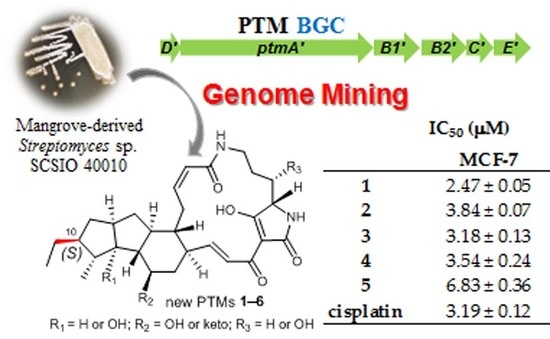
Genome Mining of Marine-Derived Streptomyces sp. SCSIO 40010 Leads to Cytotoxic New Polycyclic Tetramate Macrolactams
Abstract
1. Introduction
2. Results and Discussion
2.1. Genome Mining of a PTM Biosynthetic Gene Cluster
2.2. Biological Activities
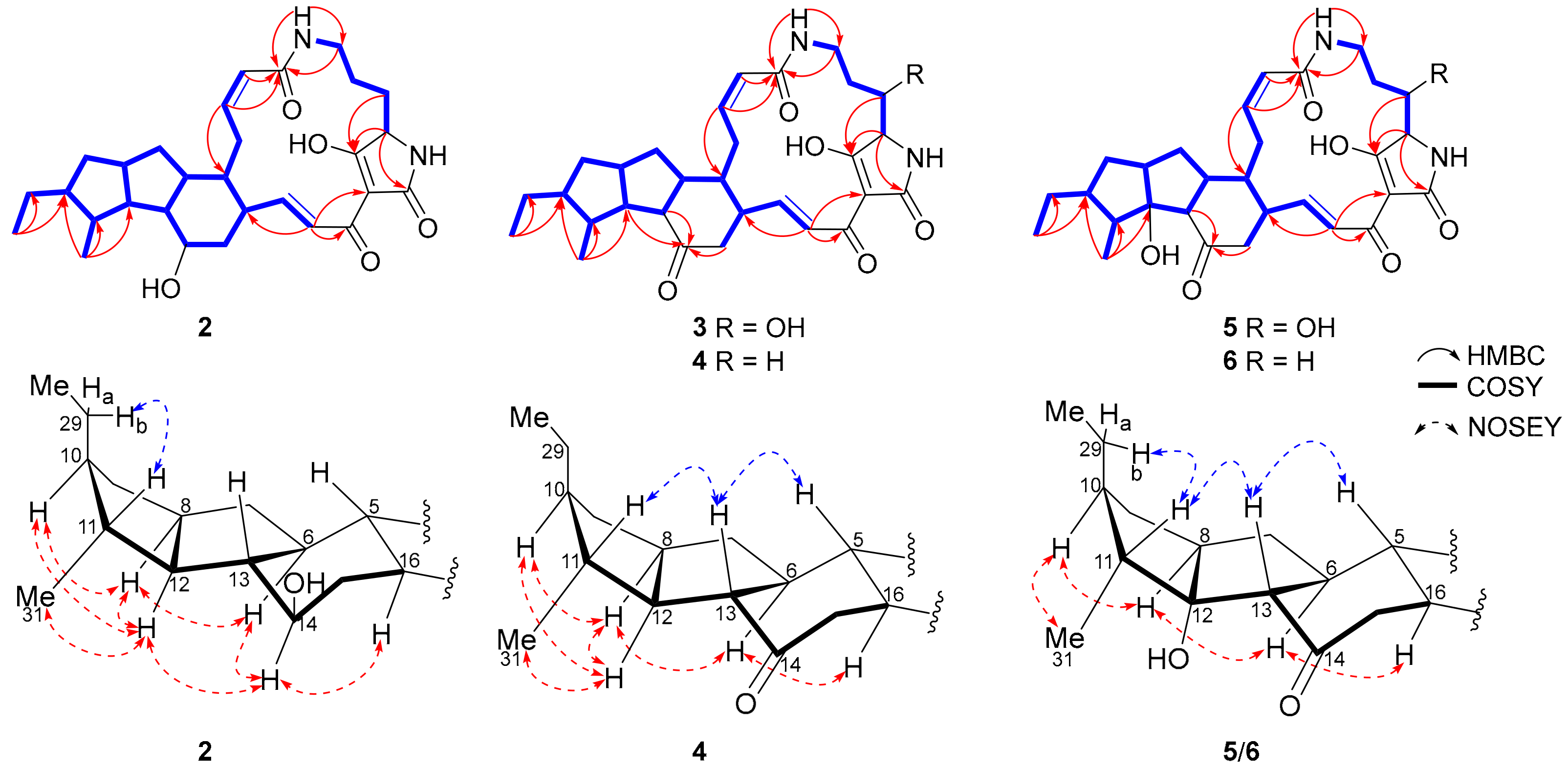
2.3. Biosynthetic Implications
2.4. Conclusion
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Strain, Screening and Culture Methods
3.3. Genome Mining and Bioinformatics Analysis
3.4. Extraction, Isolation and Purification
3.5. Physical and Chemical Properties of New Compounds 1–6
3.6. Bioactivity Assays
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Zhang, G.; Zhang, W.; Saha, S.; Zhang, C. Recent advances in discovery, biosynthesis and genome mining of medicinally relevant polycyclic tetramate macrolactams. Curr. Top. Med. Chem. 2016, 16, 1727–1739. [Google Scholar] [CrossRef] [PubMed]
- Jomon, K.; Ajisaka, M.; Sakai, H.; Kuroda, Y. New antibiotic, ikarugamycin. J. Antibiot. 1972, 25, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Calvo, A.M.; Yuen, G.Y.; Du, L.; Harris, S.D. Induction of cell wall thickening by the antifungal compound dihydromaltophilin disrupts fungal growth and is mediated by sphingolipid biosynthesis. J. Eukaryot. Microbiol. 2009, 56, 182–187. [Google Scholar] [CrossRef] [PubMed]
- Popescu, R.; Heiss, E.H.; Ferk, F.; Peschel, A.; Knasmueller, S.; Dirsch, V.M.; Krupitza, G.; Kopp, B. Ikarugamycin induces DNA damage, intracellular calcium increase, p38 MAP kinase activation and apoptosis in HL-60 human promyelocytic leukemia cells. Mutat. Res. 2011, 709–710, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Lacret, R.; Oves-Costales, D.; Gomez, C.; Diaz, C.; de la Cruz, M.; Perez-Victoria, I.; Vicente, F.; Genilloud, O.; Reyes, F. New ikarugamycin derivatives with antifungal and antibacterial properties from Streptomyces zhaozhouensis. Mar. Drugs 2015, 13, 128–140. [Google Scholar] [CrossRef]
- Xie, Y.X.; Wright, S.; Shen, Y.M.; Du, L.C. Bioactive natural products from Lysobacter. Nat. Prod. Rep. 2012, 29, 1277–1287. [Google Scholar] [CrossRef]
- Yu, F.; Zaleta-Rivera, K.; Zhu, X.; Huffman, J.; Millet, J.C.; Harris, S.D.; Yuen, G.; Li, X.-C.; Du, L. Structure and biosynthesis of deat-stable antifungal factor (HSAF), a broad-spectrum antimycotic with a novel mode of action. Antimicrob. Agents Chemother. 2007, 51, 64–72. [Google Scholar] [CrossRef]
- Li, S.; Jochum, C.C.; Yu, F.; Zaleta-Rivera, K.; Du, L.; Harris, S.D.; Yuen, G.Y. An antibiotic complex from Lysobacter enzymogenes strain C3: Antimicrobial activity and role in plant disease control. Phytopathology 2008, 98, 695–701. [Google Scholar] [CrossRef]
- Cao, S.G.; Blodgett, J.A.V.; Clardy, J. Targeted discovery of polycyclic tetramate macrolactams from an environmental Streptomyces strain. Org. Lett. 2010, 12, 4652–4654. [Google Scholar] [CrossRef]
- Elkin, S.R.; Oswald, N.W.; Reed, D.K.; Mettlen, M.; MacMillan, J.B.; Schmid, S.L. Ikarugamycin: A natural product inhibitor of clathrin-mediated endocytosis. Traffic 2016, 17, 1139–1149. [Google Scholar] [CrossRef]
- Paquette, L.A.; Macdonald, D.; Anderson, L.G.; Wright, J. A triply convergent enantioselective total synthesis of (+)-ikarugamycin. J. Am. Chem. Soc. 1989, 111, 8037–8039. [Google Scholar] [CrossRef]
- Boeckman, R.K.; Weidner, C.H.; Perni, R.B.; Napier, J.J. An enantioselective and highly convergent synthesis of (+)-ikarugamycin. J. Am. Chem. Soc. 1989, 111, 8036–8037. [Google Scholar] [CrossRef]
- Cramer, N.; Laschat, S.; Baro, A.; Schwalbe, H.; Richter, C. Enantioselective total synthesis of cylindramide. Angew. Chem. Int. Ed. 2005, 44, 820–822. [Google Scholar] [CrossRef] [PubMed]
- Henderson, J.A.; Phillips, A.J. Total synthesis of aburatubolactam A. Angew. Chem. Int. Ed. 2008, 47, 8499–8501. [Google Scholar] [CrossRef] [PubMed]
- Blodgett, J.A.V.; Oh, D.C.; Cao, S.G.; Currie, C.R.; Kolter, R.; Clardy, J. Common biosynthetic origins for polycyclic tetramate macrolactams from phylogenetically diverse bacteria. Proc. Natl. Acad. Sci. USA 2010, 107, 11692–11697. [Google Scholar] [CrossRef] [PubMed]
- Lou, L.L.; Qian, G.L.; Xie, Y.X.; Hang, J.L.; Chen, H.T.; Zaleta-Riyera, K.; Li, Y.Y.; Shen, Y.M.; Dussault, P.H.; Liu, F.Q.; et al. Biosynthesis of HSAF, a tetramic acid-containing macrolactam from Lysobacter enzymogenes. J. Am. Chem. Soc. 2011, 133, 643–645. [Google Scholar] [CrossRef]
- Luo, Y.; Huang, H.; Liang, J.; Wang, M.; Lu, L.; Shao, Z.; Cobb, R.E.; Zhao, H. Activation and characterization of a cryptic polycyclic tetramate macrolactam biosynthetic gene cluster. Nat. Commun. 2013, 4. [Google Scholar] [CrossRef]
- Zhang, G.T.; Zhang, W.J.; Zhang, Q.B.; Shi, T.; Ma, L.; Zhu, Y.G.; Li, S.M.; Zhang, H.B.; Zhao, Y.L.; Shi, R.; et al. Mechanistic insights into polycycle formation by reductive cyclization in ikarugamycin biosynthesis. Angew. Chem. Int. Ed. 2014, 53, 4840–4844. [Google Scholar] [CrossRef]
- Li, Y.Y.; Chen, H.T.; Ding, Y.J.; Xie, Y.X.; Wang, H.X.; Cerny, R.L.; Shen, Y.M.; Du, L.C. Iterative assembly of two separate polyketide chains by the same single-module bacterial polyketide synthase in the biosynthesis of HSAF. Angew. Chem. Int. Ed. 2014, 53, 7524–7530. [Google Scholar] [CrossRef]
- Saha, S.; Zhang, W.J.; Zhang, G.T.; Zhu, Y.G.; Chen, Y.C.; Liu, W.; Yuan, C.S.; Zhang, Q.B.; Zhang, H.B.; Zhang, L.P.; et al. Activation and characterization of a cryptic gene cluster reveals a cyclization cascade for polycyclic tetramate macrolactams. Chem. Sci. 2017, 8, 1607–1612. [Google Scholar] [CrossRef]
- Antosch, J.; Schaefers, F.; Gulder, T.A.M. Heterologous reconstitution of ikarugamycin biosynthesis in E. coli. Angew. Chem. Int. Ed. 2014, 53, 3011–3014. [Google Scholar] [CrossRef] [PubMed]
- Greunke, C.; Glockle, A.; Antosch, J.; Gulder, T.A. Biocatalytic total synthesis of ikarugamycin. Angew. Chem. Int. Ed. 2017, 56, 4351–4355. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, H.; Liu, Y.; Jiao, Y.; Li, S.; Shen, Y.; Du, L. Biosynthesis of the polycyclic system in the antifungal HSAF and analogues from Lysobacter enzymogenes. Angew. Chem. Int. Ed. 2018, 57, 6221–6225. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, H.; Shen, Y.; Li, Y.; Du, L. OX4 is an NADPH-dependent dehydrogenase catalyzing an extended Michael addition reaction to form the six-membered ring in the antifungal HSAF. Biochemistry 2019. [Google Scholar] [CrossRef] [PubMed]
- Olano, C.; Garcia, I.; Gonzalez, A.; Rodriguez, M.; Rozas, D.; Rubio, J.; Sanchez-Hidalgo, M.; Brana, A.F.; Mendez, C.; Salas, J.A. Activation and identification of five clusters for secondary metabolites in Streptomyces albus J1074. Microb. Biotechnol. 2014, 7, 242–256. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.J.; Zhang, G.T.; Zhang, L.P.; Liu, W.; Jiang, X.D.; Jin, H.B.; Liu, Z.W.; Zhang, H.B.; Zhou, A.H.; Zhang, C.S. New polycyclic tetramate macrolactams from marine-derived Streptomyces sp. SCSIO 40060. Tetrahedron 2018, 74, 6839–6845. [Google Scholar] [CrossRef]
- Mei, X.; Wang, L.; Wang, D.; Fan, J.; Zhu, W. Polycyclic tetramate macrolactams from the marine-derived Actinoalloteichus cyanogriseus WH1-2216-6. Chin. J. Org. Chem. 2017, 37, 2352–2360. [Google Scholar] [CrossRef]
- De Bruijn, I.; Cheng, X.; de Jager, V.; Exposito, R.G.; Watrous, J.; Patel, N.; Postma, J.; Dorrestein, P.C.; Kobayashi, D.; Raaijmakers, J.M. Comparative genomics and metabolic profiling of the genus Lysobacter. BMC Genom. 2015, 16, 991. [Google Scholar] [CrossRef]
- Zhang, W.J.; Ma, L.; Li, S.M.; Liu, Z.; Chen, Y.C.; Zhang, H.B.; Zhang, G.T.; Zhang, Q.B.; Tian, X.P.; Yuan, C.S.; et al. Indimicins A-E, Bisindole Alkaloids from the Deep-Sea-Derived Streptomyces sp SCSIO 03032. J. Nat. Prod. 2014, 77, 1887–1892. [Google Scholar] [CrossRef]
- Xu, L.; Wu, P.; Wright, S.J.; Du, L.; Wei, X. Bioactive polycyclic tetramate macrolactams from Lysobacter enzymogenes and their absolute configurations by theoretical ECD calculations. J. Nat. Prod. 2015, 78, 1841–1847. [Google Scholar] [CrossRef]
- Bae, M.A.; Yamada, K.; Ijuin, Y.; Tsuji, T.; Yazawa, K.; Tomono, Y.; Uemura, D. Aburatubolactam A, a novel inhibitor of superoxide anion generation from a marine microorganism. Heterocycl. Commun. 1996, 2, 315–318. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, H.; Song, R.; Chen, J.; Li, T.; Li, Y.; Du, L.; Shen, Y. Targeted Discovery and Combinatorial Biosynthesis of Polycyclic Tetramate Macrolactam Combamides A-E. Org. Lett. 2018, 20, 3504–3508. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, H.; Li, Y.; Du, L. Construction of a hybrid gene cluster to reveal coupled ring formation-hydroxylation in the biosynthesis of HSAF and analogues from Lysobacter enzymogenes. MedChemComm 2019, 10, 907–912. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, S.; Wong, C.P.; Ozeki, M.; Zhang, H.; Hayashi, F.; Awakawa, T.; Asamizu, S.; Onaka, H.; Abe, I. Umezawamides, new bioactive polycyclic tetramate macrolactams isolated from a combined-culture of Umezawaea sp. and mycolic acid-containing bacterium. J. Antibiot. (Tokyo) 2018, 71, 653–657. [Google Scholar] [CrossRef] [PubMed]
- Ward, A.C.; Allenby, N.E. Genome mining for the search and discovery of bioactive compounds: The Streptomyces paradigm. FEMS Microbiol. Lett. 2018, 365. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.Y.; Huffman, J.; Li, Y.; Du, L.C.; Shen, Y.M. 3-Hydroxylation of the polycyclic tetramate macrolactam in the biosynthesis of antifungal HSAF from Lysobacter enzymogenes C3. MedChemComm 2012, 3, 982–986. [Google Scholar] [CrossRef]
- Zhang, Q.; Li, H.; Li, S.; Zhu, Y.; Zhang, G.; Zhang, H.; Zhang, W.; Shi, R.; Zhang, C. Carboxyl formation from methyl via triple hydroxylations by XiaM in xiamycin A biosynthesis. Org. Lett. 2012, 14, 6142–6145. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef]
- Blin, K.; Wolf, T.; Chevrette, M.G.; Lu, X.; Schwalen, C.J.; Kautsar, S.A.; Suarez Duran, H.G.; de los Santos, E.L.; Kim, H.U.; Nave, M.; et al. antiSMASH 4.0—Improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res. 2017, 45, W36–W41. [Google Scholar] [CrossRef]
- Skehan, P.; Storeng, R.; Scudiero, D.; Monks, A.; McMahon, J.; Vistica, D.; Warren, J.T.; Bokesch, H.; Kenney, S.; Boyd, M.R. New colorimetric cytotoxicity assay for anticancer-drug screening. J. Natl. Cancer Inst. 1990, 82, 1107–1112. [Google Scholar] [CrossRef]
- Nakabayashi, H.; Taketa, K.; Miyano, K.; Yamane, T.; Sato, J. Growth of Human Hepatoma-Cell Lines with Differentiated Functions in Chemically Defined Medium. Cancer Res. 1982, 42, 3858–3863. [Google Scholar] [PubMed]



| No. | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| 2 | 5.70, dd, (2.3, 11.5) | 5.73, dd, (1.9, 11.4) | 5.72, dd, (2.2, 11.3) | 5.75, dd, (2.0, 11.5) | 5.75, dd, (2.1, 11.3) | 5.77, dd, (2.0, 11.5) |
| 3 | 5.90, td, (1.8, 11.2) | 5.9, td, (2.2, 11.1) | 5.92, td, (2.1, 11.0) | 5.92, td, (2.1, 11.1) | 5.96, td, (2.1, 11.1) | 5.96, td, (2.2, 11.1) |
| 4a | 1.89, m | 1.92, m | 2.00, m | 2.04, m | 2.03, m | 2.05, m |
| 4b | 3.52, m | 3.49, m | 3.62, m | 3.62, m | 3.65, m | 3.64, m |
| 5 | 1.28, m | 1.27, m | 1.90, m | 1.90, m | 1.87, m | 1.88, m |
| 6 | 1.64, m | 1.27, m | 2.09, m | 2.10, m | 2.56, m | 2.57, m |
| 7a | 0.86, m | 0.87, m | 1.05, m | 0.85, m | 0.98, m | 1.01, m |
| 7b | 1.96, m | 1.97, m | 2.05, m | 2.06, m | 2.11, m | 2.14, m |
| 8 | 2.35, m | 2.35, m | 2.35, m | 2.35, m | 2.03, m | 2.03, m |
| 9a | 0.80, m | 0.79, m | 0.84, m | 1.06, m | 0.68, m | 0.69, m |
| 9b | 2.01, m | 2.01, m | 2.05, m | 2.06, m | 2.02, m | 2.03, m |
| 10 | 1.33, m | 1.32, m | 1.37, m | 1.37, m | 1.53, m | 1.53, m |
| 11 | 1.27, m | 1.64, m | 1.09, m | 1.09, m | 1.21, m | 1.22, m |
| 12 | 1.75, m | 1.77, m | 2.21, m | 2.21, m | ||
| 13 | 1.10, m | 1.09, m | 2.37, m | 2.37, m | 2.32, m | 2.32, m |
| 14 | 3.25, m | 3.25, m | ||||
| 15a | 1.24, m | 1.24, m | 2.11, m | 2.11, m | 2.12, m | 2.13, m |
| 15b | 1.74, m | 1.75, m | 2.59, m | 2.59, m | 2.59, m | 2.60, m |
| 16 | 2.06, m | 2.07, m | 2.38, m | 2.40, m | 2.44, m | 2.47, m |
| 17 | 6.57, dd, (10.5, 15.5) | 6.55, dd, (10.5, 15.5) | 6.63, dd, (10.3, 15.6) | 6.61, t, (15.5) | 6.60, dd, (10.3, 15.5) | 6.59, dd, (11.5, 15.5) |
| 18 | 6.86, d, (15.5) | 6.95, d, (15.5) | 6.87, d, (15.6) | 6.96, d, (15.5) | 6.88, d, (15.5) | 6.96, d, (15.5) |
| 22 | NH, 8.68, s | NH, 8.68, s | NH, 8.95, s | NH, 8.73, brs | NH, 8.96, s | NH, 8.75, s |
| 23 | 3.86, d, (1.2) | 3.81, d, (5.7) | 3.87, d, (1.4) | 3.83, s | 3.87, d, (1.1) | 3.84, d, (6.1) |
| 25a | 3.81, dt, (1.5, 6.1) | 1.74, m | 3.81, dt, (1.4,6.2) | 1.74, m | 3.82, dt, (2.0, 6.4) | 1.74, m |
| 25b | 1.84, m | 1.86, m | 1.86, m | |||
| 26a | 1.18, m | 1.15, m | 1.18, m | 1.17, m | 1.20, m | 1.15, m |
| 26b | 1.38, m | 1.32, m | 1.39, m | 1.35, m | 1.39, m | 1.34, m |
| 27a | 2.57, m | 2.39, m | 2.59, m | 3.23, m | 2.58 m | 2.40, m |
| 27b | 3.25, m | 3.22, m | 3.26, m | 2.39, m | 3.24, m | 3.23, m |
| 28 | NH, 7.96, t, (5.7) | NH, 7.82, t, (5.3) | NH, 7.98, t, (5.6) | NH, 7.86, s | NH, 8.00, t, (5.6) | NH, 7.88, t, (5.6) |
| 29a | 1.04, m | 1.04, m | 1.03, m | 1.03, m | 0.99, m | 1.02, m |
| 29b | 1.55, m | 1.55, m | 1.53, m | 1.54, m | 1.51, m | 1.52, m |
| 30 | 0.85, t (7.4) | 0.85, t (7.4) | 0.84, t (7.4) | 0.84, t (7.4) | 0.84, t (7.4) | 0.84, t (7.4) |
| 31 | 1.06, d (6.4) | 1.06, d (6.4) | 0.96, d (6.5) | 0.96, d (6.5) | 0.94, d (6.7) | 0.94, d (6.8) |
| No. | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| 1 | 165.5, C | 165.5, C | 165.5, C | 165.6, C | 165.5, C | 165.6, C |
| 2 | 124.1, CH | 124.2, CH | 124.4, CH | 124.5, CH | 124.5, CH | 124.7, CH |
| 3 | 139.1, CH | 138.9, CH | 138.4, CH | 138.3, CH | 138.2, CH | 138.1, CH |
| 4 | 28.0, CH2 | 28.1, CH2 | 27.7, CH2 | 27.7, CH2 | 27.4, CH2 | 27.4, CH2 |
| 5 | 43.5, CH | 43.5, CH | 43.2, CH | 43.2, CH | 43.0, CH | 43.0, CH |
| 6 | 47.4, CH | 46.4, CH | 51.1, CH | 51.2, CH | 47.9, CH | 48.0, CH |
| 7 | 37.2, CH2 | 37.3, CH2 | 38.4, CH2 | 39.7, CH2 | 36.5, CH2 | 36.5, CH2 |
| 8 | 41.5, CH | 41.4, CH | 40.4, CH | 40.4, CH | 51.3, CH | 51.3, CH |
| 9 | 40.3, CH2 | 40.3, CH2 | 39.6, CH2 | 38.5, CH2 | 38.1, CH2 | 38.1, CH2 |
| 10 | 53.5, CH | 53.5, CH | 53,2, CH | 53,2, CH | 50.4, CH | 50.4, CH |
| 11 | 46.5, CH | 47.6, CH | 46.7, CH | 46.7, CH | 49.3, CH | 49.3, CH |
| 12 | 58.1, CH | 58.1, CH | 50.4, CH | 50.4, CH | 89.7, CH | 89.7, CH |
| 13 | 59.1, CH | 59.1, CH | 63.0, CH | 63.0, CH | 64.2, CH | 64.2, CH |
| 14 | 72.7, CH | 72.7, CH | 207.4, C | 207.4, C | 210.1 C | 210.2, C |
| 15 | 41.9, CH2 | 41.9, CH2 | 45.6, CH2 | 45.6, CH | 46.0, CH2 | 46.0, CH2 |
| 16 | 45.7, CH | 45.6, CH | 47.7, CH | 47.8, CH | 47.1, CH | 47.2, CH |
| 17 | 150.1, CH | 149.6, CH | 147.8, CH | 147.4, CH | 147.5, CH | 147.2, CH |
| 18 | 121.3, CH | 121.5, CH | 122.0, CH | 122.1, CH | 122.1, CH | 122.2, CH |
| 19 | 172.2, C | 171.8, C | 171.9, C | 175.1, C | 171.8, C | 171.2, C |
| 20 | 100.4, C | 100.7, C | 100.7, C | 101.1, C | 100.7, C | 101.0, C |
| 21 | 175.7, C | 175.3, C | 175.6, C | 171.4, C | 175.6, C | 175.1, C |
| 23 | 68.6, CH | 61.0, CH | 68.5, CH | 61.1, CH | 68.6, CH | 61.1, CH |
| 24 | 193.0, C | 195.8, C | 193.0, C | 195.8, C | 193.0, C | 195.9, C |
| 25 | 70.1, CH | 26.2, CH2 | 70.1, CH | 26.2, CH2 | 70.1, CH | 26.1, CH2 |
| 26 | 31.1, CH2 | 20.4, CH2 | 31.0, CH2 | 20.4, CH2 | 31.0, CH2 | 31.1, CH2 |
| 27 | 36.4, CH2 | 38.0, CH2 | 36.4, CH2 | 38.0, CH2 | 36.4, CH2 | 36.4, CH2 |
| 29 | 25.8, CH2 | 25.8, CH2 | 25.5, CH2 | 25.5, CH2 | 25.3, CH2 | 25.3, CH2 |
| 30 | 12.6, CH3 | 12.6, CH3 | 12.4, CH3 | 12.4, CH3 | 12.0, CH3 | 12.0, CH3 |
| 31 | 18.4, CH3 | 18.4, CH3 | 17.6, CH3 | 17.6, CH3 | 11.5, CH3 | 11.5, CH3 |
| IC50 (μM) | ||||
|---|---|---|---|---|
| SF-268 | MCF-7 | A549 | HepG2 | |
| 1 | 3.83 ± 0.13 | 2.47 ± 0.05 | 5.99 ± 0.15 | 3.48 ± 0.17 |
| 2 | 10.62 ± 0.45 | 3.84 ± 0.07 | 11.01 ± 1.09 | 10.34 ± 0.88 |
| 3 | 4.57 ± 0.18 | 3.18 ± 0.13 | 3.75 ± 0.62 | 6.30 ± 0.34 |
| 4 | 7.53 ± 0.27 | 3.54 ± 0.24 | 10.45 ± 0.46 | 17.86 ± 0.62 |
| 5 | 3.21 ± 0.18 | 6.83 ± 0.36 | 3.28 ± 0.04 | 3.12 ± 0.11 |
| a CP | 3.26 ± 0.29 | 3.19 ± 0.12 | 1.56 ± 0.08 | 2.42 ± 0.14 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, W.; Zhang, W.; Jin, H.; Zhang, Q.; Chen, Y.; Jiang, X.; Zhang, G.; Zhang, L.; Zhang, W.; She, Z.; et al. Genome Mining of Marine-Derived Streptomyces sp. SCSIO 40010 Leads to Cytotoxic New Polycyclic Tetramate Macrolactams. Mar. Drugs 2019, 17, 663. https://doi.org/10.3390/md17120663
Liu W, Zhang W, Jin H, Zhang Q, Chen Y, Jiang X, Zhang G, Zhang L, Zhang W, She Z, et al. Genome Mining of Marine-Derived Streptomyces sp. SCSIO 40010 Leads to Cytotoxic New Polycyclic Tetramate Macrolactams. Marine Drugs. 2019; 17(12):663. https://doi.org/10.3390/md17120663
Chicago/Turabian StyleLiu, Wei, Wenjun Zhang, Hongbo Jin, Qingbo Zhang, Yuchan Chen, Xiaodong Jiang, Guangtao Zhang, Liping Zhang, Weimin Zhang, Zhigang She, and et al. 2019. "Genome Mining of Marine-Derived Streptomyces sp. SCSIO 40010 Leads to Cytotoxic New Polycyclic Tetramate Macrolactams" Marine Drugs 17, no. 12: 663. https://doi.org/10.3390/md17120663
APA StyleLiu, W., Zhang, W., Jin, H., Zhang, Q., Chen, Y., Jiang, X., Zhang, G., Zhang, L., Zhang, W., She, Z., & Zhang, C. (2019). Genome Mining of Marine-Derived Streptomyces sp. SCSIO 40010 Leads to Cytotoxic New Polycyclic Tetramate Macrolactams. Marine Drugs, 17(12), 663. https://doi.org/10.3390/md17120663