Abstract
Dinoflagellates, a major class of marine eukaryote microalgae composing the phytoplankton, are widely recognised as producers of a large variety of toxic molecules, particularly neurotoxins, which can also act as potent bioactive pharmacological mediators. In addition, similarly to other microalgae, they are also good producers of polyunsaturated fatty acids (PUFAs), important precursors of key molecules involved in cell physiology. Among PUFA derivatives are the prostaglandins (Pgs), important physiological mediators in several physiological and pathological processes in humans, also used as “biological” drugs. Their synthesis is very expensive because of the elevated number of reaction steps required, thus the search for new Pgs production methods is of great relevance. One possibility is their extraction from microorganisms (e.g., diatoms), which have been proved to produce the same Pgs as humans. In the present study, we took advantage of the available transcriptomes for dinoflagellates in the iMicrobe database to search for the Pgs biosynthetic pathway using a bioinformatic approach. Here we show that dinoflagellates express nine Pg-metabolism related enzymes involved in both Pgs synthesis and reduction. Not all of the enzymes were expressed simultaneously in all the species analysed and their expression was influenced by culturing conditions, especially salinity of the growth medium. These results confirm the existence of a biosynthetic pathway for these important molecules in unicellular microalgae other than diatoms, suggesting a broad diffusion and conservation of the Pgs pathway, which further strengthen their importance in living organisms.
1. Introduction
Fatty acids in general and PUFAs in particular have essential functions in living organisms from all taxa, ensuring fundamental homeostatic functions [1]. Their presence is needed for the maintenance of membrane fluidity and other functions such as communication and defence, correct functioning of the cardiovascular and nervous systems, immunity and inflammation-related responses [1,2,3,4,5,6,7].
Marine eukaryotic microorganisms represent a good source of PUFAs, including long chain ω3- and ω6-PUFAs such as eicosapentaenoic acid (EPA), eicosatrienoic acid (ETrA), docosahexaenoic acid (DHA) and arachidonic acid (ARA). These are of particular interest for human health since they are beneficial for the prevention of cardiovascular diseases and chronic inflammation [1]. Microalgae, in particular diatoms, cryptophytes and dinophytes, are good producers of EPA, ETrA and DHA, but less of ARA [1,2,3,4,5,6,7]. While diatoms are rich in EPA, dinophyta are instead particularly rich in DHA. However, fatty acid composition and prevalence depends on the environmental conditions in which the microalgae live, such as temperature, light, macro- and micronutrients availability [8]. The importance of these PUFAs is also related to their function as precursors of eicosanoids, lipid mediators involved in many physiological processes such as defence, signal transduction, allelopathic competition, predator-prey interactions [9,10]. Prostaglandins are synthesized by the action of prostaglandin G/H synthase (PGHS), also known as cyclooxygenase (COX), that transforms fatty acid precursors, through two enzymatic steps, firstly into the highly unstable hydroperoxide prostaglandin G2 (PgG2), which is then rapidly reduced to prostaglandin H2 (PgH2). PgH2 is transformed in turn by specific synthases, namely prostaglandin E, F, D and I synthases (mPTGES, PTGFS, PTGDS and PTGIS) into prostaglandin E2, F2α, D2, I2 (PgE2, PgF2α, PgD2, PgI2), respectively [11]. PgE2 is transformed by the action of isomerases into PgA2 that is then converted to prostaglandin C2 and can be successively converted to PgB2. In addition, PgE2 can be reduced to PgF2α by the 9-oxo-reductase [12]. PgJ2, instead, derives from dehydration of PgD2 [13]. Depending on which precursor is used, ETrA, ARA or EPA, the Pgs produced belong to series 1, 2 or 3, respectively [14].
While their involvement in the physiology of terrestrial organisms have been extensively studied, in marine organisms only scattered studies exist that have established their presence but not yet their function [15]. Very recently, the synthesis of Pgs have been investigated in phytoplanktonic organisms, such as marine diatoms [16,17]. The same Pgs molecules present in terrestrial animals were identified in the centric diatom species Skeletonema marinoi and Thalassiosira rotula. Interestingly, the biosynthetic pathway was found to be differentially expressed during the different growth phases of the cultures, and in the case of Thalassiosira rotula, the COX gene was differentially expressed between cultures growing in different concentrations of silica [17]. Studies on the evolutionary relationships of the diatom COX protein sequence with those from other organisms, showed similarity between diatom and animal COXs. Alignment of the diatom and human sequences denoted a 29% identity and a dendrogram construction showed the clustering of the diatom and human COX sequences in sister clades [15]. The above results indicate the conservation of the biosynthetic pathway for these mediators among the different kingdoms of life, suggesting an important ecological and functional role also in simple organisms.
In mammalians, Pgs regulate important functions such as inflammation, tissue repair and immune response, and have, therefore, attracted much attention in view of high potentials as therapeutic agents. But the supply of Pgs from natural sources is difficult, and at the moment only chemical synthesis allows for their large-scale production. However, chemical synthesis is very expensive and new cheaper methods are needed for the pharmaceutical market [18].
Dinoflagellates, similarly to diatoms, are known to synthesize secondary metabolites that regulate their interactions with other microorganisms [7], which renders them very appealing as natural sources of bioactive molecules [19]. However, because of their toxicity and the tricky growing conditions they need, obtaining high biomass is difficult. Only one non-toxic species is currently grown at the industrial scale for commercial purposes, Crypthecodinium cohnii, a species producing high quantities of DHA [20]. Besides DHA, dinoflagellates are known to also produce high levels of eicosapentaenoic acid (EPA) [21], while arachidonic acid (ARA) is mostly absent.
Despite the high amount of EPAs produced, one of the precursors in the biosynthesis of Pgs, to the best of our knowledge, there are no investigations on the presence of the Pg pathway in this important group of microalgae.
The high PUFA content and the growing interest about the setting-up of dinoflagellate growth conditions to produce bioactive molecules for industrial purposes [19,22], prompted us to perform a deep transcriptome mining of all the dinoflagellate sequenced transcriptomes available in the Marine Microbial Eukaryotes Transcriptome Sequencing Project (MMETSP) iMicrobe database [23]. Here we present the results of mining of these transcriptomes highlighting the presence of enzymes involved in Pgs synthesis and metabolism in this interesting microalgal group. We also present a comparative analysis to investigate the presence and relative expression of enzymes related to Pg metabolism in transcriptomes obtained under different growth conditions for the same clone or species.
2. Results
2.1. Dinoflagellate Transcriptomes
The forty-two dinophyta transcriptomes available in the iMicrobe database have all been considered for data mining, covering 15 genera, 19 species and different combinations of growth conditions listed in Table 1.

Table 1.
List of dinoflagellate species present in the iMicrobe database. For each species, physical-chemical growth conditions and the geographical area from which they have been isolated are reported. Table abbreviations: Day portion in hours: Light portion of the day/night cycle in hours; Exp: Experimental; N.I.: Not indicated; GEO: Geographical; MMETSP: Marine Microbial Eukaryotes Transcriptome Sequencing Project.
The 42 transcriptomes differed in terms of number of assembled sequences not only among species but also among replicates of the same species or clone.
The assemblies’ sizes spanned from only 380 sequences for the species A. sanguinea to 106,664 sequences for A. tamarense-MMETSP0384 (Table 2).

Table 2.
Sequencing statistic data of the dinoflagellate transcriptomes deposited in the iMicrobe database.
The species A. tamarense, K. brevis, strain Wilson and CCMP2229, and L. polyedra showed, even among replicates, different size assemblies. A. tamarense replicates spanned from 8411 to 61,753 transcripts, K. brevis strain Wilson MMETSP0201 and 0202 differed by 11,600 transcripts, the four replicates of K. brevis strain CCMP2229 spanned from 2594 to 11,371 transcripts, and L. polyedra replicates from 709 to 2423 transcripts.
These differences, even if not very large in terms of percentages of the total number of transcripts per transcriptome, are still relevant as thousands of sequences can represent intra-clone hidden functions and underline the necessity to perform the sequencing of a larger number of replicates.
2.2. Prostaglandin Enzyme Identification
The search for the term “prostaglandin” in the Swiss-Prot annotation tables, identified nine Pgs metabolism-related functions. The identified functions were: Prostaglandin G/H synthase 2 (PTG/HS2 or COX2), prostaglandin E synthase 2 (PTGES2), hematopoietic prostaglandin D synthase (HPGDS), prostaglandin-E(2) 9-reductase (PGE2-9-OR), prostaglandin F synthase 1 and 2 (PTGFS1 and PTGFS2), 15-hydroxyprostaglandin dehydrogenase [NAD+] (15-PGDH), and prostaglandin reductase 1 and 2 (PTGR1 and PTGR2). The functions annotated as COX2, PTGES2, HPGDS, PGE2-9-OR, PTGFS1 and PTGFS2 are associated to enzymes involved in Pgs synthesis, while the ones annotated as 15-PGDH, PTGR1 and PTGR2 are associated to enzymes involved in Pgs catabolism [24,25]. The presence of both synthesising and catabolising Pgs-related functions strongly support the existence of an active Pgs metabolism in dinoflagellates. Overall, fourteen out of 19 species and 26 out of 42 transcriptomes had at least one annotated function related to Pg metabolism (Table 3).

Table 3.
Number of species and transcriptomes in which each prostaglandin metabolism related function was found annotated.
None of the selected transcriptomes expressed the complete set of identified functions (Table 4). The assemblies related to K. brevis and G. foliaceum were the ones expressing the majority of functions, since six of the nine enzymes involved in Pgs synthesis were annotated (Table 4).

Table 4.
List of the fourteen species presenting annotated enzymes related to prostaglandin metabolism. The occurrence of each enzyme per species is indicated. Abbreviations: Hematopoietic prostaglandin D synthase: HPGDS; prostaglandin reductase 1: PTGR1; prostaglandin E synthase 2: PTGES2; prostaglandin-E(2) 9-reductase: PGE2-9-OR; prostaglandin G/H synthase 2: PTG/HS2 (COX2); prostaglandin F synthase 1: PTGFS1; prostaglandin F synthase 2: PTGFS2; prostaglandin reductase 2: PTGR2; 15-hydroxyprostaglandin dehydrogenase [NAD+]: 15-PGDH.
PTGES2, HPGDS, PGE2-9-OR, PTGFS1 and PTGFS2 were widely annotated among the transcriptomes, while COX2, the enzyme that executes the initial step for the synthesis of Pgs, was annotated only in two species, A. massartii and P. glacialis (Table 4). This result suggests that COX2 may need special conditions to be expressed at detectable levels.
2.3. Clustering of Transcripts Associated to the Pgs-Related Enzymes
Most of the annotated Pgs-related enzymes were associated to more than one transcript.
To exclude redundant sequences inside the transcriptomes, all the transcripts associated to the Pg pathway were analysed with the CD-Hit software.
The analysis of all the transcripts of all the species altogether retrieved 102 clusters. Each cluster was very specific including only transcripts having the same function and coming from the same species, confirming the species specificity of each gene function and sequence equality of each transcript among different strains of a species.
The analysis was also performed for each species separately leading to the exclusion of the redundant transcripts (Table 5) and to the identification, in some species, of more than one independent protein, and thus genes, for HPGDS, PTGES2, PTGR1 and PTGR2 (Table 5).

Table 5.
CD-Hit clustering of the transcripts associated to each Pgs-related function. Table abbreviations: SP: Number of transcripts associated to each function by blast against the Swiss Prot database; CD-Hit: Number of genes associated to each function after clustering analysis. Number in merged cells indicated that before and after the clustering the gene number did not change.
According to these analyses, HPGDS, PTGES2 and PTGR1 were the most widespread functions in almost all the species considered (Figure 1).
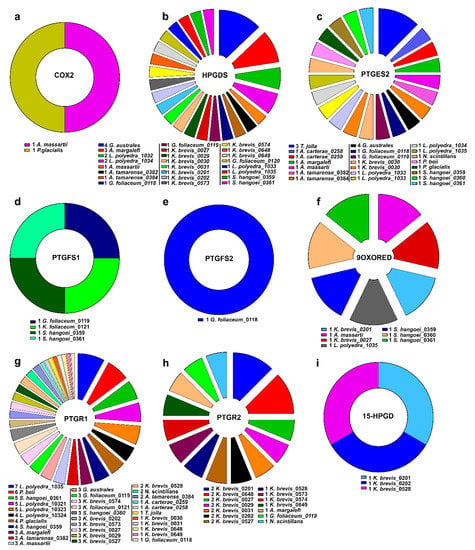
Figure 1.
Occurrence of Pgs-related functions in each dinoflagellate species.
2.4. In-Silico Analysis of Expression Levels
The in-silico expression analysis of the selected transcript for each species showed a large variability in expression levels both among different strains of the same species, culture replicates and different growing conditions. Figure 2 highlights the differences among species in relation to Pg functions, based on Fragments Per Kilobase per Million mapped reads (FPKM) values calculated for each transcriptome.
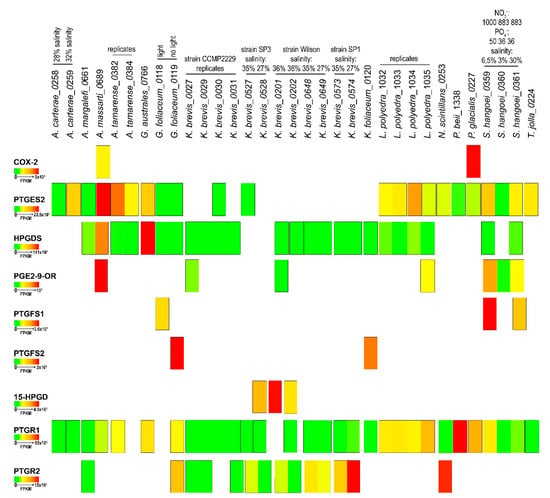
Figure 2.
Transcript expression of the prostaglandin-related functions. Heat map representation of the expression levels, in Fragments per Kilobase per Million Mapped Reads (FPKM), of the Pgs metabolism related enzymes identified in each transcriptome of the different dinoflagellate species.
The results indicated that the COX2 transcript showed the greatest expression in P. glacialis. On the other hand, PTGES2, widely expressed in the species analysed, showed the highest FPKM value in A. massartii. Interestingly, in K. brevis, of the four different strains and two replicates for each condition, PTGES2 was expressed only in one replicate for each of the strains CCMP2229 and SP3. HPGDS expression was very low and at similar expression levels in all species except for G. australes and A. massartii (Figure 2). PGE2-9-OR was highly expressed in A. massartii, which also showed the highest expression with respect to other species such as CCMP2229 and Wilson K. brevis strain, L. polyedra and S. hangoei. PTGFS1 and PTGFS2 were found expressed only in three species, G. foliaceum, K. foliaceum and S. hangoei with highest values of PTGFS1 in S. hangoei and highest values of PTGFS2 in G. foliaceum. 15-HPGD was expressed only in the Wilson and SP3 strains of K. brevis, with one replicate of the Wilson strain having a higher expression with respect to one replicate of the strain SP3. PTGR2 was expressed only in A. margalefi, G. foliaceum, N. scintillans and some strains of K. brevis, one of which showed the highest expression value. PTGR1 instead was more expressed in P. beii with respect to all other species that showed comparable expression levels.
Figure 3 reports the gene expression levels of each Pg function in each transcriptomes grouped according to the species and replicates. These analyses highlighted a lack of reproducibility in the expression levels among replicates (A. tamarense, L. polyedra, K. brevis-strain CCMP2229, K. brevis- Wilson strain, Figure 3).
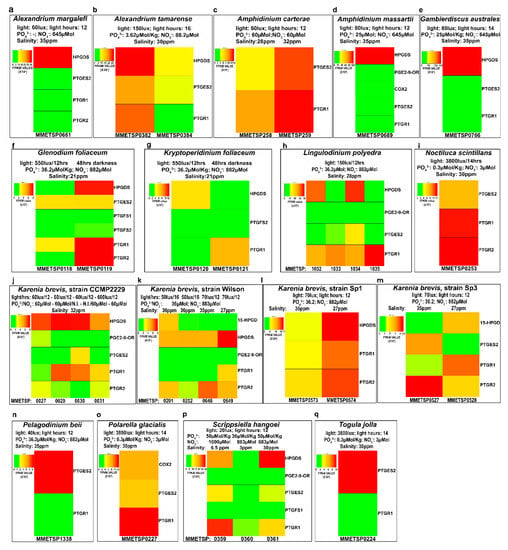
Figure 3.
Pg-related transcript expression for each species. The heat maps represent the expression of each Pg-related transcript, expressed in Fragment Per Kilobase per Million mapped reads (FPKM), for each species, grouping the strains and culture conditions.
Overall, HPGDS, when present, was the most expressed function (Figure 3a,b,d,e,f,h,j,l,p) with respect to the others.
Different light conditions induced differential expression of the Pgs-related transcripts in G. foliaceum and its basionym K. foliaceum, in which the different light conditions of growth showed appreciable differences in terms of both genes expressed and intensity of expression (Figure 3f,g).
K. brevis is one of the best represented species in the database, being present with 12 transcriptomes for four strains. Unfortunately, we could not extrapolate very good information from these 12 transcriptomes since, even among equal replicates of the same strain (Figure 3j, strain CCMP2229), reproducibility was not appreciable both as type of gene expressed and level of expression. In K. brevis strain SP1, lower salinity percentages seemed to induce higher expression of HPGDS, PTGR1 and PTGR2 (Figure 3l), while in K. brevis strain SP3 PTGES2 and PTGR2 are downregulated in conditions of lower salinity (Figure 3m).
S. hangoei (Figure 3p) showed differences among different salinity treatments, while the genus Amphidinium (Figure 3c,d) showed differences between the two species analysed, A. carterae and A. massartii. A. carterae (Figure 3c), in both conditions expressed only two genes, PTGES2 and PTGR1, while A. massartii (Figure 3d) expressed almost the complete set of genes including COX2 and the enzymes for PgE2, PgF2α, PgD2 synthesis and degradation (PTGR1).
The genus Alexandrium (Figure 3a,b) showed variability among equal replicates and differences among different species of the same genus (i.e., A. margalefi (Figure 3a) and A. tamarense (Figure 3b)).
All four L. polyedra replicates had expression of HPGDS, PGE2-9-OR, PTGES2 and PTGR1, except for PGE2-9-OR. In addition, in this case, the data showed no reproducibility (Figure 3h).
S. hangoei at higher salinity conditions had higher HPGDS expression while higher PTGR1 expression at lower percentage salinity (Figure 3p).
3. Discussion
Prostaglandins are key physiological mediators involved in many important physiological processes in animals. In terrestrial animals, they have been widely studied and their role in inflammation, development, pregnancy, sexual reproduction and defence have been established and correlated to the three series of Pgs molecules [26].
Different types of Pgs have been discovered also in marine organisms but their role in the marine environment still awaits to be discovered [15].
Recently, we identified a wide set of Pgs in diatoms—eukaryotic unicellular microalgae widely distributed in most aquatic environments. This discovery was surprising, as their presence in the vegetal kingdom is still not ascertained and their discovery in so “simple” and ancient organisms posed interesting ecological and evolutionary questions [16].
Apart from diatoms, among the phytoplanktonic microalgae, dinoflagellates are important primary producers in the marine food webs. As other microalgae, they are good PUFA producers, particularly of the ω3 type DHA [7,8,20].
Dinoflagellates are well recognised as producers of very powerful bioactive molecules. Some examples include macrolides, cyclic polyethers, spirolides and purine alkaloids, fatty acids, pigments and polysaccharides that strongly affect biological receptors and metabolic processes, such as inflammation, pain, infection and others [19]. In addition, dinoflagellates are one of the most important microalgal groups responsible for the occurrence of harmful algal blooms (HABs). During HABs they produce potent toxins such as saxitoxins, gonyautoxins, brevetoxins, yessotoxins, ciguatoxins, maitotoxins, azaspiracid toxins, and palytoxin [27]. Although these toxins can cause severe poisoning, they may have interesting pharmacological applications due to their chemical structure and mechanisms of action [27].
There is a current urgent need for antibiotics, anti-cancer and anti-inflammatory agents and dinoflagellates may provide new compounds to meet these needs [19,28]. The genus Amphidinium for example is a good producer of macrolides and polyketides with cytotoxic activity against tumour cell lines [29,30]. Karlodinium veneficum is another good example of species useful for pharmaceutical purposes, since it produces a group of toxic metabolites named karlotoxins with haemolytic, cytotoxic and ichthyotoxic activity [28]. Their mechanism of action is based on the formation of pores in the cell membrane that destroys the osmotic balance of the cells leading to cell death [28]. This cell membrane pore formation is being used to develop a new pharmacophore anti-cholesterol therapy [28].
In this context of urgent need for new biologically active molecules, the use of omics technologies applied to the discovery of new natural products from simple marine microorganisms is growing day by day. Numerous studies demonstrate the reliability of RNA-seq to find secondary metabolic pathways for potent bioactive molecules both in terms of gene sequence accuracy and level of their expression [23,31]. From this point of view, dinoflagellates are still a “hard” subject to work on because of their large and very structurally complex genomes (elevated amount of introns, repetitive redundant non-coding sequences, unusual bases, lack of recognizable promoter features and typical eukaryotic transcription factors) that render the identification of bioactive-molecules-related genes very complex [19]. Indeed, only one genome has been partially sequenced until now, the one of Symbiodinium minutum. Nonetheless, omics technologies, particularly proteomics and transcriptome sequencing, are now being applied to identify toxin related genes [32,33,34,35]. The Moore foundation has helped this process by funding the sequencing of many dinoflagellate transcriptome species [23].
Using the sequencing approach coupled to deep analysis level of data mining, we have been able to find also in dinoflagellates the enzymes involved in prostaglandin biosynthesis (Figure 4).
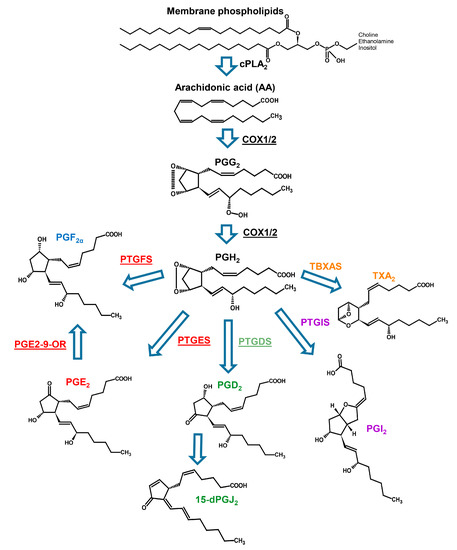
Figure 4.
Prostaglandin biosynthetic pathway. Enzymes involved in the pathway are reported as underlined text next to the arrows. For the abbreviation, refer to the text (Modified from Di Costanzo et al. 2019 [15]).
Our results suggest that dinoflagellates have the potential to synthesize at least three types of Pgs, namely PgE, PgD and PgF2α, due to the presence of prostaglandin E2, D2, F1 and F2 synthases, although COX is expressed at detectable levels only in two species, A. massartii and P. glacialis. The presence in many species of enzymes reducing PgE2, PTGR1 and PTGR2 and 15-HPGD, suggests that PgE2 may be synthesized even in the absence of proper cyclooxygenase activity. Indeed, in the diatom Phaeodactylum tricornutum, Pgs are synthetized in an enzyme independent manner [36], suggesting that this could also be the case for dinoflagellate species in which the expression of COX transcripts is not detectable. On the other hand, a very low level of expression, undetectable by sequencing methods adopted, can be the reason for the absence of the COX transcripts. This was the case for a S. marinoi strain, named FE60, that, with respect to another strain, named FE7, had an RNA-seq undetectable level of COX expression that was conversely detectable by qPCR technique [16].
In-silico expression analysis reported in the present work revealed interesting differences among different strains of the same species grown in similar (e.g., K. brevis strains) or different conditions (i.e., light versus dark; nutrient depletion; salinity). These results further demonstrate the capability of different strains of a same species, to differently express secondary metabolites [7].
Despite sequencing technology is now robust and reliable, some remarks are warranted on the potential results that are obtained from the in-silico analysis of transcriptomes. From our analysis, assemblies’ size of the transcriptomes analysed was different even among replicates of the same strain. This could be due to the occurrence of intra-clone hidden functions and could be the reason why we find the COX2 function annotated only in two species despite the annotation of the other synthetic and reducing functions of Pgs. These observations underline the necessity to sequence a statistically sufficient number of replicates to obtain robust results from bioinformatic analysis. Finally, experimental approaches to study gene expression and chemical identification of Pgs are needed to confirm the functionality of Pg pathway in dinoflagellates.
4. Materials and Methods
4.1. Transcriptomes Collection
The iMicrobe (https://www.imicrobe.us/#/projects/104) database has been questioned with the word “Dinoflagellate” to select all the sequenced transcriptomes in this group of microalgae.
Nt.fa. pep.fa, swissprot.gff3, cds.dat, contig.dat and stats.txt file were downloaded for each species listed in Table 1. Attributes listed in each assembly page were taken to create the physical-chemical information provided in Table 1. The stats.txt files were applied to extrapolated transcriptome sequences statistics to create Table 2.
4.2. Prostaglandin Enzymes Identification
The term “prostaglandin” was used to query the swissprot tables of each transcriptome.
4.3. Gene Clustering
All the id corresponding to prostaglandin functions were used to retrieve the corresponding peptide sequences from the pep.fa files. CD-Hit software (http://weizhongli-lab.org/cd-hit/) was used to cluster and compare the repetitive sequences of each transcript associated to a Pg-related function. Sequences considered head of the cluster were used for the following analysis.
4.4. Gene Expression Analysis
Gene expression levels were calculated using the contig.dat reads data using the fragments per kilobase per million mapped reads (FPKM) formula: [mapped reads pairs]/([length of transcript]/1000)/([total reads pairs]/106). If more than one transcript per Pg-related function was present, the corresponding FPKM values were added together, considering that the level of expression of a gene derives from the sum of all the genes expressing the same function at the same moment. Expression levels were represented by heat map function calculated using R scripts (https://www.r-project.org/).
5. Conclusions
The transcriptome mining approach used in this work succeeded in revealing the presence of Pg pathway in 14 out of 19 dinoflagellate species analysed. Only K. brevis possesses an almost complete set of enzymes involved in the pathway, although no annotation for COX was identified. This enzyme, catalysing the initial step of Pg biosynthesis, was found annotated only in two species, A. massartii and P. glacialis. The CD-HIT analysis excluded transcript redundancy (Table 5) and identified, in some species, more than one independent protein for HPGDS, PTGES2, PTGR1 and PTGR2 (Table 5).
In-silico analysis of gene expression levels showed differences only in G. foliaceum under different light conditions, while in S. hangoei HPGDS and PTGR1 were differentially expressed in different salinity conditions.
These findings pave the way for future investigations through experimental gene expression analysis and chemical identification, in order to confirm Pgs presence in dinoflagellates and stimulate further researches toward the understanding of their ecological role in nature.
Author Contributions
Conceptualization, V.D.D., G.R. and A.I.; methodology, V.D.D.; software, V.D.D.; investigation, V.D.D.; data curation, V.D.D.; writing—original draft preparation, V.D.D.; writing—review and editing, V.D.D., G.R. and A.I.; supervision, G.R. and A.I.; project administration, G.R.; funding acquisition, G.R. All authors have read and agree to the published version of the manuscript.
Funding
This research was funded by Ministero dell’ambiente e della tutela del territorio e del mare, Italia, project name: “Genomics for a Blue Economy,” grant number PGR00765 and Antitumor Drugs and Vaccines from the Sea (ADVISE) project _(PG/2018/0494374).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Wiktorowska-Owczarek, A.; Berezińska, M.; Nowak, J.Z. PUFAs: Structures, Metabolism and Functions. Adv. Clin. Exp. Med. 2015, 24, 931–941. [Google Scholar] [CrossRef] [PubMed]
- Ikawa, M. Algal polyunsaturated fatty acids and effects on plankton ecology and other organisms. UNH Center Freshw. Biol. Res. 2004, 6, 17–44. [Google Scholar]
- Gladyshev, M.I.; Sushchik, N.N.; Makhutova, O.N. Production of EPA and DHA in aquatic ecosystems and their transfer to the land. Prostaglandins Other Lipid Mediat. 2013, 107, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Guschina, I.A.; Harwood, J.L. Lipids and lipid metabolism in eukaryotic algae. Prog. Lipid Res. 2006, 45, 160–186. [Google Scholar] [CrossRef] [PubMed]
- Uttaro, A.D. Biosynthesis of polyunsaturated fatty acids in lower eukaryotes. IUBMB Life 2006, 58, 563–571. [Google Scholar] [CrossRef]
- Andreou, A.; Brodhun, F.; Feussner, I. Biosynthesis of oxylipins in non-mammals. Prog. Lipid Res. 2009, 48, 148–170. [Google Scholar] [CrossRef]
- Peltomaa, E.; Hällfors, H.; Taipale, S.J. Comparison of diatoms and dinoflagellates from different habitats as sources of PUFAs. Mar. Drugs 2019, 17, 233. [Google Scholar] [CrossRef]
- Jónasdóttir, S.H. Fatty acid profiles and production in marine phytoplankton. Mar Drugs 2019, 17, 151. [Google Scholar] [CrossRef]
- Capra, V.; Rovati, G.E.; Mangano, P.; Buccellati, C.; Murphy, R.C.; Sala, A. Transcellular biosynthesis of eicosanoid lipid mediators. BBA—Mol. Cell Biol. Lipids 2015, 1851, 377–382. [Google Scholar] [CrossRef]
- Stonik, V.; Stonik, I. Low-Molecular-Weight metabolites from diatoms: Structures, biological roles and biosynthesis. Mar. Drugs 2015, 13, 3672–3709. [Google Scholar] [CrossRef]
- Clària, J. Cyclooxygenase-2 biology. Curr. Pharm. Des. 2003, 9, 2177–2190. [Google Scholar] [CrossRef] [PubMed]
- Piper, P.J. Introduction to the biosynthesis and metabolism of prostaglandins. Postgrad. Med. J. 1977, 53, 643–646. [Google Scholar] [CrossRef] [PubMed]
- Kawamoto, Y.; Nakamura, Y.; Naito, Y.; Torii, Y.; Kumagai, T.; Osawa, T.; Ohigashi, H.; Satoh, K.; Imagawa, M.; Uchida, K. Cyclopentenone prostaglandins as potential inducers of phase II detoxification enzymes. 15-deoxy-delta(12,14)-prostaglandin j2-induced expression of glutathione S-transferases. J. Biol. Chem. 2000, 275, 11291–11299. [Google Scholar] [CrossRef]
- Wada, M.; DeLong, C.J.; Hong, Y.H.; Rieke, C.J.; Song, I.; Sidhu, R.S.; Yuan, C.; Warnock, M.; Schmaier, A.H.; Yokoyama, C.; et al. Enzymes and receptors of prostaglandin pathways with arachidonic acid-derived versus eicosapentaenoic acid-derived substrates and products. J. Biol. Chem. 2007, 282, 22254–22266. [Google Scholar] [CrossRef]
- Di Costanzo, F.; Di Dato, V.; Ianora, A.; Romano, G. Prostaglandins in marine organisms: A review. Mar. Drugs 2019, 17, 428. [Google Scholar] [CrossRef]
- Di Dato, V.; Orefice, I.; Amato, A.; Fontanarosa, C.; Amoresano, A.; Cutignano, A.; Ianora, A.; Romano, G. Animal-like prostaglandins in marine microalgae. ISME J. 2017, 11, 1722–1726. [Google Scholar] [CrossRef]
- Di Dato, V.; Di Costanzo, F.; Barbarinaldi, R.; Perna, A.; Ianora, A.; Romano, G. Unveiling the presence of biosynthetic pathways for bioactive compounds in the Thalassiosira rotula transcriptome. Sci. Rep. 2019, 9, 9893. [Google Scholar] [CrossRef]
- Yilmaz, E. Biotechnological production of prostaglandin. Biotechnol. Adv. 2001, 19, 387–397. [Google Scholar] [CrossRef]
- Assunção, J.; Guedes, A.C.; Malcata, F.X. Biotechnological and pharmacological applications of biotoxins and other bioactive molecules from dinoflagellates. Mar. Drugs 2017, 15, 393. [Google Scholar] [CrossRef]
- Mendes, A.; Reis, A.; Vasconcelos, R.; Guerra, P.; Lopes da Silva, T. Crypthecodinium cohnii with emphasis on DHA production: A review. J. Appl. Phycol. 2009, 21, 199–214. [Google Scholar] [CrossRef]
- Řezanka, T.; Lukavský, J.; Nedbalová, L.; Sigler, K. Lipidomic profile in three species of dinoflagellates (Amphidinium carterae, Cystodinium sp., and Peridinium aciculiferum) containing very long chain polyunsaturated fatty acids. Phytochemistry 2017, 139, 88–97. [Google Scholar] [CrossRef] [PubMed]
- Beuzenberg, V.; Mountfort, D.; Holland, P.; Shi, F.; MacKenzie, L. Optimization of growth and production of toxins by three dinoflagellates in photobioreactor cultures. J. Appl. Phycol. 2012, 24, 1023–1033. [Google Scholar] [CrossRef]
- Keeling, P.J.; Burki, F.; Wilcox, H.M.; Allam, B.; Allen, E.E.; Amaral-Zettler, L.A.; Armbrust, E.V.; Archibald, J.M.; Bharti, A.K.; Bell, C.J.; et al. The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): Illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol. 2014, 12, e1001889. [Google Scholar] [CrossRef]
- Tai, H.-H.; Ensor, C.M.; Tong, M.; Zhou, H.; Yan, F. Prostaglandin catabolizing enzymes. Prostaglandins Other Lipid Mediat. 2002, 68–69, 483–493. [Google Scholar] [CrossRef]
- Mesa, J.; Alsina, C.; Oppermann, U.; Parés, X.; Farrés, J.; Porté, S. Human prostaglandin reductase 1 (PGR1): Substrate specificity, inhibitor analysis and site-directed mutagenesis. Chem. Biol. Interact. 2015, 234, 105–113. [Google Scholar] [CrossRef]
- Versteeg, H.H.; van Bergen en Henegouwen, P.M.; van Deventer, S.J.; Peppelenbosch, M.P. Cyclooxygenase-dependent signalling: Molecular events and consequences. FEBS Lett. 1999, 445, 1–5. [Google Scholar] [CrossRef]
- Wang, D.-Z. Neurotoxins from marine dinoflagellates: A brief review. Mar. Drugs 2008, 6, 349–371. [Google Scholar] [CrossRef]
- Waters, A.L.; Hill, R.T.; Place, A.R.; Hamann, M.T. The expanding role of marine microbes in pharmaceutical development. Curr. Opin. Biotechnol. 2010, 21, 780–786. [Google Scholar] [CrossRef]
- Kobayashi, J.; Kubota, T. Bioactive macrolides and polyketides from marine dinoflagellates of the genus Amphidinium. J. Nat. Prod. 2007, 70, 451–460. [Google Scholar] [CrossRef]
- Martínez, K.A.; Lauritano, C.; Druka, D.; Romano, G.; Grohmann, T.; Jaspars, M.; Martín, J.; Díaz, C.; Cautain, B.; de la Cruz, M.; et al. Amphidinol 22, a new cytotoxic and antifungal amphidinol from the dinoflagellate Amphidinium carterae. Mar Drugs 2019, 17, 385. [Google Scholar] [CrossRef]
- Han, R.; Rai, A.; Nakamura, M.; Suzuki, H.; Takahashi, H.; Yamazaki, M.; Saito, K. De novo deep transcriptome analysis of medicinal plants for gene discovery in biosynthesis of plant natural products. In Methods in Enzymology; Elsevier: Amsterdam, The Netherland, 2016; Volume 576, pp. 19–45. ISBN 978-0-12-804539-8. [Google Scholar]
- Wang, D.-Z.; Zhang, S.-F.; Zhang, Y.; Lin, L. Paralytic shellfish toxin biosynthesis in cyanobacteria and dinoflagellates: A molecular overview. J. Proteom. 2016, 135, 132–140. [Google Scholar] [CrossRef] [PubMed]
- Jaeckisch, N.; Yang, I.; Wohlrab, S.; Glöckner, G.; Kroymann, J.; Vogel, H.; Cembella, A.; John, U. Comparative genomic and transcriptomic characterization of the toxigenic marine dinoflagellate Alexandrium ostenfeldii. PLoS ONE 2011, 6, e28012. [Google Scholar] [CrossRef] [PubMed]
- Lauritano, C.; De Luca, D.; Ferrarini, A.; Avanzato, C.; Minio, A.; Esposito, F.; Ianora, A. De novo transcriptome of the cosmopolitan dinoflagellate Amphidinium carterae to identify enzymes with biotechnological potential. Sci. Rep. 2017, 7, 11701. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, S.-F.; Lin, L.; Wang, D.-Z. Whole transcriptomic analysis provides insights into molecular mechanisms for toxin biosynthesis in a toxic dinoflagellate Alexandrium catenella (ACHK-T). Toxins (Basel) 2017, 9, 213. [Google Scholar] [CrossRef]
- Lupette, J.; Jaussaud, A.; Vigor, C.; Oger, C.; Galano, J.-M.; Réversat, G.; Vercauteren, J.; Jouhet, J.; Durand, T.; Maréchal, E. Non-enzymatic synthesis of bioactive isoprostanoids in the diatom Phaeodactylum following oxidative stress. Plant Physiol. 2018, 178, 1344–1357. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).