Comparative Genomic Insights into Secondary Metabolism Biosynthetic Gene Cluster Distributions of Marine Streptomyces
Abstract
1. Introduction
2. Results and Discussion
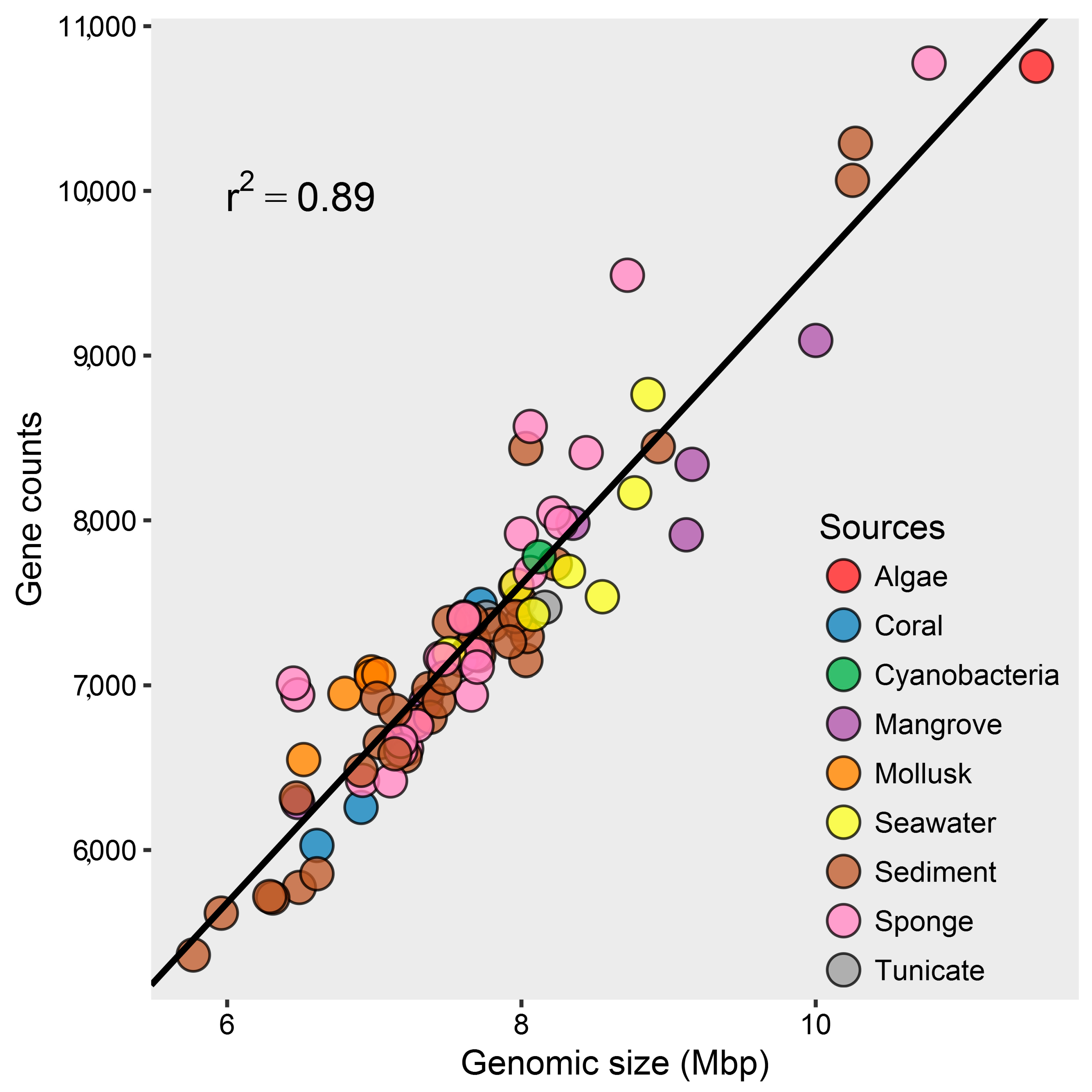
2.1. Genomic Characteristics and Annotation Results of Marine Streptomyces
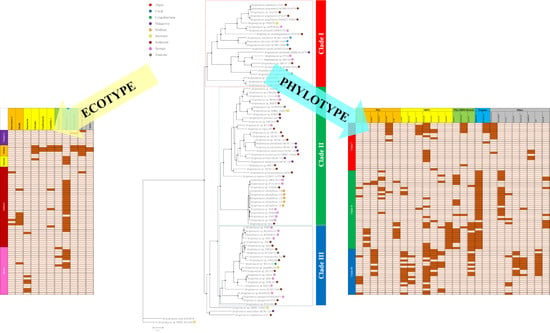
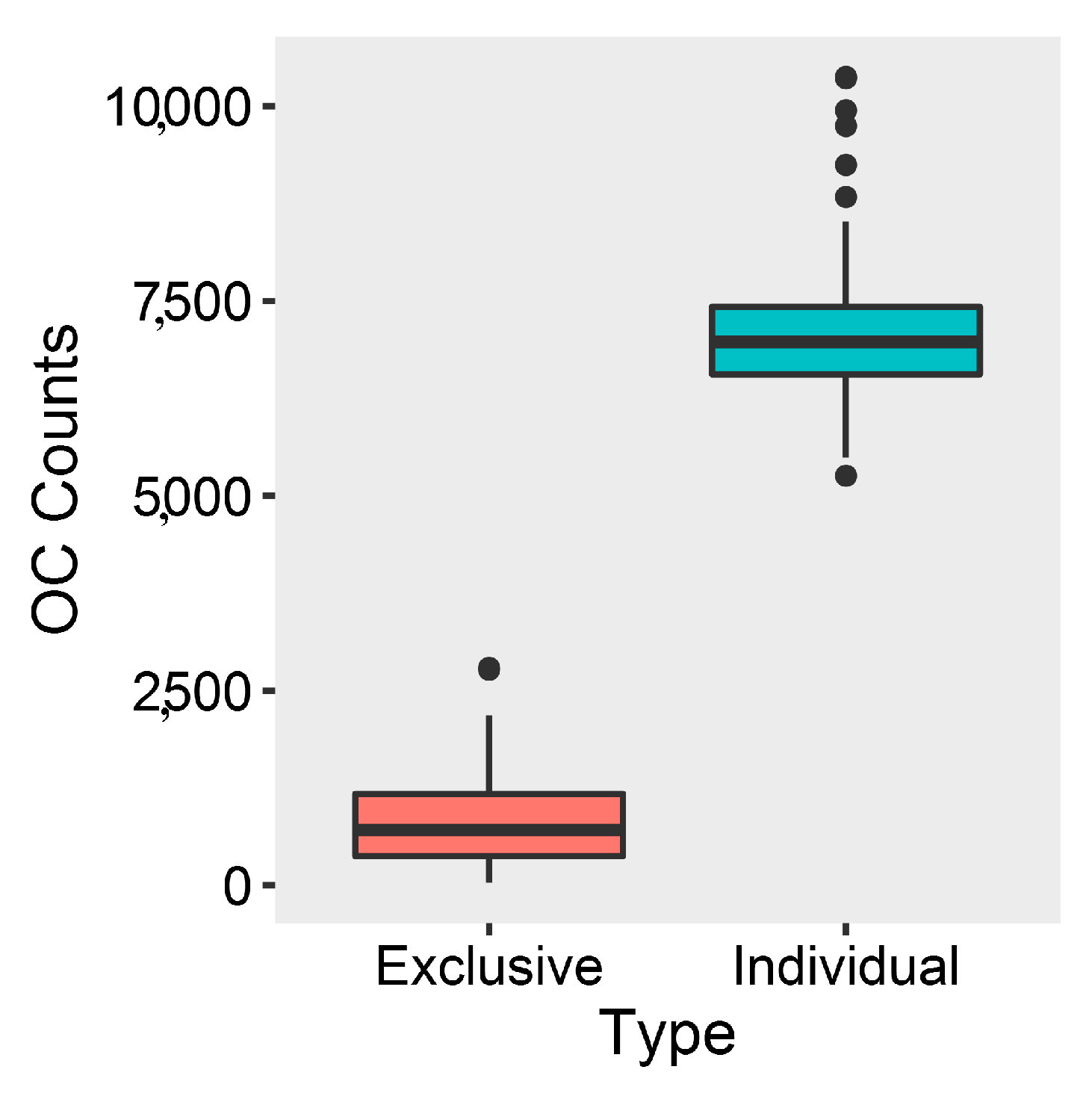
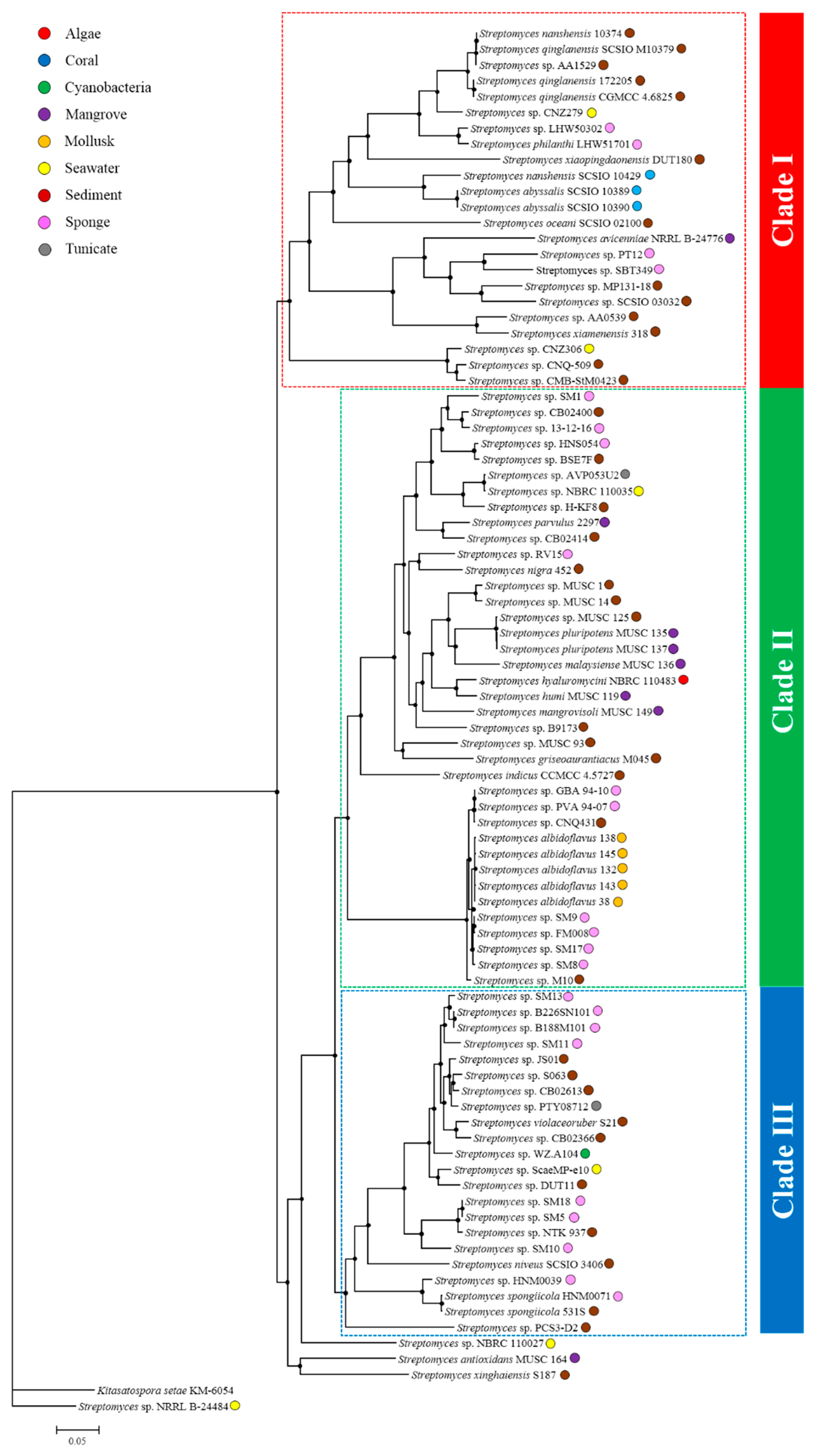
2.2. Comparative Genomics and Phylogenomic Relationship of Marine Streptomyces
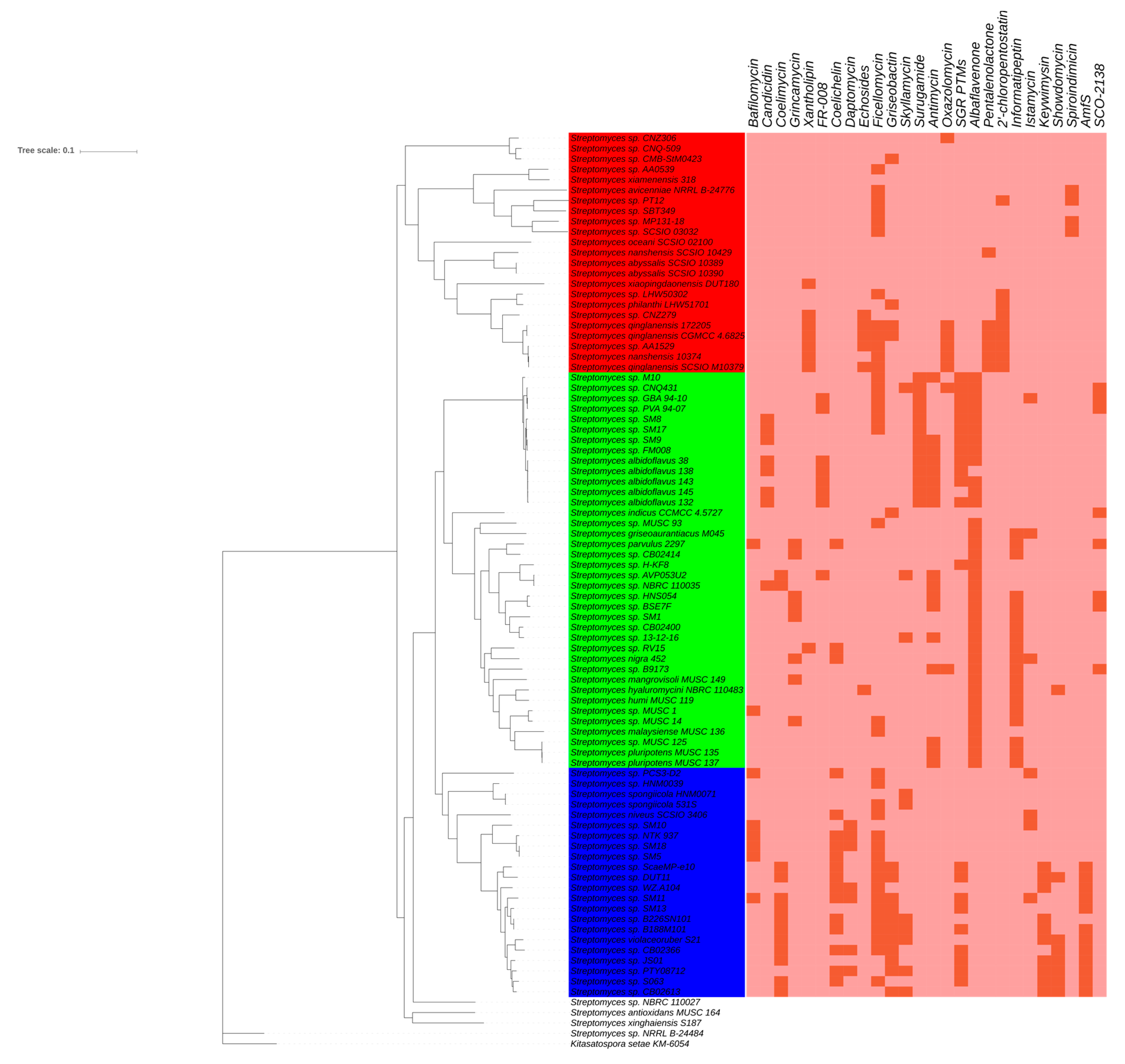
2.3. Phylotype-Associated SMBGCs
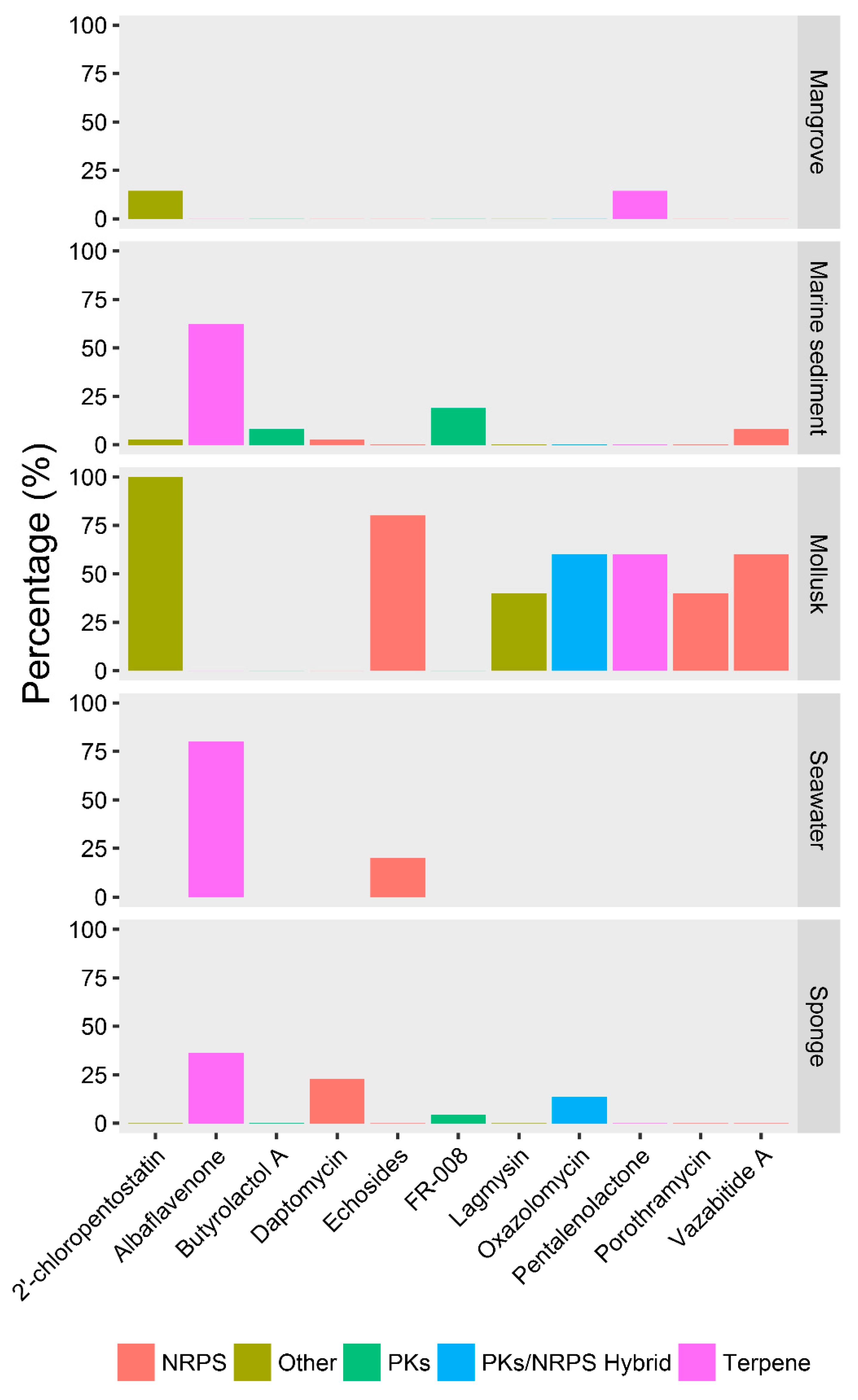
2.4. Ecotype-Associated SMBGCs
3. Materials and Methods
3.1. Obtain, Assess, and Annotate Marine Streptomyces Genomes
3.2. Comparative Genomic Analysis of Marine Streptomyces Genomes
3.3. Phylogenomic Analysis and Genomic Similarity Calculation of Marine Streptomyces
3.4. Statistical Analysis and Visualization
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Sekurova, O.N.; Schneider, O.; Zotchev, S.B. Novel bioactive natural products from bacteria via bioprospecting, genome mining and metabolic engineering. Microb. Biotechnol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Malik, V. Microbial secondary metabolism. Trends Biochem. Sci. 1980, 5, 68–72. [Google Scholar] [CrossRef]
- Van der Meij, A.; Worsley, S.F.; Hutchings, M.I.; van Wezel, G.P. Chemical ecology of antibiotic production by actinomycetes. FEMS Microbiol. Rev. 2017, 41, 392–416. [Google Scholar] [CrossRef] [PubMed]
- Mascuch, S.; Kubanek, J. A marine chemical defense partnership. Science 2019, 364, 1034–1035. [Google Scholar] [CrossRef] [PubMed]
- Demain, A.L.; Sanchez, S. Microbial drug discovery: 80 years of progress. J. Antibiot. 2009, 62, 5–16. [Google Scholar] [CrossRef] [PubMed]
- Purves, K.; Macintyre, L.; Brennan, D.; Hreggviethsson, G.O.; Kuttner, E.; Asgeirsdottir, M.E.; Young, L.C.; Green, D.H.; Edrada-Ebel, R.; Duncan, K.R. Using Molecular Networking for Microbial Secondary Metabolite Bioprospecting. Metabolites 2016, 6, 2. [Google Scholar] [CrossRef]
- Machado, H.; Sonnenschein, E.C.; Melchiorsen, J.; Gram, L. Genome mining reveals unlocked bioactive potential of marine Gram-negative bacteria. BMC Genom. 2015, 16, 158. [Google Scholar] [CrossRef]
- Kamjam, M.; Sivalingam, P.; Deng, Z.; Hong, K. Deep Sea Actinomycetes and Their Secondary Metabolites. Front. Microbiol. 2017, 8, 760. [Google Scholar] [CrossRef]
- Jackson, S.A.; Crossman, L.; Almeida, E.L.; Margassery, L.M.; Kennedy, J.; Dobson, A. Diverse and Abundant Secondary Metabolism Biosynthetic Gene Clusters in the Genomes of Marine Sponge Derived Streptomyces spp. Isolates. Mar. Drugs 2018, 16, 67. [Google Scholar] [CrossRef]
- Fenical, W.; Jensen, P.R. Developing a new resource for drug discovery: Marine actinomycete bacteria. Nat. Chem. Bio. 2006, 2, 666–673. [Google Scholar] [CrossRef]
- Ziemert, N.; Alanjary, M.; Weber, T. The evolution of genome mining in microbes–a review. Nat. Prod. Rep. 2016, 33, 988–1005. [Google Scholar] [CrossRef]
- Palazzotto, E.; Weber, T. Omics and multi-omics approaches to study the biosynthesis of secondary metabolites in microorganisms. Curr. Opin. Microbiol. 2018, 45, 109–116. [Google Scholar] [CrossRef]
- Foulston, L. Genome mining and prospects for antibiotic discovery. Curr. Opin. Microbiol. 2019, 51, 1–8. [Google Scholar] [CrossRef]
- Cimermancic, P.; Medema, M.H.; Claesen, J.; Kurita, K.; Wieland Brown, L.C.; Mavrommatis, K.; Pati, A.; Godfrey, P.A.; Koehrsen, M.; Clardy, J.; et al. Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell 2014, 158, 412–421. [Google Scholar] [CrossRef]
- Weissman, K.J. The structural biology of biosynthetic megaenzymes. Nat. Chem. Bio. 2015, 11, 660–670. [Google Scholar] [CrossRef]
- Eisenreich, W.; Bacher, A.; Arigoni, D.; Rohdich, F. Biosynthesis of isoprenoids via the non-mevalonate pathway. Cell. Mol. Life Sci. 2004, 61, 1401–1426. [Google Scholar] [CrossRef]
- Zurbriggen, A.; Kirst, H.; Melis, A. Isoprene production via the mevalonic acid pathway in Escherichia coli (Bacteria). BioEnergy Res. 2012, 5, 814–828. [Google Scholar] [CrossRef]
- Jensen, P.R.; Fenical, W. Strategies for the discovery of secondary metabolites from marine bacteria: Ecological perspectives. Annu. Rev. Microbiol. 1994, 48, 559–584. [Google Scholar] [CrossRef]
- O’Brien, J.; Wright, G.D. An ecological perspective of microbial secondary metabolism. Curr. Opin. Biotechnol. 2011, 22, 552–558. [Google Scholar] [CrossRef]
- Jensen, P.R.; Williams, P.G.; Oh, D.C.; Zeigler, L.; Fenical, W. Species-specific secondary metabolite production in marine actinomycetes of the genus Salinispora. Appl. Environ. Microb. 2007, 73, 1146–1152. [Google Scholar] [CrossRef]
- Adamek, M.; Alanjary, M.; Sales-Ortells, H.; Goodfellow, M.; Bull, A.T.; Winkler, A.; Wibberg, D.; Kalinowski, J.; Ziemert, N. Comparative genomics reveals phylogenetic distribution patterns of secondary metabolites in Amycolatopsis species. BMC Genom. 2018, 19, 426. [Google Scholar] [CrossRef]
- Kämpfer, P. The Prokaryotes: Archaea. Bacteria: Firmicutes, Actinomycetes; The family Streptomycetaceae, part I: Taxonomy; Rosenberg, E., DeLong, E.F., Lory, S., Stackbrandt, E., Thompson, F., Eds.; Springer: Berlin, Germany, 2006; Volume 3, pp. 538–604. [Google Scholar]
- Parte, A.C. LPSN–List of Prokaryotic names with Standing in Nomenclature (bacterio.net), 20 years on. Int. J. Syst. Evol. Microbiol. 2018, 68, 1825–1829. [Google Scholar] [CrossRef]
- Hwang, K.S.; Kim, H.U.; Charusanti, P.; Palsson, B.O.; Lee, S.Y. Systems biology and biotechnology of Streptomyces species for the production of secondary metabolites. Biotechnol. Adv. 2014, 32, 255–268. [Google Scholar] [CrossRef]
- Liu, R.; Deng, Z.; Liu, T. Streptomyces species: Ideal chassis for natural product discovery and overproduction. Metab. Eng. 2018, 50, 74–84. [Google Scholar] [CrossRef]
- Hakvag, S.; Fjaervik, E.; Josefsen, K.D.; Ian, E.; Ellingsen, T.E.; Zotchev, S.B. Characterization of Streptomyces spp. isolated from the sea surface microlayer in the Trondheim Fjord, Norway. Mar. Drugs 2008, 6, 620–635. [Google Scholar] [CrossRef]
- Zhu, H.; Jiang, S.; Yao, Q.; Wang, Y.; Chen, M.; Chen, Y.; Guo, J. Streptomyces fenghuangensis sp. nov., isolated from seawater. Int. J Syst. Evol. Microbiol. 2011, 61, 2811–2815. [Google Scholar] [CrossRef][Green Version]
- Macherla, V.R.; Liu, J.; Bellows, C.; Teisan, S.; Nicholson, B.; Lam, K.S.; Potts, B.C. Glaciapyrroles A, B, and C, pyrrolosesquiterpenes from a Streptomyces sp. isolated from an Alaskan marine sediment. J. Nat. Prod. 2005, 68, 780–783. [Google Scholar] [CrossRef]
- Zhao, X.Q.; Li, W.J.; Jiao, W.C.; Li, Y.; Yuan, W.J.; Zhang, Y.Q.; Klenk, H.P.; Suh, J.W.; Bai, F.W. Streptomyces xinghaiensis sp. nov., isolated from marine sediment. Int. J. Syst. Evol. Microbiol. 2009, 59, 2870–2874. [Google Scholar] [CrossRef][Green Version]
- Braña, A.F.; Sarmiento-Vizcaíno, A.; Pérez-Victoria, I.; Martín, J.; Otero, L.; Palacios-Gutiérrez, J.J.; Fernández, J.; Mohamedi, Y.; Fontanil, T.; Salmón, M. Desertomycin G, a New Antibiotic with Activity against Mycobacterium tuberculosis and Human Breast Tumor Cell Lines Produced by Streptomyces althioticus MSM3, Isolated from the Cantabrian Sea Intertidal Macroalgae Ulva sp. Mar. Drugs 2019, 17, 114. [Google Scholar] [CrossRef]
- Girão, M.; Ribeiro, I.; Ribeiro, T.; Azevedo, I.C.; Pereira, F.; Urbatzka, R.; Leão, P.N.; Carvalho, M.F. Actinobacteria isolated from Laminaria ochroleuca: A source of new bioactive compounds. Front. Microbiol. 2019, 10, 683. [Google Scholar] [CrossRef]
- Xiao, J.; Wang, Y.; Luo, Y.; Xie, S.J.; Ruan, J.S.; Xu, J. Streptomyces avicenniae sp. nov., a novel actinomycete isolated from the rhizosphere of the mangrove plant Avicennia mariana. Int. J. Syst. Evol. Microbiol. 2009, 59, 2624–2628. [Google Scholar] [CrossRef]
- Yan, L.-L.; Han, N.-N.; Zhang, Y.-Q.; Yu, L.-Y.; Chen, J.; Wei, Y.-Z.; Li, Q.-P.; Tao, L.; Zheng, G.-H.; Yang, S.-E. Antimycin A 18 produced by an endophytic Streptomyces albidoflavus isolated from a mangrove plant. J. Antibiot. 2010, 63, 259. [Google Scholar] [CrossRef]
- Khan, S.T.; Komaki, H.; Motohashi, K.; Kozone, I.; Mukai, A.; Takagi, M.; Shin-ya, K. Streptomyces associated with a marine sponge Haliclona sp.; biosynthetic genes for secondary metabolites and products. Env. Microbiol. 2011, 13, 391–403. [Google Scholar] [CrossRef]
- Huang, X.; Zhou, S.; Huang, D.; Chen, J.; Zhu, W. Streptomyces spongiicola sp. nov., an actinomycete derived from marine sponge. Int. J. Syst. Evol. Microbiol. 2016, 66, 738–743. [Google Scholar] [CrossRef][Green Version]
- Hodges, T.W.; Slattery, M.; Olson, J.B. Unique actinomycetes from marine caves and coral reef sediments provide novel PKS and NRPS biosynthetic gene clusters. Mar. Biotechnol. 2012, 14, 270–280. [Google Scholar] [CrossRef]
- Braña, A.; Sarmiento-Vizcaíno, A.; Osset, M.; Pérez-Victoria, I.; Martín, J.; de Pedro, N.; de la Cruz, M.; Díaz, C.; Vicente, F.; Reyes, F. Lobophorin K, a new natural product with cytotoxic activity produced by Streptomyces sp. M-207 associated with the deep-sea coral Lophelia pertusa. Mar. Drugs 2017, 15, 144. [Google Scholar] [CrossRef]
- Shaala, L.A.; Youssef, D.T.; Badr, J.M.; Harakeh, S.M. Bioactive 2(1H)-Pyrazinones and Diketopiperazine Alkaloids from a Tunicate-Derived Actinomycete Streptomyces sp. Molecules 2016, 21, 1116. [Google Scholar] [CrossRef]
- Sung, A.A.; Gromek, S.M.; Balunas, M.J. Upregulation and Identification of Antibiotic Activity of a Marine-Derived Streptomyces sp. via Co-Cultures with Human Pathogens. Mar. Drugs 2017, 15, 250. [Google Scholar] [CrossRef]
- Lin, Z.; Flores, M.; Forteza, I.; Henriksen, N.M.; Concepcion, G.P.; Rosenberg, G.; Haygood, M.G.; Olivera, B.M.; Light, A.R.; Cheatham III, T.E.; et al. Totopotensamides, polyketide-cyclic peptide hybrids from a mollusk-associated bacterium Streptomyces sp. J. Nat. Prod. 2012, 75, 644–649. [Google Scholar] [CrossRef]
- Lin, Z.; Koch, M.; Pond, C.D.; Mabeza, G.; Seronay, R.A.; Concepcion, G.P.; Barrows, L.R.; Olivera, B.M.; Schmidt, E.W. Structure and activity of lobophorins from a turrid mollusk-associated Streptomyces sp. J. Antibiot. 2014, 67, 121. [Google Scholar] [CrossRef]
- Bentley, S.D.; Chater, K.F.; Cerdeno-Tarraga, A.M.; Challis, G.L.; Thomson, N.R.; James, K.D.; Harris, D.E.; Quail, M.A.; Kieser, H.; Harper, D.; et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 2002, 417, 141–147. [Google Scholar] [CrossRef]
- Studholme, D.J. Genome Update. Let the consumer beware: Streptomyces genome sequence quality. Microb. Biotechnol. 2016, 9, 3–7. [Google Scholar] [CrossRef]
- Bush, M.J.; Chandra, G.; Bibb, M.J.; Findlay, K.C.; Buttner, M.J. Genome-Wide Chromatin Immunoprecipitation Sequencing Analysis Shows that WhiB Is a Transcription Factor That Cocontrols Its Regulon with WhiA To Initiate Developmental Cell Division in Streptomyces. MBio 2016, 7, e00523-e16. [Google Scholar] [CrossRef]
- Doroghazi, J.R.; Metcalf, W.W. Comparative genomics of actinomycetes with a focus on natural product biosynthetic genes. BMC Genom. 2013, 14, 611. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, H.; Wang, Y.; Cui, H.; Xie, Z.; Pu, Y.; Pei, S.; Li, F.; Qin, S. Genomic sequence-based discovery of novel angucyclinone antibiotics from marine Streptomyces sp. W007. FEMS Microbiol. Lett. 2012, 332, 105–112. [Google Scholar] [CrossRef]
- Seipke, R.F. Strain-level diversity of secondary metabolism in Streptomyces albus. PLoS ONE 2015, 10, e0116457. [Google Scholar] [CrossRef]
- Zotchev, S.B. Marine actinomycetes as an emerging resource for the drug development pipelines. J. Biotechnol. 2012, 158, 168–175. [Google Scholar] [CrossRef]
- Xu, M.J.; Wang, J.H.; Bu, X.L.; Yu, H.L.; Li, P.; Ou, H.Y.; He, Y.; Xu, F.D.; Hu, X.Y.; Zhu, X.M.; et al. Deciphering the streamlined genome of Streptomyces xiamenensis 318 as the producer of the anti-fibrotic drug candidate xiamenmycin. Sci. Rep. 2016, 6, 18977. [Google Scholar] [CrossRef]
- Ian, E.; Malko, D.B.; Sekurova, O.N.; Bredholt, H.; Ruckert, C.; Borisova, M.E.; Albersmeier, A.; Kalinowski, J.; Gelfand, M.S.; Zotchev, S.B. Genomics of sponge-associated Streptomyces spp. closely related to Streptomyces albus J1074: Insights into marine adaptation and secondary metabolite biosynthesis potential. PLoS ONE 2014, 9, e96719. [Google Scholar] [CrossRef]
- Tian, X.; Zhang, Z.; Yang, T.; Chen, M.; Li, J.; Chen, F.; Yang, J.; Li, W.; Zhang, B.; Zhang, Z.; et al. Comparative Genomics Analysis of Streptomyces Species Reveals Their Adaptation to the Marine Environment and Their Diversity at the Genomic Level. Front. Microbiol. 2016, 7, 998. [Google Scholar] [CrossRef]
- Ventura, M.; Canchaya, C.; Tauch, A.; Chandra, G.; Fitzgerald, G.F.; Chater, K.F.; van Sinderen, D. Genomics of Actinobacteria: Tracing the evolutionary history of an ancient phylum. Microbiol. Mol. Biol. Rev. 2007, 71, 495–548. [Google Scholar] [CrossRef]
- Kim, M.; Oh, H.S.; Park, S.C.; Chun, J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 2014, 64, 346–351. [Google Scholar] [CrossRef]
- Ross, A.B.; Kamal-Eldin, A.; Aman, P. Dietary alkylresorcinols: Absorption, bioactivities, and possible use as biomarkers of whole-grain wheat- and rye-rich foods. Nutr. Rev. 2004, 62, 81–95. [Google Scholar] [CrossRef]
- Hayashi, M.; Yamada, H.; Mitamura, T.; Horii, T.; Yamamoto, A.; Moriyama, Y. Vacuolar H+-ATPase localized in plasma membranes of malaria parasite cells, Plasmodium falciparum, is involved in regional acidification of parasitized erythrocytes. J. Biol. Chem. 2000, 275, 34353–34358. [Google Scholar] [CrossRef]
- Whitton, B.; Okamoto, H.; Packham, G.; Crabb, S.J. Vacuolar ATPase as a potential therapeutic target and mediator of treatment resistance in cancer. Cancer Med. 2018, 7, 3800–3811. [Google Scholar] [CrossRef]
- Keller, C.W.; Schmidt, J.; Lunemann, J.D. Immune and myodegenerative pathomechanisms in inclusion body myositis. Ann. Clin. Transl. Neur. 2017, 4, 422–445. [Google Scholar] [CrossRef]
- Szczeblewski, P.; Laskowski, T.; Kubacki, B.; Dziergowska, M.; Liczmanska, M.; Grynda, J.; Kubica, P.; Kot-Wasik, A.; Borowski, E. Analytical studies on ascosin, candicidin and levorin multicomponent antifungal antibiotic complexes. The stereostructure of ascosin A2. Sci. Rep. 2017, 7, 40158. [Google Scholar] [CrossRef]
- Gomez-Escribano, J.P.; Song, L.; Fox, D.J.; Yeo, V.; Bibb, M.J.; Challis, G.L. Structure and biosynthesis of the unusual polyketide alkaloid coelimycin P1, a metabolic product of the cpk gene cluster of Streptomyces coelicolor M145. Chem. Sci. 2012, 3, 2716–2720. [Google Scholar] [CrossRef]
- Lai, Z.; Yu, J.; Ling, H.; Song, Y.; Yuan, J.; Ju, J.; Tao, Y.; Huang, H. Grincamycins I–K, Cytotoxic Angucycline Glycosides Derived from Marine-Derived Actinomycete Streptomyces lusitanus SCSIO LR32. Planta Med. 2018, 84, 201–207. [Google Scholar] [CrossRef]
- Matsumoto, N.; Tsuchida, T.; Maruyama, M.; Kinoshita, N.; Homma, Y.; Iinuma, H.; Sawa, T.; Hamada, M.; Takeuchi, T.; Heida, N.; et al. Lactonamycin, a new antimicrobial antibiotic produced by Streptomyces rishiriensis MJ773-88K4. I. Taxonomy, fermentation, isolation, physico-chemical properties and biological activities. J. Antibiot. 1999, 52, 269–275. [Google Scholar] [CrossRef]
- Boonlarppradab, C.; Kauffman, C.A.; Jensen, P.R.; Fenical, W. Marineosins A and B, cytotoxic spiroaminals from a marine-derived actinomycete. Org. Lett. 2008, 10, 5505–5508. [Google Scholar] [CrossRef]
- Gullon, S.; Olano, C.; Abdelfattah, M.S.; Brana, A.F.; Rohr, J.; Mendez, C.; Salas, J.A. Isolation, characterization, and heterologous expression of the biosynthesis gene cluster for the antitumor anthracycline steffimycin. Appl. Environ. Microb. 2006, 72, 4172–4183. [Google Scholar] [CrossRef]
- Kelemen, G.H.; Brian, P.; Flardh, K.; Chamberlin, L.; Chater, K.F.; Buttner, M.J. Developmental regulation of transcription of whiE, a locus specifying the polyketide spore pigment in Streptomyces coelicolor A3(2). J. Bacteriol. 1998, 180, 2515–2521. [Google Scholar]
- Wu, S.; Huang, T.; Xie, D.; Wo, J.; Wang, X.; Deng, Z.; Lin, S. Xantholipin B produced by the stnR inactivation mutant Streptomyces flocculus CGMCC 4.1223 WJN-1. J. Antibiot. 2017, 70, 90–95. [Google Scholar] [CrossRef]
- Zhou, Y.; Li, J.; Zhu, J.; Chen, S.; Bai, L.; Zhou, X.; Wu, H.; Deng, Z. Incomplete beta-ketone processing as a mechanism for polyene structural variation in the FR-008/candicidin complex. Chem. Biol. 2008, 15, 629–638. [Google Scholar] [CrossRef][Green Version]
- Williams, J.C.; Sheldon, J.R.; Imlay, H.D.; Dutter, B.F.; Draelos, M.M.; Skaar, E.P.; Sulikowski, G.A. Synthesis of the Siderophore Coelichelin and Its Utility as a Probe in the Study of Bacterial Metal Sensing and Response. Org. Lett. 2019, 21, 679–682. [Google Scholar] [CrossRef]
- Sader, H.S.; Flamm, R.K.; Farrell, D.J.; Jones, R.N. Daptomycin activity against uncommonly isolated streptococcal and other gram-positive species groups. Antimicrob. Agents Chemother. 2013, 57, 6378–6380. [Google Scholar] [CrossRef]
- Zhu, J.; Chen, W.; Li, Y.Y.; Deng, J.J.; Zhu, D.Y.; Duan, J.; Liu, Y.; Shi, G.Y.; Xie, C.; Wang, H.X.; et al. Identification and catalytic characterization of a nonribosomal peptide synthetase-like (NRPS-like) enzyme involved in the biosynthesis of echosides from Streptomyces sp. LZ35. Gene 2014, 546, 352–358. [Google Scholar] [CrossRef]
- He, X.; Li, M.; Song, S.; Wu, X.; Zhang, J.; Wu, G.; Yue, R.; Cui, H.; Song, S.; Ma, C.; et al. Ficellomycin: An aziridine alkaloid antibiotic with potential therapeutic capacity. Appl. Microbiol. Biot. 2018, 102, 4345–4354. [Google Scholar] [CrossRef]
- Patzer, S.I.; Braun, V. Gene cluster involved in the biosynthesis of griseobactin, a catechol-peptide siderophore of Streptomyces sp. ATCC 700974. J. Bacteriol. 2010, 192, 426–435. [Google Scholar] [CrossRef]
- Pohle, S.; Appelt, C.; Roux, M.; Fiedler, H.P.; Sussmuth, R.D. Biosynthetic gene cluster of the non-ribosomally synthesized cyclodepsipeptide skyllamycin: Deciphering unprecedented ways of unusual hydroxylation reactions. J. Am. Chem. Soc. 2011, 133, 6194–6205. [Google Scholar] [CrossRef]
- Giltrap, A.M.; Haeckl, F.P.J.; Kurita, K.L.; Linington, R.G.; Payne, R.J. Synthetic Studies Toward the Skyllamycins: Total Synthesis and Generation of Simplified Analogues. J. Org. Chem. 2018, 83, 7250–7270. [Google Scholar] [CrossRef]
- Wang, X.; Shaaban, K.A.; Elshahawi, S.I.; Ponomareva, L.V.; Sunkara, M.; Copley, G.C.; Hower, J.C.; Morris, A.J.; Kharel, M.K.; Thorson, J.S. Mullinamides A and B, new cyclopeptides produced by the Ruth Mullins coal mine fire isolate Streptomyces sp. RM-27-46. J. Antibiot. 2014, 67, 571–575. [Google Scholar] [CrossRef]
- Hamilton, B.T.; Moore, S.E.; Williams, T.B.; Darby, N.; Vinson, M.R. Comparative effects of rotenone and antimycin on macroinvertebrate diversity in two streams in Great Basin National Park, Nevada. N. Am. J. Fish. Manag. 2009, 29, 1620–1635. [Google Scholar] [CrossRef]
- Angelov, P.; Chau, Y.K.; Fryer, P.J.; Moloney, M.G.; Thompson, A.L.; Trippier, P.C. Biomimetic synthesis, antibacterial activity and structure-activity properties of the pyroglutamate core of oxazolomycin. Org. Biomol. Chem. 2012, 10, 3472–3485. [Google Scholar] [CrossRef]
- Luo, Y.; Huang, H.; Liang, J.; Wang, M.; Lu, L.; Shao, Z.; Cobb, R.E.; Zhao, H. Activation and characterization of a cryptic polycyclic tetramate macrolactam biosynthetic gene cluster. Nat. Commun. 2013, 4, 2894. [Google Scholar] [CrossRef]
- Lin, X.; Cane, D.E. Biosynthesis of the sesquiterpene antibiotic albaflavenone in Streptomyces coelicolor. Mechanism and stereochemistry of the enzymatic formation of epi-isozizaene. J. Am. Chem. Soc. 2009, 131, 6332–6333. [Google Scholar] [CrossRef]
- Uyeda, M.; Mizukami, M.; Yokomizo, K.; Suzuki, K. Pentalenolactone I and hygromycin A, immunosuppressants produced by Streptomyces filipinensis and Streptomyces hygroscopicus. Biosci. Biotech. Bioch. 2001, 65, 1252–1254. [Google Scholar] [CrossRef]
- Gao, Y.; Xu, G.; Wu, P.; Liu, J.; Cai, Y.S.; Deng, Z.; Chen, W. Biosynthesis of 2′-Chloropentostatin and 2’-Amino-2’-Deoxyadenosine Highlights a Single Gene Cluster Responsible for Two Independent Pathways in Actinomadura sp. Strain ATCC 39365. Appl. Environ. Microbiol. 2017, 83, e00078-17. [Google Scholar] [CrossRef]
- Riley, P.A. Melanin. Int. J. Biochem. Cell Biol. 1997, 29, 1235–1239. [Google Scholar] [CrossRef]
- Numao, N.; Hemmi, H.; Naujokaitis, S.A.; Rabinovitz, M.; Beisler, J.A. Showdomycin analogues: Synthesis and antitumor evaluation. J. Med. Chem. 1981, 24, 515–520. [Google Scholar] [CrossRef]
- Kasanah, N.; Triyanto, T. Bioactivities of Halometabolites from Marine Actinobacteria. Biomolecules 2019, 9, 225. [Google Scholar] [CrossRef]
- Ueda, K.; Oinuma, K.; Ikeda, G.; Hosono, K.; Ohnishi, Y.; Horinouchi, S.; Beppu, T. AmfS, an extracellular peptidic morphogen in Streptomyces griseus. J. Bacteriol. 2002, 184, 1488–1492. [Google Scholar] [CrossRef]
- Tanaka, Y.; Komaki, H.; Yazawa, K.; Mikami, Y.; Nemoto, A.; Tojyo, T.; Kadowaki, K.; Shigemori, H.; Kobayashi, J. Brasilinolide A, a new macrolide antibiotic produced by Nocardia brasiliensis: Producing strain, isolation and biological activity. J. Antibiot. 1997, 50, 1036–1041. [Google Scholar] [CrossRef]
- Kotake, C.; Yamasaki, T.; Moriyama, T.; Shinoda, M.; Komiyama, N.; Furumai, T.; Konishi, M.; Oki, T. Butyrolactols A and B, new antifungal antibiotics. J. Antibiot. 1992, 45, 1442–1450. [Google Scholar] [CrossRef][Green Version]
- Futamura, Y.; Sawa, R.; Umezawa, Y.; Igarashi, M.; Nakamura, H.; Hasegawa, K.; Yamasaki, M.; Tashiro, E.; Takahashi, Y.; Akamatsu, Y. Discovery of incednine as a potent modulator of the anti-apoptotic function of Bcl-xL from microbial origin. J. Am. Chem. Soc. 2008, 130, 1822–1823. [Google Scholar] [CrossRef]
- Cheng, Y.B.; Jensen, P.R.; Fenical, W. Cytotoxic and Antimicrobial Napyradiomycins from Two Marine-Derived, MAR 4 Streptomyces Strains. Eur. J. Org. Chem. 2013, 2013, 3751–3757. [Google Scholar] [CrossRef]
- Xia, M.; Suchland, R.J.; Carswell, J.A.; Van Duzer, J.; Buxton, D.K.; Brown, K.; Rothstein, D.M.; Stamm, W.E. Activities of rifamycin derivatives against wild-type and rpoB mutants of Chlamydia trachomatis. Antimicrob. Agents Chemother. 2005, 49, 3974–3976. [Google Scholar] [CrossRef]
- Shindo, K.; Kamishohara, M.; Odagawa, A.; Matsuoka, M.; Kawai, H. Vicenistatin, a novel 20-membered macrocyclic lactam antitumor antibiotic. J. Antibiot. 1993, 46, 1076–1081. [Google Scholar] [CrossRef]
- Koba, M.; Konopa, J. Actinomycin D and its mechanisms of action. Postepy Hig. Med. Dosw. 2005, 59, 290–298. [Google Scholar]
- Tsunakawa, M.; Kamei, H.; Konishi, M.; Miyaki, T.; Oki, T.; Kawaguchi, H. Porothramycin, a new antibiotic of the anthramycin group: Production, isolation, structure and biological activity. J. Antibiot. 1988, 41, 1366–1373. [Google Scholar] [CrossRef]
- Revathi, S.; Malathy, N.S. Antibacterial Activity of Rhizome of Curcuma aromatica and Partial Purification of Active Compounds. Indian J. Pharm. Sci. 2013, 75, 732–735. [Google Scholar]
- Kodani, S.; Bicz, J.; Song, L.; Deeth, R.J.; Ohnishi-Kameyama, M.; Yoshida, M.; Ochi, K.; Challis, G.L. Structure and biosynthesis of scabichelin, a novel tris-hydroxamate siderophore produced by the plant pathogen Streptomyces scabies 87.22. Org. Biomol. Chem. 2013, 11, 4686–4694. [Google Scholar] [CrossRef]
- Hasenbohler, A.; Kneifel, H.; Konig, W.A.; Zahner, H.; Zeiler, H.J. Metabolic products of microorganisms. 134. Stenothricin, a new inhibitor of the bacterial cell wall synthesis (author’s transl). Arch. Microbiol. 1974, 99, 307–321. [Google Scholar]
- Hu, F.P.; Young, J.M.; Fletcher, M.J. Preliminary description of biocidal (syringomycin) activity in fluorescent plant pathogenic Pseudomonas species. J. Appl. Microbiol. 1998, 85, 365–371. [Google Scholar] [CrossRef]
- Fu, C.; Keller, L.; Bauer, A.; Bronstrup, M.; Froidbise, A.; Hammann, P.; Herrmann, J.; Mondesert, G.; Kurz, M.; Schiell, M.; et al. Biosynthetic Studies of Telomycin Reveal New Lipopeptides with Enhanced Activity. J. Am. Chem. Soc. 2015, 137, 7692–7705. [Google Scholar] [CrossRef]
- Yashiro, T.; Sakata, F.; Sekimoto, T.; Shirai, T.; Hasebe, F.; Matsuda, K.; Kurosawa, S.; Suzuki, S.; Nagata, K.; Kasakura, K.; et al. Immunosuppressive effect of a non-proteinogenic amino acid from Streptomyces through inhibiting allogeneic T cell proliferation. Biosci. Biotechnol. Biochem. 2019, 83, 1111–1116. [Google Scholar] [CrossRef]
- Tichenor, M.S.; MacMillan, K.S.; Trzupek, J.D.; Rayl, T.J.; Hwang, I.; Boger, D.L. Systematic exploration of the structural features of yatakemycin impacting DNA alkylation and biological activity. J. Am. Chem. Soc. 2007, 129, 10858–10869. [Google Scholar] [CrossRef]
- Moore, B.S.; Floss, H.G. Biosynthetic studies on the origin of the cyclohexanecarboxylic acid moiety of ansatrienin A and omega-cyclohexyl fatty acids. J. Nat. Prod. 1994, 57, 382–386. [Google Scholar] [CrossRef]
- Greule, A.; Izore, T.; Iftime, D.; Tailhades, J.; Schoppet, M.; Zhao, Y.; Peschke, M.; Ahmed, I.; Kulik, A.; Adamek, M.; et al. Kistamicin biosynthesis reveals the biosynthetic requirements for production of highly crosslinked glycopeptide antibiotics. Nat. Commun. 2019, 10, 2613. [Google Scholar] [CrossRef]
- Sang, F.; Li, D.; Sun, X.; Cao, X.; Wang, L.; Sun, J.; Sun, B.; Wu, L.; Yang, G.; Chu, X.; et al. Total synthesis and determination of the absolute configuration of rakicidin A. J. Am. Chem. Soc. 2014, 136, 15787–15791. [Google Scholar] [CrossRef]
- Wang, L.; Yun, B.S.; George, N.P.; Wendt-Pienkowski, E.; Galm, U.; Oh, T.J.; Coughlin, J.M.; Zhang, G.; Tao, M.; Shen, B. Glycopeptide antitumor antibiotic zorbamycin from Streptomyces flavoviridis ATCC 21892: Strain improvement and structure elucidation. J. Nat. Prod. 2007, 70, 402–406. [Google Scholar] [CrossRef]
- Finlay, J.; Miller, L.; Poupard, J.A. A review of the antimicrobial activity of clavulanate. J. Antimicrob. Ch. 2003, 52, 18–23. [Google Scholar] [CrossRef]
- Umemura, M.; Kim, J.H.; Aoyama, H.; Hoshino, Y.; Fukumura, H.; Nakakaji, R.; Sato, I.; Ohtake, M.; Akimoto, T.; Narikawa, M.; et al. The iron chelating agent, deferoxamine detoxifies Fe(Salen)-induced cytotoxicity. J. Pharm. Sci. 2017, 134, 203–210. [Google Scholar] [CrossRef]
- Rateb, M.E.; Zhai, Y.; Ehrner, E.; Rath, C.M.; Wang, X.; Tabudravu, J.; Ebel, R.; Bibb, M.; Kyeremeh, K.; Dorrestein, P.C.; et al. Legonaridin, a new member of linaridin RiPP from a Ghanaian Streptomyces isolate. Org. Biomol. Chem. 2015, 13, 9585–9592. [Google Scholar] [CrossRef]
- Lee, E.R.; Blount, K.F.; Breaker, R.R. Roseoflavin is a natural antibacterial compound that binds to FMN riboswitches and regulates gene expression. RNA Biol. 2009, 6, 187–194. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Lagesen, K.; Hallin, P.; Rodland, E.A.; Staerfeldt, H.H.; Rognes, T.; Ussery, D.W. RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007, 35, 3100–3108. [Google Scholar] [CrossRef]
- Lowe, T.M.; Chan, P.P. tRNAscan-SE On-line: Integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016, 44, W54–W57. [Google Scholar] [CrossRef]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014, 42, D206–D214. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef]
- Moriya, Y.; Itoh, M.; Okuda, S.; Yoshizawa, A.C.; Kanehisa, M. KAAS: An automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007, 35, W182–W185. [Google Scholar] [CrossRef]
- Wu, S.; Zhu, Z.; Fu, L.; Niu, B.; Li, W. WebMGA: A customizable web server for fast metagenomic sequence analysis. BMC Genom. 2011, 12, 444. [Google Scholar] [CrossRef]
- Lee, I.; Ouk Kim, Y.; Park, S.C.; Chun, J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 2016, 66, 1100–1103. [Google Scholar] [CrossRef]
- Amin, A.; Ahmed, I.; Khalid, N.; Osman, G.; Khan, I.U.; Xiao, M.; Li, W.J. Streptomyces caldifontis sp. nov., isolated from a hot water spring of Tatta Pani, Kotli, Pakistan. Antonie van Leeuwenhoek 2017, 110, 77–86. [Google Scholar] [CrossRef]
- Cao, T.; Mu, S.; Lu, C.; Zhao, S.; Li, D.; Yan, K.; Xiang, W.; Liu, C. Streptomyces amphotericinicus sp. nov., an amphotericin-producing actinomycete isolated from the head of an ant (Camponotus japonicus Mayr). Int. J. Syst. Evol. Microbiol. 2017, 67, 4967–4973. [Google Scholar] [CrossRef]
- Take, A.; Inahashi, Y.; Omura, S.; Takahashi, Y.; Matsumoto, A. Streptomyces boninensis sp. nov., isolated from soil from a limestone cave in the Ogasawara Islands. Int. J. Syst. Evol. Microbiol. 2018, 68, 1795–1799. [Google Scholar] [CrossRef]
- Xu, L.; Wu, Y.H.; Zhou, P.; Cheng, H.; Liu, Q.; Xu, X.W. Investigation of the thermophilic mechanism in the genus Porphyrobacter by comparative genomic analysis. BMC Genom. 2018, 19, 385. [Google Scholar] [CrossRef]
- Lechner, M.; Findeiss, S.; Steiner, L.; Marz, M.; Stadler, P.F.; Prohaska, S.J. Proteinortho: Detection of (co-)orthologs in large-scale analysis. BMC Bioinform. 2011, 12, 124. [Google Scholar] [CrossRef]
- Jordan, I.K.; Rogozin, I.B.; Wolf, Y.I.; Koonin, E.V. Essential genes are more evolutionarily conserved than are nonessential genes in bacteria. Genome Res. 2002, 12, 962–968. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Felsenstein, J. Evolutionary trees from DNA sequences: A maximum likelihood approach. J. Mol. Evol. 1981, 17, 368–376. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2014, 32, 268–274. [Google Scholar] [CrossRef]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Ihaka, R.; Gentleman, R. R: A language for data analysis and graphics. J. Comput. Graph. Stat. 1996, 5, 299–314. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]

| Clade | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 |
|---|---|---|---|---|---|---|---|---|---|
| I | 0% | 13% | 0% | 4% | 0% | 9% | 56% | 18% | 0% |
| II | 3% | 0% | 0% | 16% | 13% | 5% | 37% | 23% | 3% |
| III | 0% | 0% | 4% | 0% | 0% | 4% | 46% | 42% | 4% |
| SMBGC | p Value | Activity | |
|---|---|---|---|
| PKs | |||
| Alkylresorcinol | 0.003 | Prevention of tumor [54] | |
| Bafilomycin | 0.003 | Antiprozoan [55] Antitumor [56] Immunosuppressant [57] | |
| Candicidin | 0.005 | Antifungus [58] | |
| Coelimycin | 1.60 × 10−6 | Yellow pigment [59] | |
| Grincamycin | 0.005 | Antitumor [60] | |
| Lactonamycin | 0.005 | Antibacteria [61] | |
| Marineosin | 0.004 | Cytotoxicity [62] | |
| Steffimycin | 2.43 × 10−7 | Antitumor [63] | |
| Spore pigment | 0.0002 | Regulate sporulation [64] | |
| Xantholipin | 0.0003 | Antibacteria [65] Cytotoxicity [65] | |
| FR-008 | 0.005 | Antifungus [66] | |
| NRPS | |||
| Coelichelin | 3.22 × 10−8 | Siderophore [67] | |
| Daptomycin | 2.84 × 10−5 | Antibacteria [68] | |
| Echosides | 0.006 | Antivirus [69] | |
| Ficellomycin | 0.003 | Antibacteria [70] | |
| Griseobactin | 4.72 × 10−5 | Siderophore [71] | |
| Skyllamycin | 0.003 | Antitumor [72] Antibacteria [73] | |
| Surugamide | 0.0001 | Antibacteria [74] | |
| PKs/NRPS hybrid | |||
| Antimycin | 4.24 × 10−6 | Piscicide [75] | |
| Oxazolomycin | 0.005 | Antibacteria [76] | |
| SGR PTMs | 0.003 | Antifungal [77] Antioxidant [77] | |
| Terpene | |||
| Albaflavenone | 2.20 × 10−16 | Antibacteria [78] | |
| Pentalenolactone | 0.002 | Immunosuppressant [79] | |
| Other | |||
| 2′-chloropentostatin | 2.2 × 10−16 | Antivirus [80] | |
| Informatipeptin | 5.50 × 10−7 | Unknown | |
| Keywimysin | 1.72 × 10−7 | Unknown | |
| Melanin | 4.58 × 10−6 | Antioxidant [81] | |
| Showdomycin | 0.0002 | Antitumor [82] | |
| Spiroindimicin | 0.004 | Cytotoxicity [83] | |
| AmfS | 4.52 × 10−9 | Morphogen [84] | |
| SCO-2138 | 0.006 | Unknown | |
| SMBGC | p Value | Activity | |
|---|---|---|---|
| PKs | |||
| Brasilinolide | 0.007 | Antifungus [85] | |
| Butyrolactol A | 0.0008 | Antifungus [86] | |
| Incednine | 0.007 | Antiapotosis [87] | |
| Micromonolactam | 0.0009 | Unknown | |
| Napyradiomycin | 0.007 | Antibacteria [88] Cytotoxicity [88] | |
| Rifamycin | 0.007 | Antibacteria [89] | |
| Vicenistatin | 0.007 | Antitumor [90] | |
| FR-008 | 5.14 × 10−11 | Antifungus [66] | |
| NRPS | |||
| Actinomycin | 0.009 | Antibacteria [91] Antitumor [91] | |
| Daptomycin | 0.007 | Antibacteria [68] | |
| Echosides | 4.07 × 10−8 | Antivirus [69] | |
| Porothramycin | 6.81 × 10−5 | Antibacteria [92] Antitumor [92] | |
| Rhizomide | 0.008 | Antibacteria [93] | |
| Scabichelin | 0.009 | Siderophore [94] | |
| Stenothricin | 0.003 | Antibacteria [95] | |
| Syringomycin | 0.0005 | Antifungus [96] | |
| Telomycin | 0.0005 | Antibacteria [97] | |
| Vazabitide A | 0.002 | Immunosuppressant [98] | |
| Yatakemycin | 1.21 × 10−6 | Cytotoxicity [99] | |
| PKs/NRPS hybrid | |||
| Ansatrienin | 7.44 × 10−11 | Antibacteria [100] | |
| Kistamicin A | 0.003 | Antibacteira [101] | |
| Oxazolomycin | 6.60 × 10−5 | Antibacteria [76] | |
| Rakicidin | 1.07 × 10−5 | Antitumor [102] | |
| Zorbamycin | 1.21 × 10−6 | Antitumor [103] | |
| Terpene | |||
| Albaflavenone | 0.003 | Antibacteria [78] | |
| Pentalenolactone | 8.94 × 10−7 | Immunosuppressant [79] | |
| Other | |||
| 2′-chloropentostatin | 1.50 × 10−9 | Antivirus [80] | |
| Clavulanic acid | 0.003 | Antibacteria [104] | |
| Desferrioxamine | 0.009 | Antitumor [105] | |
| Lagmysin | 6.81 × 10−5 | Unknown | |
| Legonaridin | 1.21 × 10−6 | Cytotoxicity [106] | |
| Marinophenazines | 0.005 | Unknown | |
| Roseoflavin | 0.0002 | Antibacteria [107] | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, L.; Ye, K.-X.; Dai, W.-H.; Sun, C.; Xu, L.-H.; Han, B.-N. Comparative Genomic Insights into Secondary Metabolism Biosynthetic Gene Cluster Distributions of Marine Streptomyces. Mar. Drugs 2019, 17, 498. https://doi.org/10.3390/md17090498
Xu L, Ye K-X, Dai W-H, Sun C, Xu L-H, Han B-N. Comparative Genomic Insights into Secondary Metabolism Biosynthetic Gene Cluster Distributions of Marine Streptomyces. Marine Drugs. 2019; 17(9):498. https://doi.org/10.3390/md17090498
Chicago/Turabian StyleXu, Lin, Kai-Xiong Ye, Wen-Hua Dai, Cong Sun, Lian-Hua Xu, and Bing-Nan Han. 2019. "Comparative Genomic Insights into Secondary Metabolism Biosynthetic Gene Cluster Distributions of Marine Streptomyces" Marine Drugs 17, no. 9: 498. https://doi.org/10.3390/md17090498
APA StyleXu, L., Ye, K.-X., Dai, W.-H., Sun, C., Xu, L.-H., & Han, B.-N. (2019). Comparative Genomic Insights into Secondary Metabolism Biosynthetic Gene Cluster Distributions of Marine Streptomyces. Marine Drugs, 17(9), 498. https://doi.org/10.3390/md17090498