]
- Kleindienst, S.; Paul, J.H.; Joye, S.B. Using dispersants after oil spills: Impacts on the composition and activity of microbial communities. Nat. Rev. Microbiol. 2015, 13, 388–396. [Google Scholar] [CrossRef]
- Speight, J.G.; El-Gendy, N.S. Introduction to petroleum biotechnology; Elsevier: Amsterdam, The Netherlands, 2017; ISBN 9780128051511. [Google Scholar]
- Fuentes, S.; Méndez, V.; Aguila, P.; Seeger, M. Bioremediation of petroleum hydrocarbons: Catabolic genes, microbial communities, and applications. Appl. Microbiol. Biotechnol. 2014, 98, 4781–4794. [Google Scholar] [CrossRef]
- Fuentes, S.; Barra, B.; Gregory Caporaso, J.; Seeger, M. From rare to dominant: A fine-tuned soil bacterial bloom during petroleum hydrocarbon bioremediation. Appl. Environ. Microbiol. 2016, 82, 888–896. [Google Scholar] [CrossRef] [PubMed]
- Varjani, S.J.; Gnansounou, E. Microbial dynamics in petroleum oilfields and their relationship with physiological properties of petroleum oil reservoirs. Bioresour. Technol. 2017, 245, 1258–1265. [Google Scholar] [CrossRef] [PubMed]
- Coulon, F.; McKew, B.A.; Osborn, A.M.; McGenity, T.J.; Timmis, K.N. Effects of temperature and biostimulation on oil-degrading microbial communities in temperate estuarine waters. Environ. Microbiol. 2007, 9, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Liu, W.; Tian, S.; Wang, W.; Qi, Q.; Jiang, P.; Gao, X.; Li, F.; Li, H.; Yu, H. Petroleum Hydrocarbon-Degrading Bacteria for the Remediation of Oil Pollution Under Aerobic Conditions: A Perspective Analysis. Front. Microbiol. 2018, 9, 1–11. [Google Scholar] [CrossRef]
- Fathepure, B.Z. Recent studies in microbial degradation of petroleum hydrocarbons in hypersaline environments. Front. Microbiol. 2014, 5. [Google Scholar] [CrossRef]
- Oren, A. Aerobic Hydrocarbon-Degrading Archaea. In Taxonomy, Genomics and Ecophysiology of Hydrocarbon-Degrading Microbes; Springer International Publishing: Cham, Switzerland, 2017; pp. 1–12. [Google Scholar]
- Gutierrez, T.; Singleton, D.R.; Berry, D.; Yang, T.; Aitken, M.D.; Teske, A. Hydrocarbon-degrading bacteria enriched by the Deepwater Horizon oil spill identified by cultivation and DNA-SIP. ISME J. 2013, 7, 2091–2104. [Google Scholar] [CrossRef]
- Yakimov, M.M.; Timmis, K.N.; Golyshin, P.N. Obligate oil-degrading marine bacteria. Curr. Opin. Biotechnol. 2007, 18, 257–266. [Google Scholar] [CrossRef]
- McGenity, T.J.; Folwell, B.D.; McKew, B.A.; Sanni, G.O. Marine crude-oil biodegradation: A central role for interspecies interactions. Aquat. Biosyst. 2012, 8, 10. [Google Scholar] [CrossRef]
- Head, I.M.; Jones, D.M.; Röling, W.F.M. Marine microorganisms make a meal of oil. Nat. Rev. Microbiol. 2006, 4, 173–182. [Google Scholar] [CrossRef]
- Barbato, M.; Mapelli, F.; Magagnini, M.; Chouaia, B.; Armeni, M.; Marasco, R.; Crotti, E.; Daffonchio, D.; Borin, S. Hydrocarbon pollutants shape bacterial community assembly of harbor sediments. Mar. Pollut. Bull. 2016, 104, 211–220. [Google Scholar] [CrossRef]
- Barbato, M.; Mapelli, F.; Chouaia, B.; Crotti, E.; Daffonchio, D.; Borin, S. Draft genome sequence of the hydrocarbon-degrading bacterium Alcanivorax dieselolei KS-293 isolated from surface seawater in the Eastern Mediterranean Sea. Genome Announc. 2015, 3. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Shao, Z. The long-chain alkane metabolism network of Alcanivorax dieselolei. Nat. Commun. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Shao, Z. Biochemical characterization of a haloalkane dehalogenase DadB from Alcanivorax dieselolei B-5. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- Scoma, A.; Barbato, M.; Hernandez-Sanabria, E.; Mapelli, F.; Daffonchio, D.; Borin, S.; Boon, N. Microbial oil-degradation under mild hydrostatic pressure (10 MPa): Which pathways are impacted in piezosensitive hydrocarbonoclastic bacteria? Sci. Rep. 2016, 6, 1–14. [Google Scholar] [CrossRef]
- Barbato, M.; Scoma, A.; Mapelli, F.; De Smet, R.; Banat, I.M.; Daffonchio, D.; Boon, N.; Borin, S. Hydrocarbonoclastic alcanivorax isolates exhibit different physiological and expression responses to N-dodecane. Front. Microbiol. 2016, 7, 1–14. [Google Scholar] [CrossRef]
- Sass, A.M.; McKew, B.A.; Sass, H.; Fichtel, J.; Timmis, K.N.; McGenity, T.J. Diversity of Bacillus-like organisms isolated from deep-sea hypersaline anoxic sediments. Saline Systems 2008, 4, 1–11. [Google Scholar] [CrossRef]
- Antunes, A.; Eder, W.; Fareleira, P.; Santos, H.; Huber, R. Salinisphaera shabanensis gen. nov., sp. nov., a novel, moderately halophilic bacterium from the brine-seawater interface of the Shaban Deep, Red Sea. Extremophiles 2003, 7, 29–34. [Google Scholar] [CrossRef]
- Palau, M.; Boujida, N.; Manresa, À.; Miñana-Galbis, D. Complete genome sequence of Marinobacter flavimaris LMG 23834T, which is potentially useful in bioremediation. Genome Announc. 2018, 6, 1–2. [Google Scholar] [CrossRef]
- Marquez, M.C. Marinobacter hydrocarbonoclasticus Gauthier et al. 1992 and Marinobacter aquaeolei Nguyen et al. 1999 are heterotypic synonyms. Int. J. Syst. Evol. Microbiol. 2005, 55, 1349–1351. [Google Scholar] [CrossRef]
- Handley, K.M.; Lloyd, J.R. Biogeochemical implications of the ubiquitous colonization of marine habitats and redox gradients by Marinobacter species. Front. Microbiol. 2013, 4, 1–10. [Google Scholar] [CrossRef]
- Gomes, M.B.; Gonzales-Limache, E.E.; Sousa, S.T.P.; Dellagnezze, B.M.; Sartoratto, A.; Silva, L.C.F.; Gieg, L.L.M.; Valoni, E.; Souza, R.S.; Torres, A.P.R.; et al. Exploring the potential of halophilic bacteria from oil terminal environments for biosurfactant production and hydrocarbon degradation under high-salinity conditions. Int. Biodeterior. Biodegradation 2018, 126, 231–242. [Google Scholar] [CrossRef]
- Kodama, Y.; Stiknowati, L.I.; Ueki, A.; Ueki, K.; Watanabe, K. Thalassospira tepidiphila sp. nov., a polycyclic aromatic hydrocarbon-degrading bacterium isolated from seawater. Int. J. Syst. Evol. Microbiol. 2008, 58, 711–715. [Google Scholar] [CrossRef] [PubMed]
- Budiyanto, F.; Thukair, A.; Al-Momani, M.; Musa, M.M.; Nzila, A. Characterization of Halophilic Bacteria Capable of Efficiently Biodegrading the High-Molecular-Weight Polycyclic Aromatic Hydrocarbon Pyrene. Environ. Eng. Sci. 2018, 35, 616–626. [Google Scholar] [CrossRef]
- Castillo-Carvajal, L.C.; Sanz-Martín, J.L.; Barragán-Huerta, B.E. Biodegradation of organic pollutants in saline wastewater by halophilic microorganisms: A review. Environ. Sci. Pollut. Res. 2014, 21, 9578–9588. [Google Scholar] [CrossRef] [PubMed]
- Paniagua-Michel, J.; Babu, Z. Fathepure Microbial Consortia and Biodegradation of Petroleum Hydrocarbons in Marine Environments. In Microbial Action on Hydrocarbons; Kumar, V., Kumar, M., Prasad, R., Eds.; Springer Nature: Singapore, 2019; pp. 1–20. [Google Scholar]
- Erdoǧmuş, S.F.; Mutlu, B.; Korcan, S.E.; Güven, K.; Konuk, M. Aromatic hydrocarbon degradation by halophilic archaea isolated from ÇamaltI Saltern, Turkey. Water. Air. Soil Pollut. 2013, 224. [Google Scholar] [CrossRef]
- Al-Mailem, D.M.; Eliyas, M.; Radwan, S.S. Oil-bioremediation potential of two hydrocarbonoclastic, diazotrophic Marinobacter strains from hypersaline areas along the Arabian Gulf coasts. Extremophiles 2013, 17, 463–470. [Google Scholar] [CrossRef] [PubMed]
- Edbeib, M.F.; Wahab, R.A.; Huyop, F. Halophiles: Biology, adaptation, and their role in decontamination of hypersaline environments. World J. Microbiol. Biotechnol. 2016, 32, 1–23. [Google Scholar] [CrossRef]
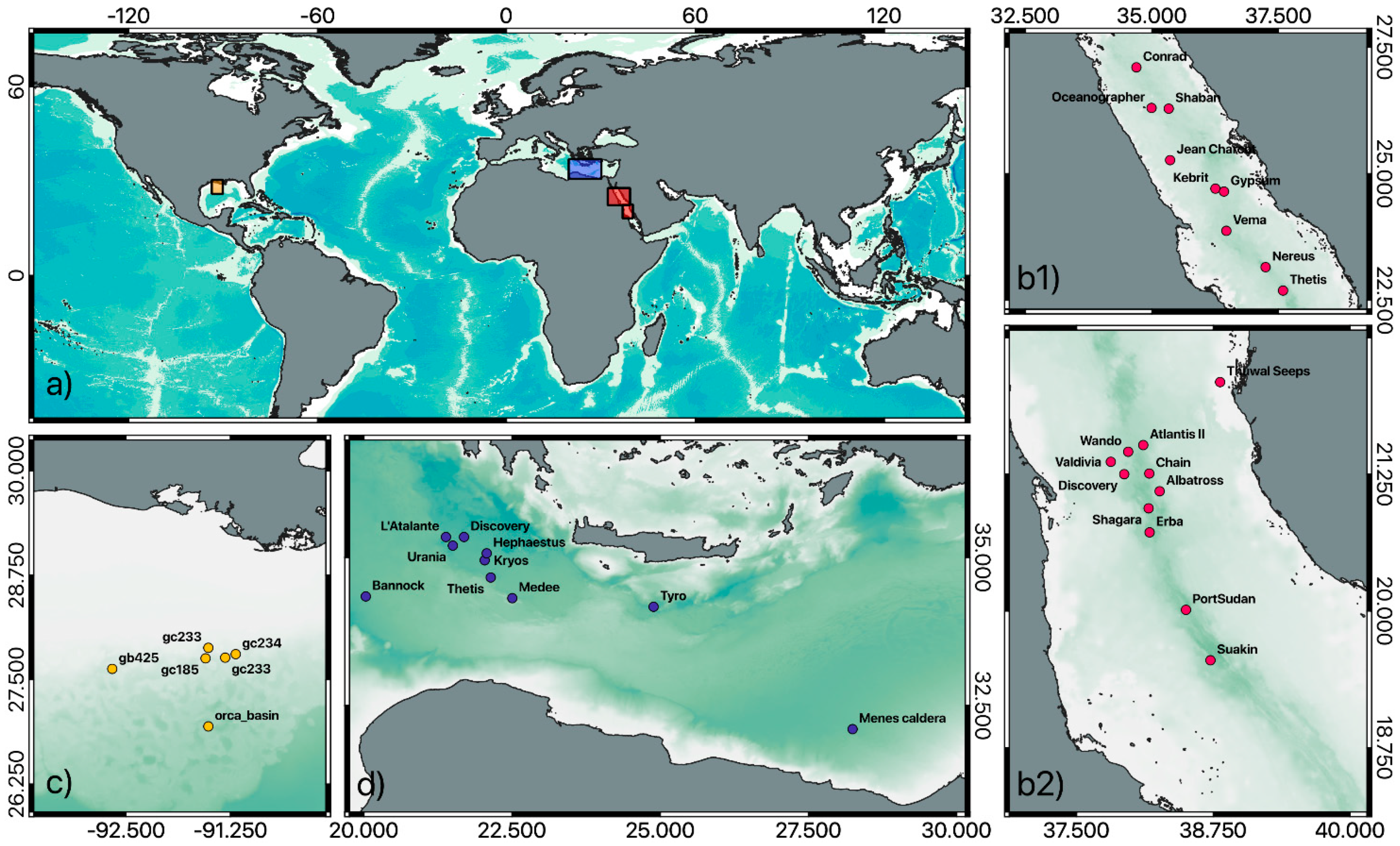
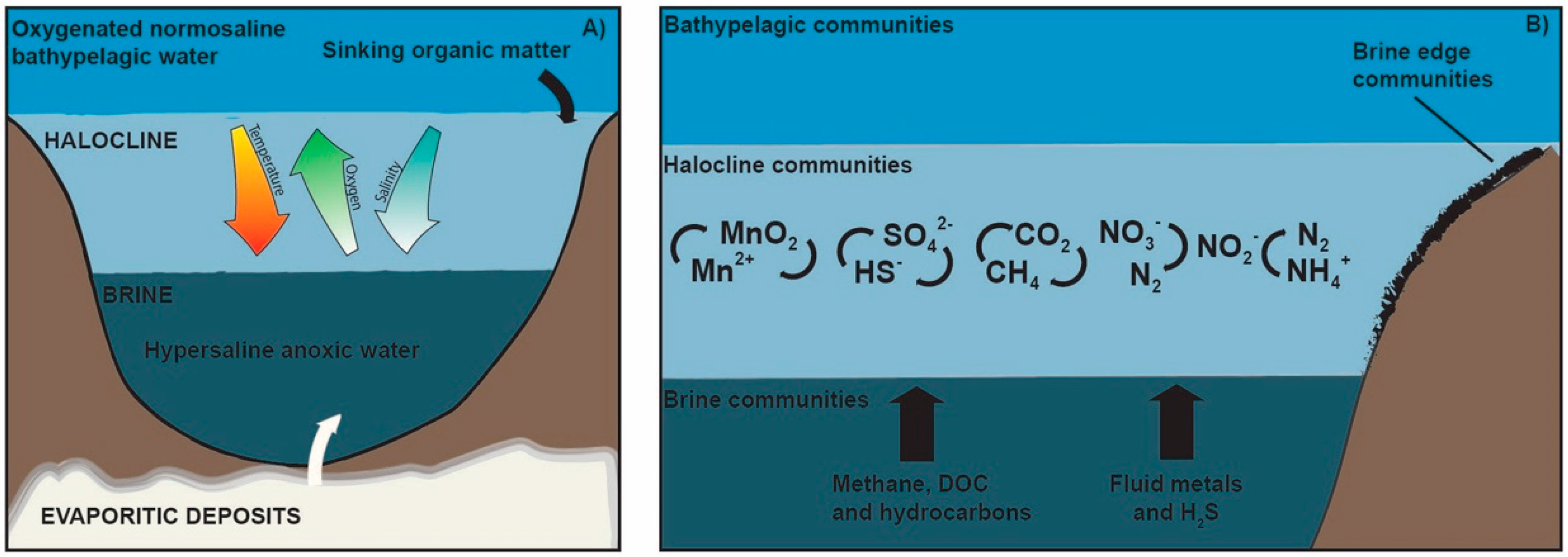
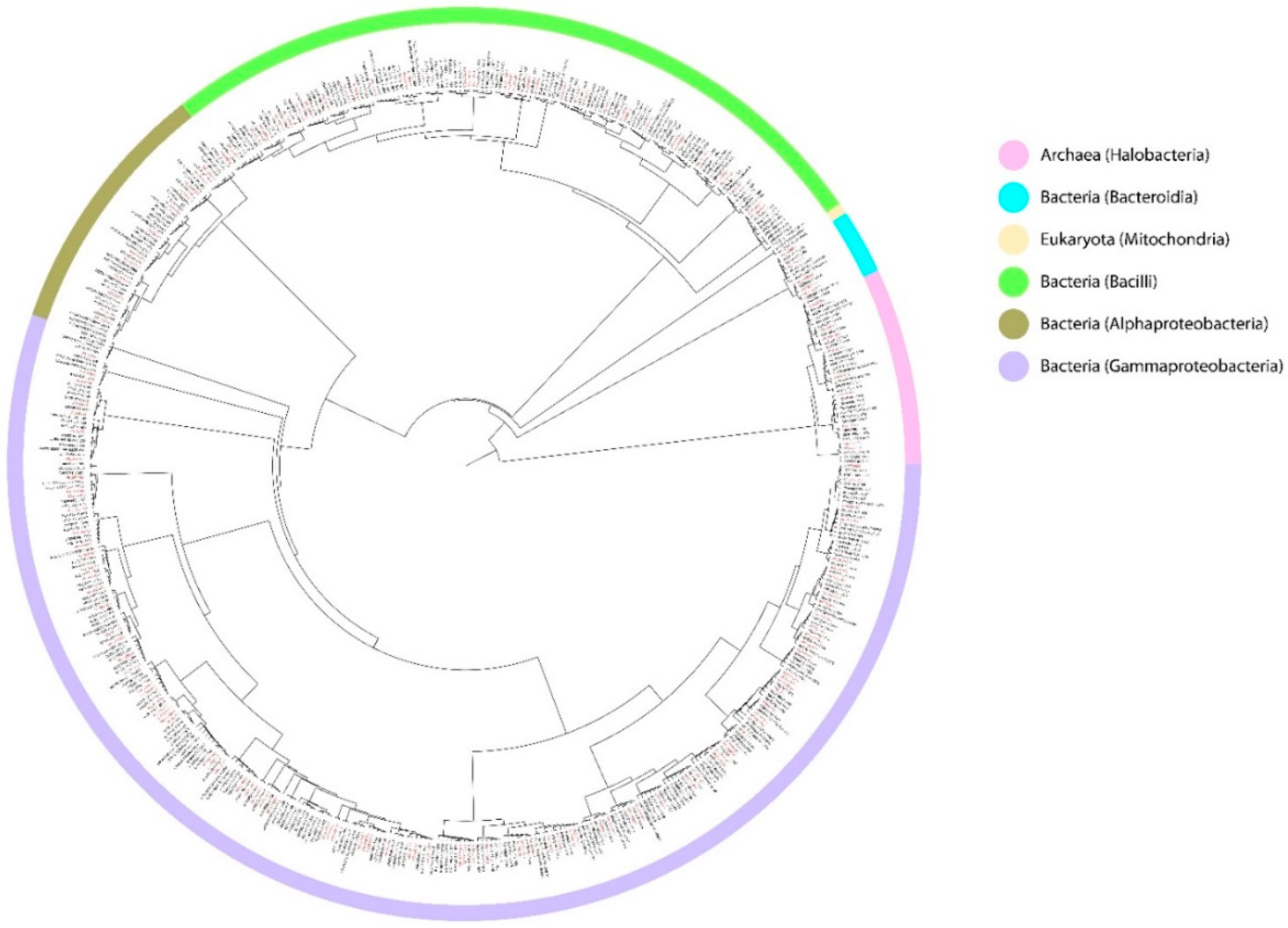
| Environmental Parameters | Ranges | DHABs | Location | References |
|---|---|---|---|---|
| Temperature | Min: 14 °C Max: 68 °C | La Medee Atlantis II | Mediterranean Sea Red Sea | [31] [32] |
| Depth | Min 630 m Max: 3580 m | GC233 Discovery | Gulf of Mexico Mediterranean Sea | [33] [17] |
| Na+ | Min: 1751 mM Max 5300 mM | GC233 Tyro | Gulf of Mexico Mediterranean Sea | [24] [34] |
| Cl- | Min: 2092 mM Max: 10,154.3 mM | GC233 Discovery | Gulf of Mexico Mediterranean Sea | [24] [17] |
| Mg2+ | Min: 8.7 mM Max: 5143 mM | GB425 Discovery | Gulf of Mexico Mediterranean Sea | [24] [17] |
| K+ | Min: 17.2 mM Max: 471 mM | Orca La Medee | Gulf of Mexico Mediterranean Sea | [35] [31] |
| Ca2+ | Min: 1 mM Max: 150 mM | Discovery, Kyros Atlantis II | Mediterranean Sea Red Sea | [17] [36] |
| SO42− | Min: <1 mM Max: 333.1 mM | GB425; GC233 L’Atalante | Gulf of Mexico Mediterranean Sea | [24] [17] |
| Sulfide | Min: 0.002 mM Max: 16 mM | GC233 Urania | Gulf of Mexico Mediterranean Sea | [24] [16] |
| Marine Prokaryotes | Product | Bioactivity | Environmental Sources | Ref. |
|---|---|---|---|---|
| Alteromonas macleodii | Dithiolopyrrolone | Antibiotic and antitumor | Erba and Nereus DHABs | [84,104] |
| Alteromonas sp. B-10-31 | Marinostatins B-1, C1, and C2 | Serine protease inhibitor | Coastal seawater | [105] |
| Bacillus sp. | Macrolactins A–F | Cytotoxic, antimicrobial, antiviral | Deep sea | [106,107,108,109] |
| Bacillus halodurans | Enfuvirtide | Antiviral | Nereus DHAB | [84,110] |
| Bacillus MK-PNG-276A | Loloatins A–D | Antimicrobial | Great barrier reef | [111] |
| Bacillus sp. | Bogorol A | Antimicrobial | Seawater | [112] |
| Bacillus sp. CND-914 | Halobacillin | Antitumor | Deep-sea sediments | [113] |
| Bacillus sp. MIX-62 | Mixirins A–C | Antitumor | [114] | |
| Bacteroidetes rapidithrix HC35 | Ariakemicins A and B | Antimicrobial, cytotoxic | Sea mud | [115] |
| Erythrobacter sp. | Erythrazoles A and B Erythrolic acids A–E | Cytotoxic | Mangrove sediments | [116,117] |
| Halobacteroides lacunaris TB21 | R-LPS | Immunomodulator | Thetis DHAB | [118] |
| Halomonas LOB-5 | Loihichelins A–F | n.a. | Deep sea hydrothermal vents | [119] |
| Halomonas meridiana | n.a. | Antitumor | Nereus DHAB | [84] |
| Halomonas sp. GWS-BW-H8hM | 3-(4′-Hydroxyphenyl)-4-phenylpyrrole-2,5-dicarboxylic acid (HPPD-1 and HPPD-2) | Cytotoxic | Seawater | [120] |
| Halomonas sp. GWS-BW-H8hM | 2-Amino-6-hydroxyphenoxazin-3-one2-Amino-8-benzoyl-phenoxazin-3-one2-Amino-8-(4-hydroxybenzoyl)-6-hydroxyphenoxazin-3-one | Antimicrobial, cytotoxic | Seawater | [121] |
| Pseudoalteromonas carrageenovora IAM 12662 | LPS | Antiviral | Erba DHAB | [84,122] |
| Pseudoalteromonas haloplanktis TAC125 | Peptides | Antioxidant | Antarctic coastal sea water | [123] |
| Pseudoalteromonas mariniglutinosa | n.a. | Antitumor | Erba and Nereus DHABs | [84] |
| Pseudoalteromonas rava SANK 73390 | Thiomarinols A–H and J | Antimicrobial | Seawater | [124,125] |
| Streptomyces aureoverticillatus (NPS001583) | Aureoverticillactam | Antitumor | Marine sediments | [126] |
| Streptomyces C42 | Champacyclin | Antimicrobial | Deep sea | [127] |
| Streptomyces CNH-990 | Marmycins A and B | Cytotoxic | Seawater | [128,129] |
| Streptomyces drozdowiczii SCSIO 10141 | Marfomycins A, B, and E | Anti-infective | Deep sea | [130] |
| Streptomyces drozdowiczii NTK 97 | Frigocyclinone | Antimicrobial | Antarctica | [131] |
| Streptomyces Merv 8102 | Essramycin | Antimicrobial | Marine animals, plants, and sediments | [132] |
| Streptomyces niveus SCSIO 3406 | Marfuraquinocins | Cytotoxic antimicrobial | Deep sea | [133] |
| Streptomyces scopuliridis SCSIO ZJ46 | Desotamide B | Antimicrobial | Deep-sea sediments | [134] |
| Streptomyces sioyaensis SA-1758 | Altemicidin | Cytotoxic, antimicrobial | Sea mud | [135] |
| Streptomyces sp. 12A35 | Lobophorins H and I | Antimicrobial | Deep sea | [136] |
| Streptomyces sp. ART5 | Articoside | Cytotoxic, | Arctic deep sea | [137] |
| Streptomyces sp. CNB-982 | Cyclomarins A–C | anti-inflammatory | Marine sediments | [138,139] |
| Streptomyces sp. CNQ-418 | Marinopyrroles A–F | Antimicrobial, cytotoxic, anti-apoptotic | Deep-sea sediments | [140,141] |
| Streptomyces sp. CNQ-85 | Daryamides A–C (2E,4E)-7-Methylocta-2,4-dienoic acid amide 26 | Antitumor, antifungal | Seawater | [142] |
| Streptomyces sp. CNR-698 | Ammosamides A–D | Cytotoxic | Deep sea | [143,144,145] |
| Streptomyces sp. M045 | Chinikomycins A and B | Antitumor | Seawater | [146] |
| Streptomyces sp. MDF-04-17-069 | Tartrolon D | Cytotoxic | Marine sediments | [147] |
| Streptomyces sp. Mei37 | Mansouramycins A–D | Antimicrobial, cytotoxic | Marine sediments | [148] |
| Streptomyces sp. NTK 935 | Benzoxacystol | Antiproliferative | Deep sea | [149] |
| Streptomyces sp. SCSIO 03032 | Spiroindimicins A–D | Antitumor | Deep sea | [150] |
| Streptomyces sp. SCSIO 11594 | Dehydroxyaquayamycin, Marangucycline B | Antibacterial, antitumor | Deep sea | [151] |
| Streptomyces xiamenensis M1-94P | Xiamenmycin C and D | Anti-fibrotic | Deep-sea sediments | [152] |
| Streptomycete sp. | Piperazimycins A–C | Antitumor | Marine sediments | [153] |
| Zunongwangia profunda SM-A87 | EPS | Antioxidant | Nereus DHAB | [154] |
| Enzyme | Biological Source | Specific Adaptations | Function and/or Applications | Ref. |
|---|---|---|---|---|
| Aldehyde dehydrogenase (EC 1.2.1.3–7) | Bacillus halodurans from Nereus interface; Atlantis II Red Sea brine pool; Cytophaga sp. KUC-1 from Antarctic seawater and Halobacterium salinarum | Slight halophile; thermo- and psychrophilic | Biotransformation of a large number of drugs and other xenobiotics generates aldehydes as intermediates or as products resulting from oxidative deaminations | [172,173,174,175] |
| Protease (EC 3.4.21–25) | Salinivibrio costicola* and Pseudoalteromonas ruthenica* from Erba DHAB. Bacillus circulans BM15 and PseudoAlteromonas sp. 129-1. Bacillus sp. NPST-AK1, Halobacterium halobium (ATCC 43214), Bacillus licheniformis, Bacillus halophilus, Pseudoalteromonas strain EB27, Halomonas meridiana DSM 5425, Bacillus sp. (Ve2-20-91 (HM047794)), and Bacillus caseinilyticus | Haloalkaliphilic and thermotolerant alkaline | Protein hydrolysis finds a broad variety of potential applications in diverse biotechnological processes such as in the feed, food, pharmacology (anticancer and antihemolytic activity) and cosmetic (keratin-based preparation) industries, and cleaning processes (e.g., detergent additive) | [176,177,178,179,180,181,182,183,184,185] |
| Cellulase (EC 3.2.1.4) | Cytophaga hutchinsonii, Halorhabdus tiamatea from Shaban DHAB, Bacillus sp. SR22 from seawater, Bacillus sp., Vibrio sp., Rhodococcus sp., Clostridium and Streptomyces from mangrove Halorhabdus utahensis from Great Salt Lake | Halo-alkali tolerant and thermotolerant | Breakdown of cellulose-producing polysaccharides; potential application in the food, animal feed, beer and wine, textile and laundry, and pulp and paper industries, agriculture, biofuel, pharmaceutical industries, and waste management | [186,187,188,189,190,191] |
| Chitinase (EC 3.2.1.14), chitin deacetylase (EC 3.5.1.41) | Bacillus thuringiensis HBK-51 from soil. PseudoAlteromonas sp. DC14, Vibrio cholerae, Vibrio parahaemolyticus, and Arthrobacter sp. AW19M34-1 from seawater | Halo-alkali tolerant and thermotolerant | Hydrolysis of chitin and hence N-acetyl chitobiose production which in turn can be useful in fermentation research and biomedicine. There have also been applications in the cosmetic and pharmaceutic fields | [192,193,194] |
| Esterase (EC 3.1.1.1) | Zunongwangia profunda* from Atlantis II and Nereus interface and brine pools. Alcanivorax dieselolei B5(T) from Erba interface. Bacillus cereus AGP-03 from hot spring. Archaeoglobus fulgidus | Thermo-halotolerant and metal resistant; cold-active and organic solvent-tolerant | Leather manufacturing, flavor development in the dairy industry, oil biodegradation, and the synthesis of pharmaceuticals and chemicals | [195,196,197,198,199,200] |
| Ferredoxin oxidoreductase (EC 1.2.7.1) | Halorhabdus tiamatea SARL4BT* from Shaban DHAB. Desulfovibrio sp. from Atlantis II DHAB. Methanosarcina barkeri | Low-oxygen tolerant | Oxidation/reduction processes which are applied in the asymmetric oxyfunctionalization of steroids and other pharmaceuticals, synthesis and modification of polymers, oxidative degradation of pollutants, oxyfunctionalization of hydrocarbons, and the construction of biosensors for diverse clinical applications | [189,201,202] |
| Lipase (EC 3.1.1.3) | Idiomarina sp. W33, HaloBacillus sp., and Archaeoglobus fulgidus. Marinobacter alkaliphilus ABN-IAUF-1. Bacillus sp., Arthrobacter sp., Pseudomonas sp., and Psychrobacter sp. from Antarctic marine sediments. Oceano Bacillus sp. PUMB02 from seawater | Halo- alkalitolerant and hyperthermophilic | Hydrolysis of acylglycerols to release fatty acids and lower acylglycerols or glycerol. Lipase enzymes are exploited in the food, beverage, detergent, biofuel production, animal feed, textiles, leather, paper processing, and cosmetic industries | [203,204,205,206,207,208] |
| Mercuric reductase (EC 1.16.1.1) | Atlantis II deep-sea brine. Chromohalobacter israelensis* from Erba and Atlantis II DHABs. Bacillus firmus* from Discovery DHAB | Extreme halophilic and thermophilic | This enzyme can convert toxic mercury ions into relatively inert elemental mercury. It is very useful in waste-water treatments | [209,210] |
| Nitrilase (EC 3.5.5.1) | Red Sea Atlantis II brine | Thermostable and heavy metal tolerant | Nitrilase can hydrolyze a single cyano group in dinitriles or polynitriles, yelding cyanocarboxilic acids, which are used in different kinds of industries, including the food and pharmacology industries; also used for bioremediative purposes | [211] |
| Pullulanase (EC 3.2.1.41) | Bacillus sp. and Streptomyces sp. | Alkaliphilic | Utilized to hydrolyze the α-1,6 glucosidic linkages in starch, enabling a complete and efficient conversion of the branched polysaccharides into small fermentable sugars during the saccharification process | [212] |
| Xylanase (EC 3.2.1.8) and β-Xylosidase (EC 3.2.1.37) | Staphylococcus sp., Arthrobacter sp., Streptomyces sp., and Vibrio sp. XY-214 from seawater. Oceanospirillum linum CL8 and Halorhabdus utahensis from Great Salt Lake. Halorhabdus tiamatea SARL4BT* from Shaban DHAB. Pseudoalteromonas mariniglutinosa* from Erba and Nereus DHAB. Marinimicrobium haloxylanilyticum* from Kebrit DHAB. Zunongwangia profunda* from Nereus and Atlantis II DHABs. Halomonas meridiana* from Bannock, Erba, and Nereus DHABs. Bacillus halodurans* from Nereus interface | Alkali-halotolerant and psychrophilic | Commercial exploitation in the areas of the food, feed, and paper and pulp industries; also used to increase sugar recovery from agricultural residues for biofuel production | [189,213,214,215,216,217,218] |
| α-agarase (EC 3.2.1.158) and β-agarase (EC 3.2.1.81) | Alteromonas macleodii* from Erba, Discovery, and Nereus DHABs. Alteromonas sp. GNUM-1, Alteromonas agarlyticus, Alteromonas sp. strain C-1, Vibrio sp. PO-303, Altermonas sp. SY37-12, and Cytophaga flevensis from seawater and marine sediments | Moderate halophile | Degradation of agar-degrading bacteria used as oriental food; wide applications in the food industry, cosmetics, and medical fields, and as a tool enzyme for biological, physiological, and cytological studies | [219,220,221] |
| α-amylase (EC 3.2.1.1) | PontiBacillus chungwhensis* from Discovery DHAB. Halomonas meridiana* from Nereus, Erba, and Bannock DHABs. Zunongwangia profunda* from Atlantis II and Nereus DHABs. Cytophaga sp. HaloBacillus sp., Bacillus sp. GM8901, Bacillus sp. TSCVKK, and Methanococcus jannaschii. Halobacterium sp. from hypersaline environment. Alteromonas haloplanctis from Antarctic seawater | Moderate halophile and alkali- tolerant; hyperthermophilic | α-amylase has implications in the food, pharmaceutical, and chemical industries; multifunctional amylase exhibits transglycosylation and hydrolysis activities to produce isomaltooligosaccharides, maltooligosaccharides and glucose | [222,223,224,225,226,227,228,229,230,231,232] |
| β-glucosidases (EC 3.2.1.21) | Halorhabdus tiamatea SARL4BT* from Shaban DHAB. Alteromonas sp. L82 from the Mariana Trench. Cytophaga hutchinsonii | Low-oxygen tolerant, cold-adapted, and salt-tolerant | β-glucosidases convert cellobiose and short cellodextrins into glucose. β-glucosidases are widely used in the production of biofuels and ethanol from cellulosic agricultural wastes, in the production of wine, and in the flavor industry. They can cleave phenolic and phytoestrogen glucosides from fruits and vegetables for extracting medicinally important compounds and enhancing the quality of beverages | [189,191,233,234] |
| κ-Carragenases (EC 3.2.1.83) | Pseudoalteromonas carrageenovora* from Erba sediments. Bacillus sp. Alteronomonas sp., Cytophaga sp., and PseudoAlteromonas sp. Pseudomonas sp., Vibrio sp. NJ-2, and Vibrio parahaemolyticus from seawater | Alkali-halotolerant | Production of oligosaccharides with potential applications in the biomedical field, in bioethanol production, in the textile industry, and as a detergent additive | [235,236,237] |
| Cyclodextrin glycosyltransferase (EC 2.4.1.19) | Bacillus lehensis* from Discovery DHAB | Alkali-halotolerant | Cyclodextrins produced by this enzyme have broad, non-toxic applications in the pharmaceutical, cosmetic, and food industries | [66,238] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Varrella, S.; Tangherlini, M.; Corinaldesi, C. Deep Hypersaline Anoxic Basins as Untapped Reservoir of Polyextremophilic Prokaryotes of Biotechnological Interest. Mar. Drugs 2020, 18, 91. https://doi.org/10.3390/md18020091
Varrella S, Tangherlini M, Corinaldesi C. Deep Hypersaline Anoxic Basins as Untapped Reservoir of Polyextremophilic Prokaryotes of Biotechnological Interest. Marine Drugs. 2020; 18(2):91. https://doi.org/10.3390/md18020091
Chicago/Turabian StyleVarrella, Stefano, Michael Tangherlini, and Cinzia Corinaldesi. 2020. "Deep Hypersaline Anoxic Basins as Untapped Reservoir of Polyextremophilic Prokaryotes of Biotechnological Interest" Marine Drugs 18, no. 2: 91. https://doi.org/10.3390/md18020091
APA StyleVarrella, S., Tangherlini, M., & Corinaldesi, C. (2020). Deep Hypersaline Anoxic Basins as Untapped Reservoir of Polyextremophilic Prokaryotes of Biotechnological Interest. Marine Drugs, 18(2), 91. https://doi.org/10.3390/md18020091






