Biosynthetic Potential of a Novel Antarctic Actinobacterium Marisediminicola antarctica ZS314T Revealed by Genomic Data Mining and Pigment Characterization
Abstract
1. Introduction
2. Results and Discussion
2.1. General Feature of the Genome
2.2. Prediction of Secondary Metabolite Biosynthetic Gene Clusters
2.2.1. Cluster 1: T3PKS
2.2.2. Cluster 2: Terpene
2.2.3. Cluster 3: Terpene
2.2.4. Cluster 4: NRPS
2.2.5. Cluster 5: Oligosaccharide
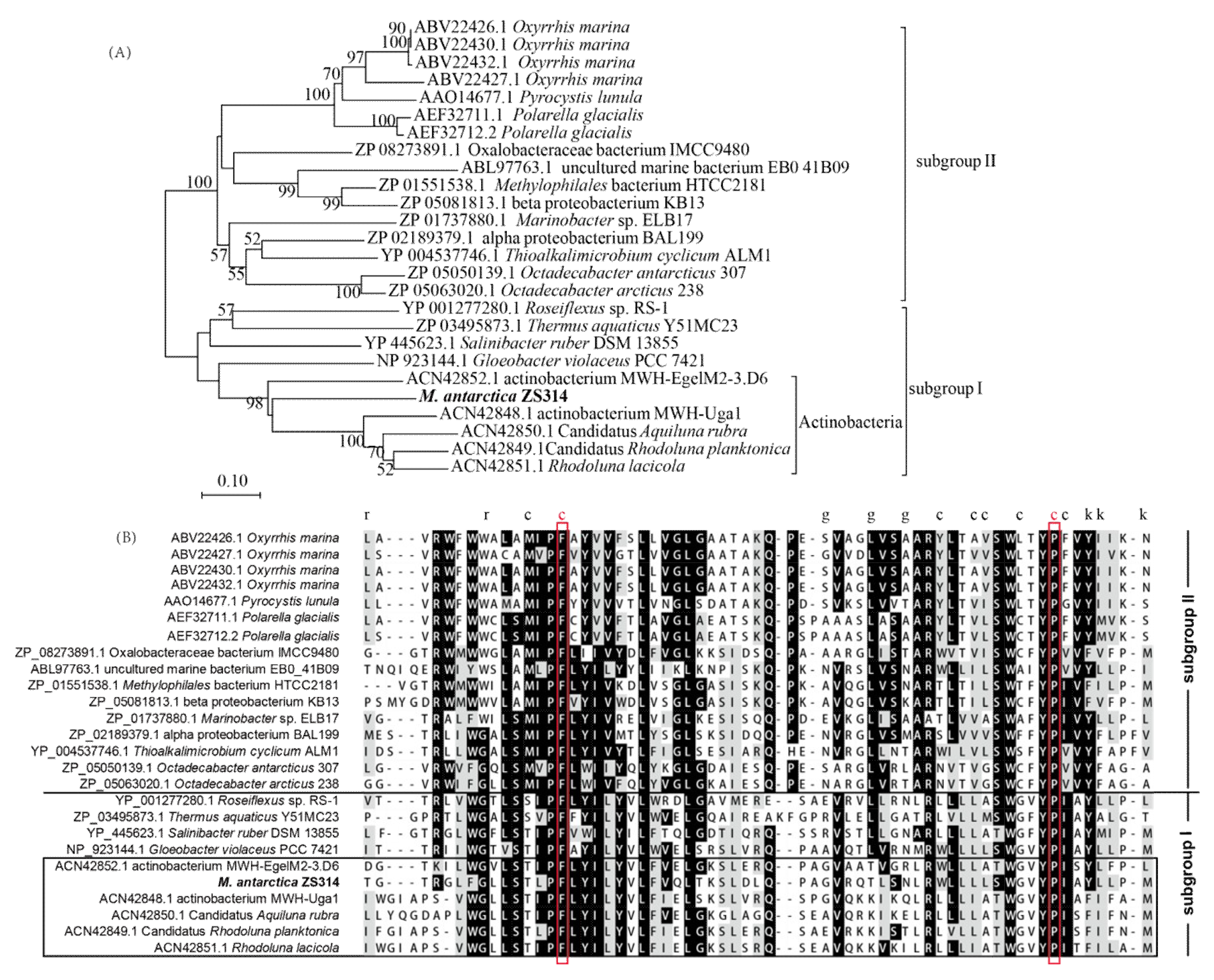
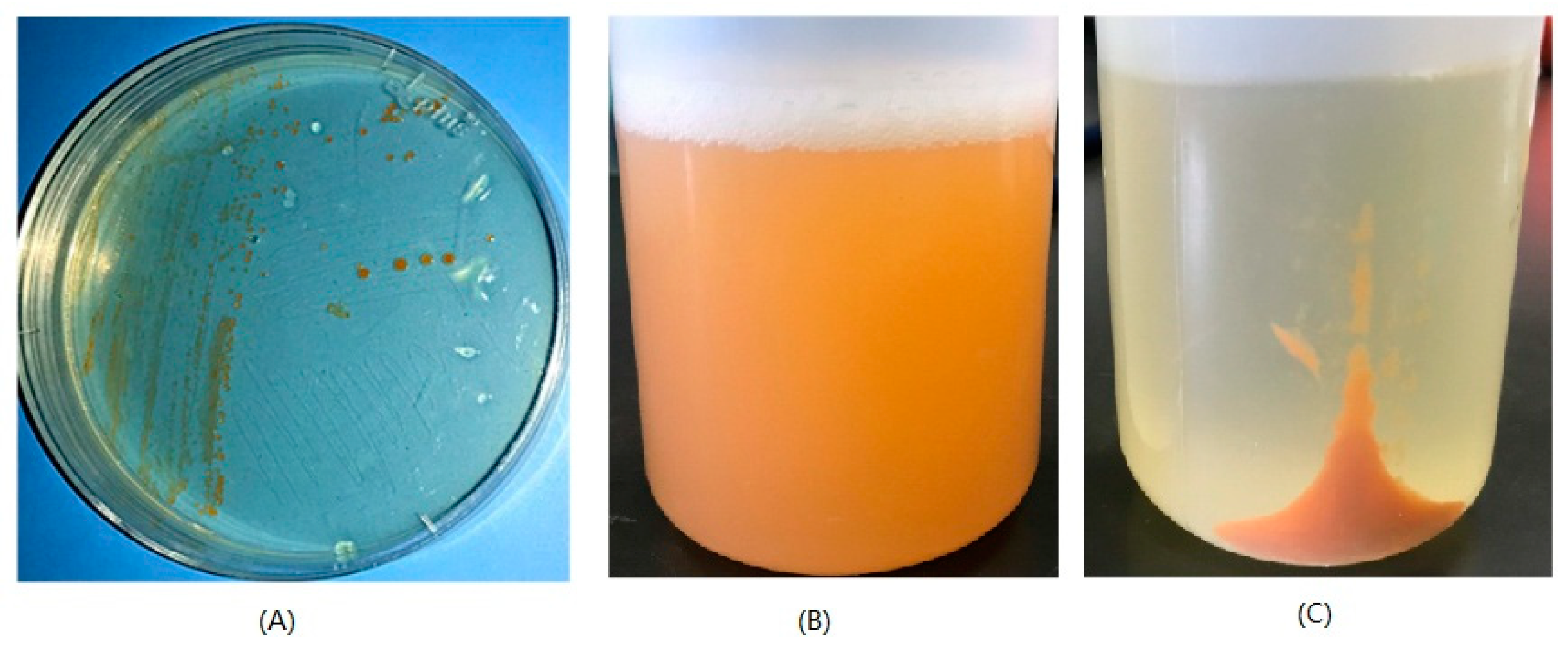
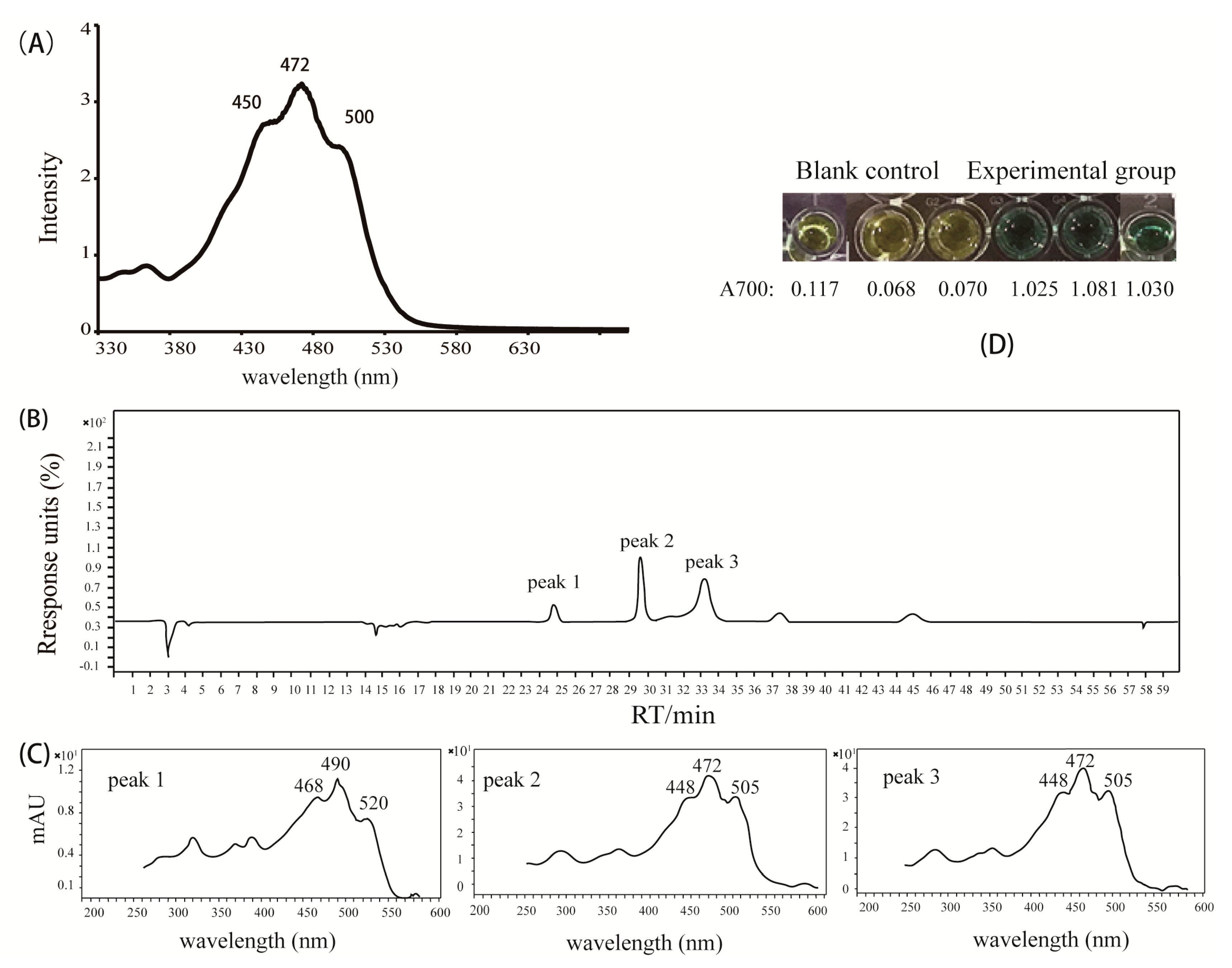
2.3. Analysis of the Pigments from M. antarctica ZS314T
3. Materials and Methods
3.1. Strain and Culture Conditions
3.2. Genome Sequencing and Data Mining of M. antarctica ZS314T
3.3. Extraction and Analysis of Reddish Orange Pigments
3.4. Anti-Oxidation Assay of Reddish Orange Pigments
3.5. Data Accessibility
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Azman, A.S.; Othman, I.; Velu, S.S.; Chan, K.G.; Lee, L.H. Mangrove rare actinobacteria: Taxonomy, natural compound, and discovery of bioactivity. Front. Microbiol. 2015, 6, 856. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Li, L.; Yu, Y.; Bo, C. Complete genome of Brachybacterium sp. P6-10-X1 isolated from deep-sea sediments of the Southern Ocean. Mar. Genom. 2017, 35, 27–29. [Google Scholar] [CrossRef]
- Xiong, Z.Q.; Wang, J.F.; Hao, Y.Y.; Wang, Y. Recent advances in the discovery and development of marine microbial natural products. Mar. Drugs 2013, 11, 700–717. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, K.; Gupta, R.K. Diversity and isolation of rare actinomycetes: an overview. Crit. Rev. Microbiol. 2013, 39, 256–294. [Google Scholar] [CrossRef] [PubMed]
- Rampelotto, P.H. Polar microbiology: Recent advances and future perspectives. Biology (Base) 2014, 3, 81–84. [Google Scholar] [CrossRef]
- Li, H.R.; Yu, Y.; Luo, W.; Zeng, Y.X. Marisediminicola antarctica gen. nov., sp. nov., an actinobacterium isolated from the Antarctic. Int. J. Syst. Evol. Microbiol. 2010, 60, 2535–2539. [Google Scholar] [CrossRef] [PubMed]
- Jagannadham, M.V.; Chattopadhyay, M.K.; Subbalakshmi, C.; Vairamani, M.; Narayanan, K.; Rao, C.M.; Shivaji, S. Carotenoids of an Antarctic psychrotolerant bacterium, Sphingobacterium antarcticus, and a mesophilic bacterium, Sphingobacterium multivorum. Arch. Microbiol. 2000, 173, 418–424. [Google Scholar] [CrossRef]
- Joanna, F.; Květoslava, B. Potential role of carotenoids as antioxidants in human health and disease. Nutrients 2014, 6, 466–488. [Google Scholar]
- Roman, N.; Stafsnes, M.H.; Trygve, A.; Audun, G.Y.; Per, B.; Trygve, B. Biosynthetic pathway for γ-cyclic sarcinaxanthin in Micrococcus luteus: heterologous expression and evidence for diverse and multiple catalytic functions of C(50) carotenoid cyclases. J. Bacteriol. 2010, 192, 5688–5699. [Google Scholar]
- Wang, F.; Jiang, J.G.; Chen, Q. Progress on molecular breeding and metabolic engineering of biosynthesis pathways of C-30, C-35, C-40, C-45, C-50 carotenoids. Biotechnol. Adv. 2007, 25, 211–222. [Google Scholar] [CrossRef]
- Takaichi, S.; Inoue, K.; Akaike, M.; Kobayashi, M.; Ohoka, H.; Madigan, M.T. The major carotenoid in all known species of heliobacteria is the C30 carotenoid 4,4’-diaponeurosporene, not neurosporene. Arch. Microbiol. 1997, 168, 277–281. [Google Scholar] [CrossRef] [PubMed]
- Tao, L.; Yao, H.; Cheng, Q. Genes from a Dietzia sp. for synthesis of C40 and C50 β-cyclic carotenoids. Gene 2007, 386, 90–97. [Google Scholar] [CrossRef] [PubMed]
- Funabashi, M.; Funa, N.; Horinouchi, S. Phenolic lipids synthesized by type III polyketide synthase confer penicillin resistance on Streptomyces griseus. J. Biol. Chem. 2008, 283, 13983–13991. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Xu, F.; Zeng, J.; Zhan, J. Type III polyketide synthases in natural product biosynthesis. IUBMB Life 2012, 64, 285–295. [Google Scholar] [CrossRef] [PubMed]
- Kozubek, A.; Pietr, S.; Czerwonka, A. Alkylresorcinols are abundant lipid components in different strains of Azotobacter chroococcum and Pseudomonas spp. J. Bacteriol. 1996, 178, 4027–4030. [Google Scholar] [CrossRef]
- Krubasik, P.; Kobayashi, M.; Sandmann, G. Expression and functional analysis of a gene cluster involved in the synthesis of decaprenoxanthin reveals the mechanisms for C50 carotenoid formation. FEBS J. 2001, 268, 3702–3708. [Google Scholar] [CrossRef]
- Sandmann, G. Molecular evolution of carotenoid biosynthesis from bacteria to plants. Physiol. Plant 2002, 116, 431–440. [Google Scholar] [CrossRef]
- Balashov, S.P.; Imasheva, E.S.; Boichenko, V.A.; Antón, J.; Wang, J.M.; Lanyi, J.K. Xanthorhodopsin: A proton pump with a light-harvesting carotenoid antenna. Science 2005, 309, 2061–2064. [Google Scholar] [CrossRef]
- Lanyi, J.K.; Balashov, S.P. Xanthorhodopsin: A bacteriorhodopsin-like proton pump with a carotenoid antenna. Biochim. Biophys. Acta-Bioenerg. 2008, 1777, 684–688. [Google Scholar] [CrossRef]
- Rodrigobaños, M.; Garbayo, I.; Vílchez, C.; Bonete, M.J.; Martínezespinosa, R.M. Carotenoids from haloarchaea and their potential in biotechnology. Mar. Drugs 2015, 13, 5508–5532. [Google Scholar] [CrossRef]
- Estrada, A.F.; Maier, D.; Scherzinger, D.; Avalos, J.; Al-Babili, S. Novel apocarotenoid intermediates in Neurospora crassa mutants imply a new biosynthetic reaction sequence leading to neurosporaxanthin formation. Fungal Genet Biol. 2008, 45, 1497–1505. [Google Scholar] [CrossRef] [PubMed]
- Hannibal, L.; Lorquin, J.; Dortoli, N.A.; Garcia, N.; Chaintreuil, C.; Massonboivin, C.; Dreyfus, B.; Giraud, E. Isolation and characterization of canthaxanthin biosynthesis genes from the photosynthetic bacterium Bradyrhizobium sp. strain ORS278. J. Bacteriol. 2000, 182, 3850–3853. [Google Scholar] [CrossRef] [PubMed]
- Graham, J.E.; Bryant, D.A. The biosynthetic pathway for Myxol-2’fucoside (myxoxanthophyll) in the cyanobacterium Synechococcus sp. strain PCC 7002. J. Bacteriol. 2009, 191, 3292–3300. [Google Scholar] [CrossRef] [PubMed]
- Moriya, Y.; Shigemizu, D.; Hattori, M.; Tokimatsu, T.; Kotera, M.; Goto, S.; Kanehisa, M. PathPred: an enzyme-catalyzed metabolic pathway prediction server. Nucleic Acids Res. 2010, 38, W138–W143. [Google Scholar] [CrossRef] [PubMed]
- Ullrich, A.; Rohrschneider, M.; Scheuermann, G.; Stadler, P.F.; Flamm, C. In silico evolution of early metabolism. Artif Life 2011, 17, 87–108. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Thiele, I.; Palsson, B.O. A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat. Protoc. 2010, 5, 93–121. [Google Scholar] [CrossRef]
- Vollmers, J.; Voget, S.; Dietrich, S.; Gollnow, K.; Smits, M.; Meyer, K.; Brinkhoff, T.; Simon, M.; Daniel, R. Poles Apart: Arctic and Antarctic Octadecabacter strains share high genome plasticity and a new type of xanthorhodopsin. PLOS ONE 2013, 8, e63422. [Google Scholar] [CrossRef]
- Imasheva, E.S.; Balashov, S.P.; Choi, A.R.; Jung, K.H.; Lanyi, J.K. Reconstitution of Gloeobacter violaceus rhodopsin with a light-harvesting carotenoid antenna. Biochemistry 2009, 48, 10948–10955. [Google Scholar] [CrossRef]
- Mccranie, E.K.; Bachmann, B.O. Bioactive oligosaccharide natural products. Nat. Prod. Rep. 2014, 31, 1026–1042. [Google Scholar] [CrossRef]
- Weitnauer, G.; Mühlenweg, A.; Trefzer, A.; Hoffmeister, D.; Süßmuth, R.D.; Jung, G.; Welzel, K.; Vente, A.; Girreser, U.; Bechthold, A. Biosynthesis of the orthosomycin antibiotic avilamycin A: deductions from the molecular analysis of the avi biosynthetic gene cluster of Streptomyces viridochromogenes Tü57 and production of new antibiotics. Chem. Biol. 2001, 8, 569–581. [Google Scholar] [CrossRef]
- Sarvari, S.; Moakedi, F. High carotenoid production by a halotolerant bacterium, Kocuria sp. strain QWT-12 and anticancer activity of its carotenoid. EXCLI J. 2017, 16, 840–851. [Google Scholar]
- Britton, G.; Liaaenjensen, S.; Hanspeter, P. Carotenoids, Volume 5: Nutrition and health. Carotenoids 2009, 24, 497–522. [Google Scholar]
- Luo, R.; Liu, B.; Xie, Y.; Li, Z.; Huang, W.; Yuan, J.; He, G.; Chen, Y.; Pan, Q.; Liu, Y.; et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 2012, 1, 18. [Google Scholar] [CrossRef] [PubMed]
- Boetzer, M.; Pirovano, W. SSPACE-LongRead: scaffolding bacterial draft genomes using long read sequence information. BMC Bioinform. 2014, 15, 211. [Google Scholar] [CrossRef] [PubMed]
- Delcher, A.L.; Bratke, K.A.; Powers, E.C.; Salzberg, S.L. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 2007, 23, 673–679. [Google Scholar] [CrossRef] [PubMed]
- Pruitt, K.D.; Tatusova, T.; Maglott, D.R. NCBI reference sequences (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007, 35, D61–D65. [Google Scholar] [CrossRef] [PubMed]
- Boeckmann, B.; Bairoch, A.; Apweiler, R.; Blatter, M.C.; Estreicher, A.; Gasteiger, E.; Martin, M.J.; Michoud, K.; O’Donovan, C.; Phan, I.; et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003, 31, 365–370. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 1999, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Tatusov, R.L.; Fedorova, N.D.; Jackson, J.D.; Jacobs, A.R.; Kiryutin, B.; Koonin, E.V.; Krylov, D.M.; Mazumder, R.; Mekhedov, S.L.; Nikolskaya, A.N. The COG database: An updated version includes eukaryotes. BMC Bioinform. 2003, 4, 41. [Google Scholar] [CrossRef]
- Harris, M.A.; Clark, J.; Ireland, A.; Lomax, J.; Ashburner, M.; Foulger, R.; Eilbeck, K.; Lewis, S.; Marshall, B.; Mungall, C.; et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004, 32, D258–D261. [Google Scholar] [PubMed]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef] [PubMed]
- Lagesen, K.; Hallin, P.; Rodland, E.; Staerfeldt, H.; Rognes, T.; Ussery, D. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007, 35, 3100–3108. [Google Scholar] [CrossRef] [PubMed]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Wolf, T.; Chevrette, M.G.; Lu, X.; Schwalen, C.J.; Kautsar, S.A.; Suarez Duran, H.G.; de Los Santos, E.L.C.; Kim, H.U.; Nave, M.; et al. antiSMASH 4.0-improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res. 2017, 45, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, M.; Schein, J.I. Circos: an information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Bolanos de la Torre, A.A.S.; Henderson, T.; Nigam, S.P.; Owusu-Apenten, K.R. A universally calibrated microplate ferric reducing antioxidant power (FRAP) assay for foods and applications to Manuka honey. Food Chem. 2015, 174, 119–123. [Google Scholar] [CrossRef] [PubMed]
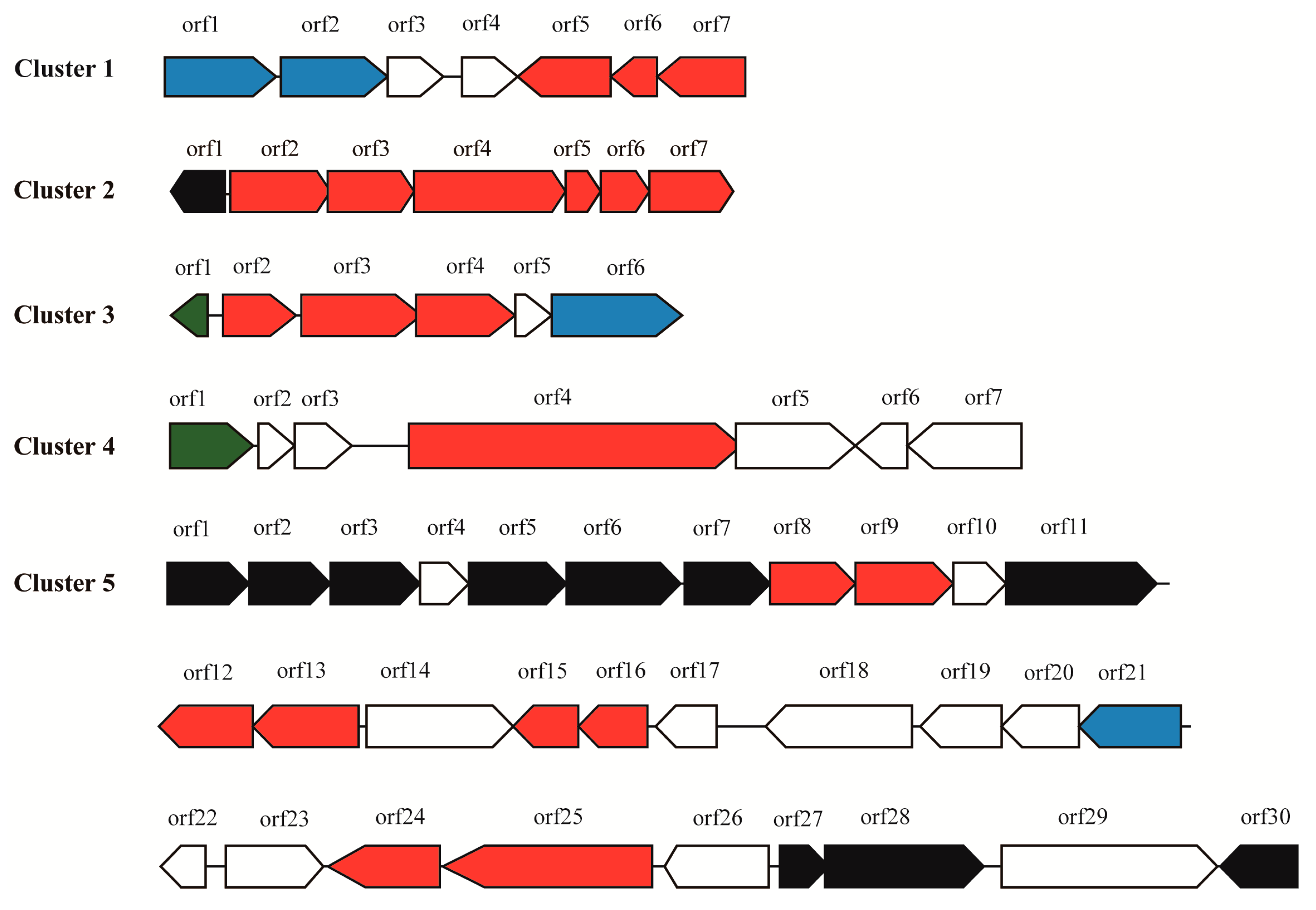
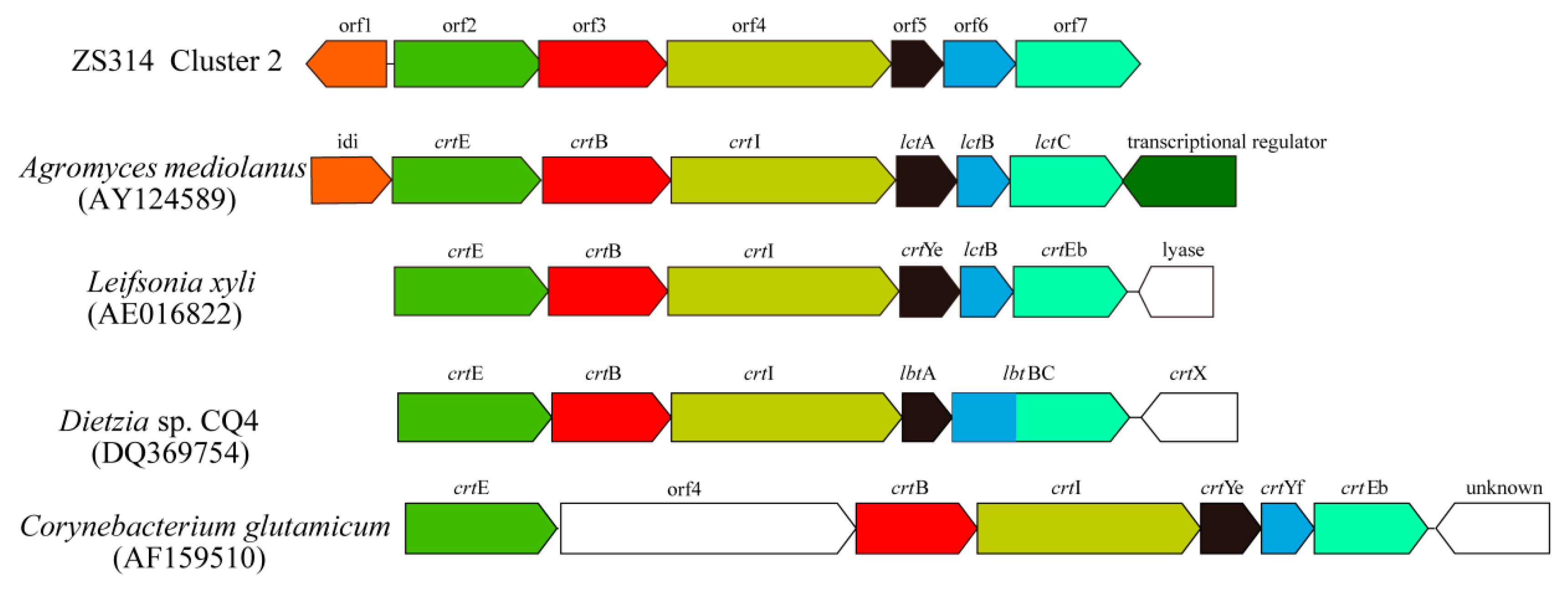
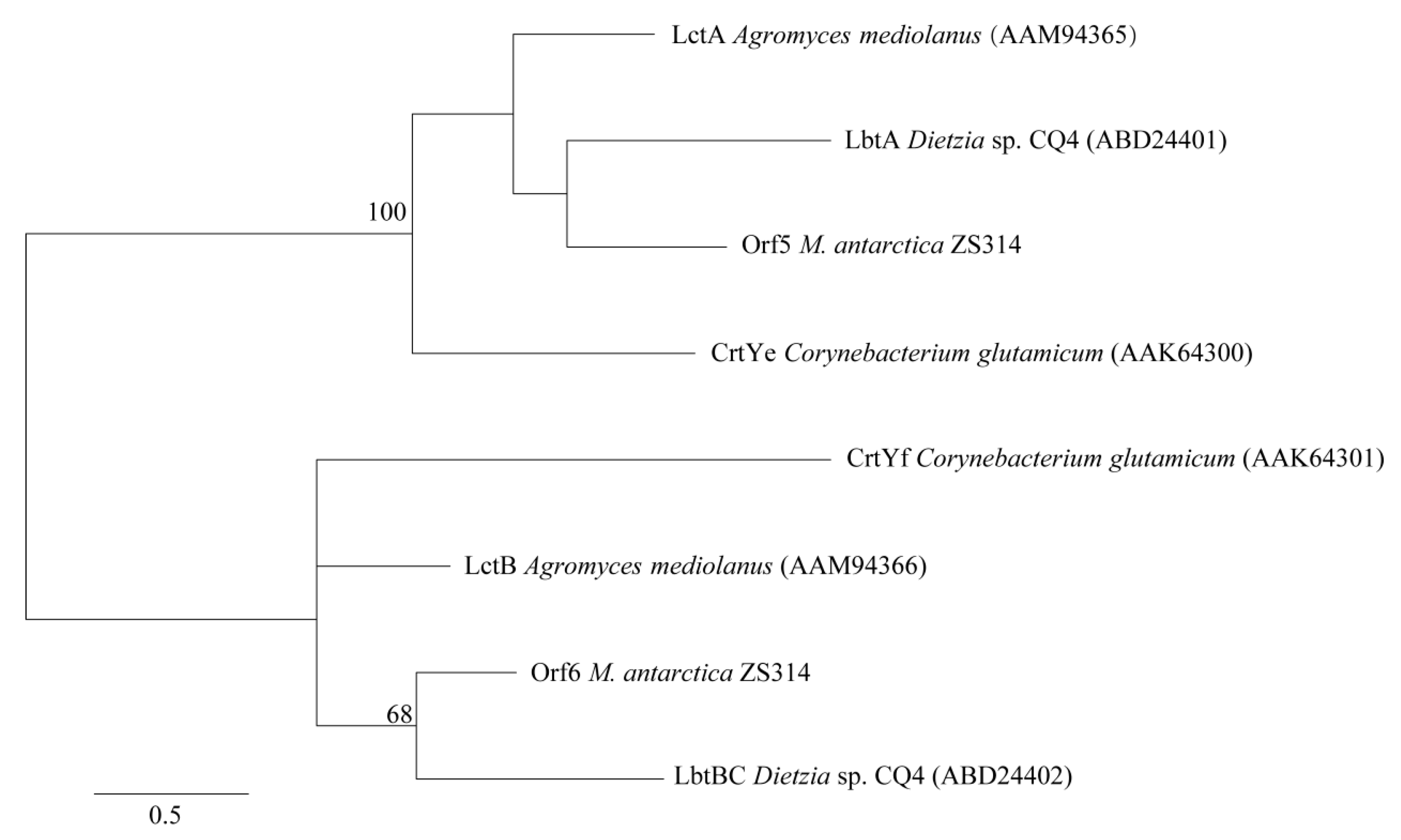
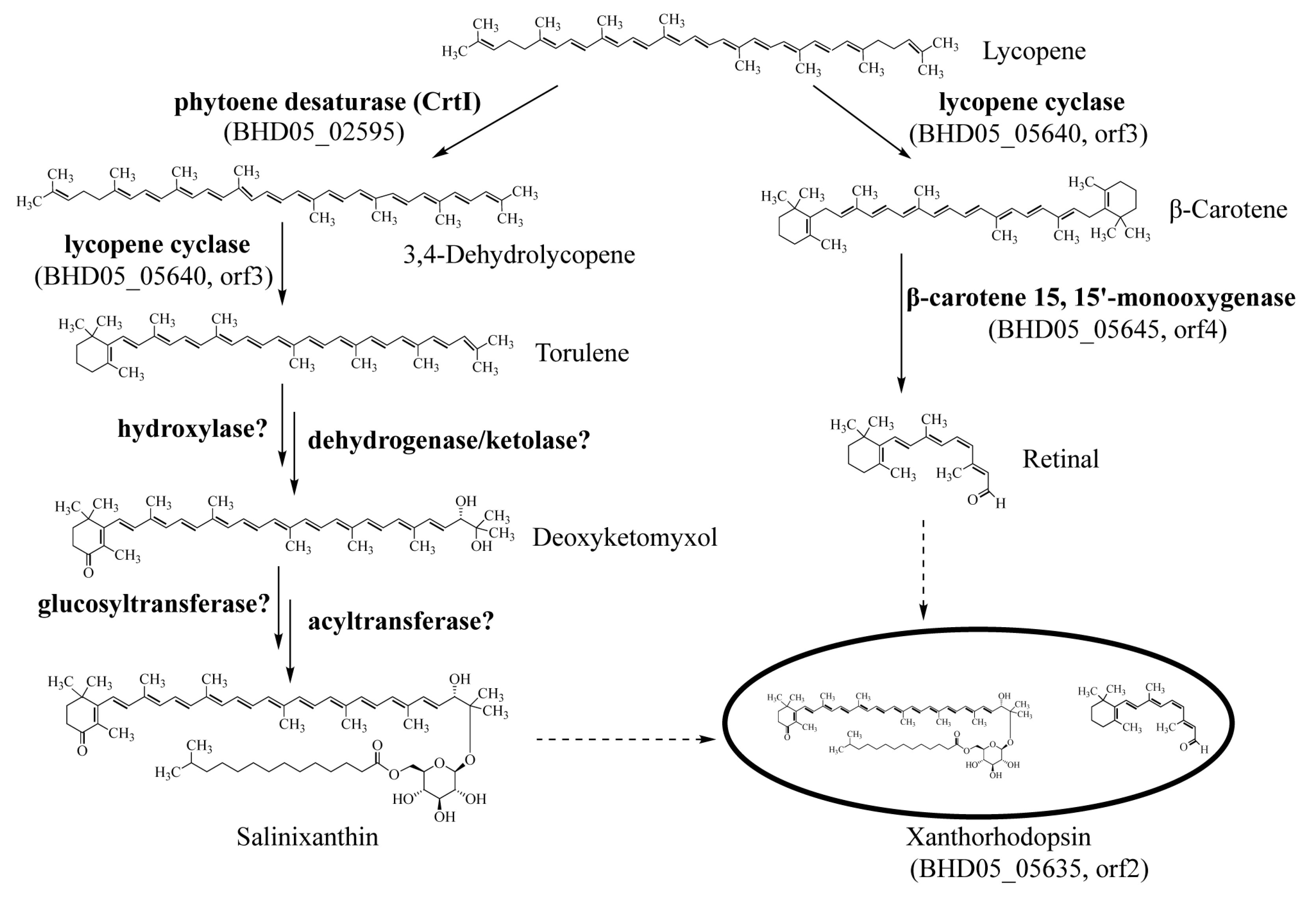
| Cluster ID | Type | Gene Number | Most Similar Known Gene Cluster | Percentage of Similar Genes |
|---|---|---|---|---|
| 1 | T3PKS | 7 | Alkylresorcinol | 100% |
| 2 | Terpene | 7 | Carotenoid | 50% |
| 3 | Terpene | 6 | NA * | 0 |
| 4 | NRPS | 7 | NA * | 0 |
| 5 | Oligosaccharide | 30 | NA * | 0 |
| Cluster 2 M. antarctica ZS314T | Corynebacterium glutamicum | Dietzia sp. CQ4 | Leifsonia xyli | Agromyces mediolanus | |||||
|---|---|---|---|---|---|---|---|---|---|
| Gene | Annotation | Gene | Identity | Gene | Identity | Gene | Identity | Gene | Identity |
| orf2 | Geranylgeranyl pyrophosphate synthase | crtE | 34% | crtE | 34% | crtE | 44% | crtE | 43% |
| orf3 | Phytoene synthase | crtB | 46% | crtB | 49% | crtB | 58% | crtB | 61% |
| orf4 | Phytoene desaturase | crtI | 52% | crtI | 61% | crtI | 61% | crtI | 58% |
| orf5 | Lycopene cyclase | crtYe | 41% | lbtA | 40% | crtYe | 65% | lctA | 50% |
| orf6 | Lycopene cyclase | crtYf | 38% | lbtBC1 | 46% | lctB | 35% | lctB | 43% |
| orf7 | Lycopene elongase | crtEb | 56% | lbtBC2 | 59% | crtEb | 61% | lctC | 64% |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liao, L.; Su, S.; Zhao, B.; Fan, C.; Zhang, J.; Li, H.; Chen, B. Biosynthetic Potential of a Novel Antarctic Actinobacterium Marisediminicola antarctica ZS314T Revealed by Genomic Data Mining and Pigment Characterization. Mar. Drugs 2019, 17, 388. https://doi.org/10.3390/md17070388
Liao L, Su S, Zhao B, Fan C, Zhang J, Li H, Chen B. Biosynthetic Potential of a Novel Antarctic Actinobacterium Marisediminicola antarctica ZS314T Revealed by Genomic Data Mining and Pigment Characterization. Marine Drugs. 2019; 17(7):388. https://doi.org/10.3390/md17070388
Chicago/Turabian StyleLiao, Li, Shiyuan Su, Bin Zhao, Chengqi Fan, Jin Zhang, Huirong Li, and Bo Chen. 2019. "Biosynthetic Potential of a Novel Antarctic Actinobacterium Marisediminicola antarctica ZS314T Revealed by Genomic Data Mining and Pigment Characterization" Marine Drugs 17, no. 7: 388. https://doi.org/10.3390/md17070388
APA StyleLiao, L., Su, S., Zhao, B., Fan, C., Zhang, J., Li, H., & Chen, B. (2019). Biosynthetic Potential of a Novel Antarctic Actinobacterium Marisediminicola antarctica ZS314T Revealed by Genomic Data Mining and Pigment Characterization. Marine Drugs, 17(7), 388. https://doi.org/10.3390/md17070388





