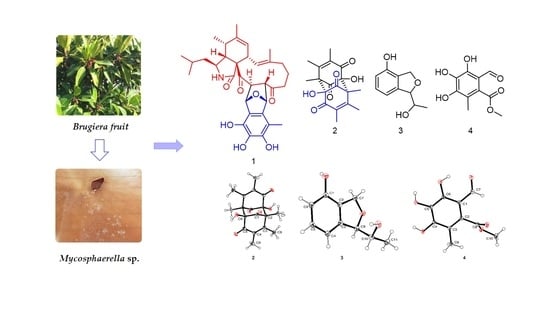
Secondary Metabolites with α-Glucosidase Inhibitory Activity from the Mangrove Fungus Mycosphaerella sp. SYSU-DZG01
Abstract
1. Introduction
2. Results
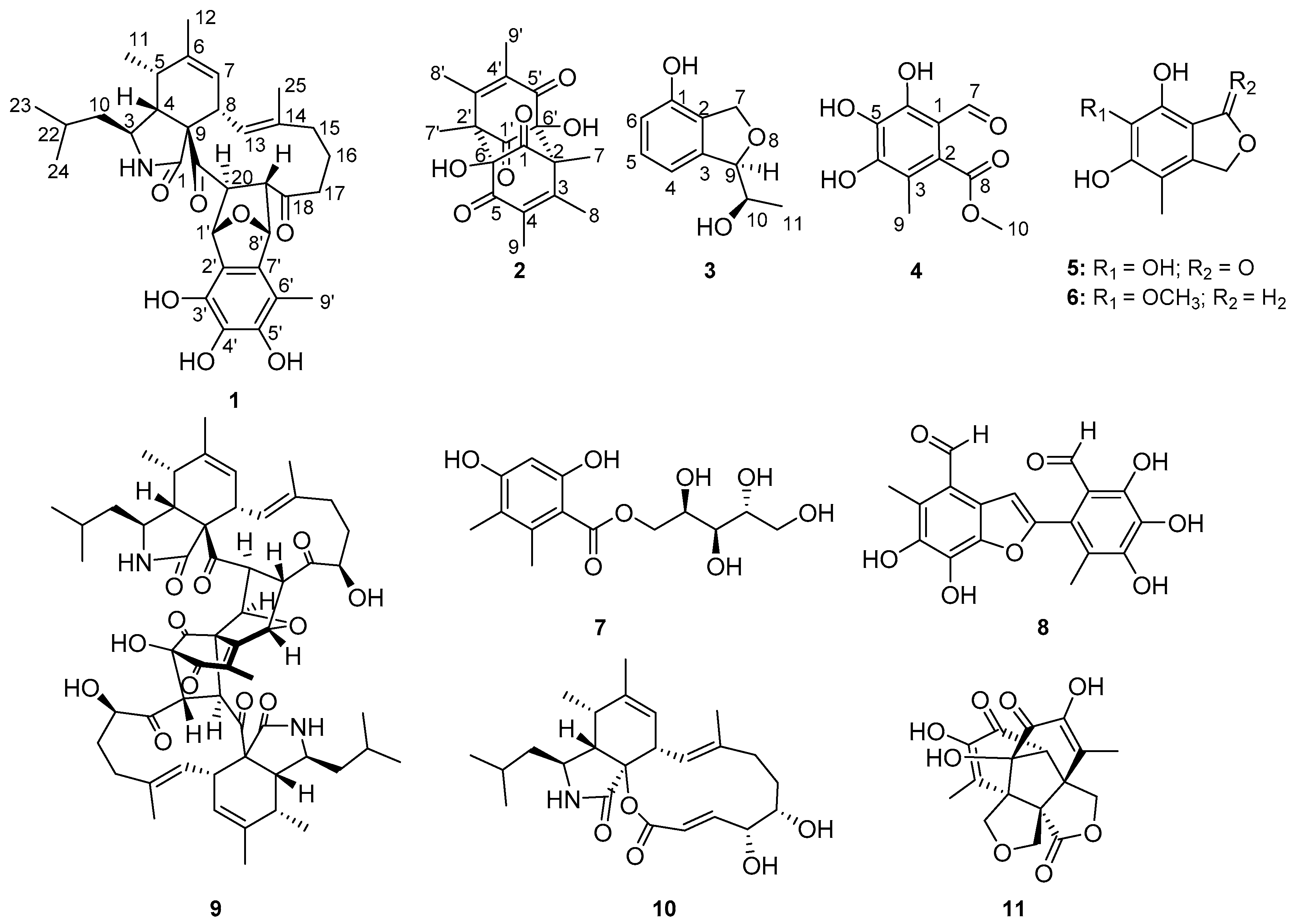
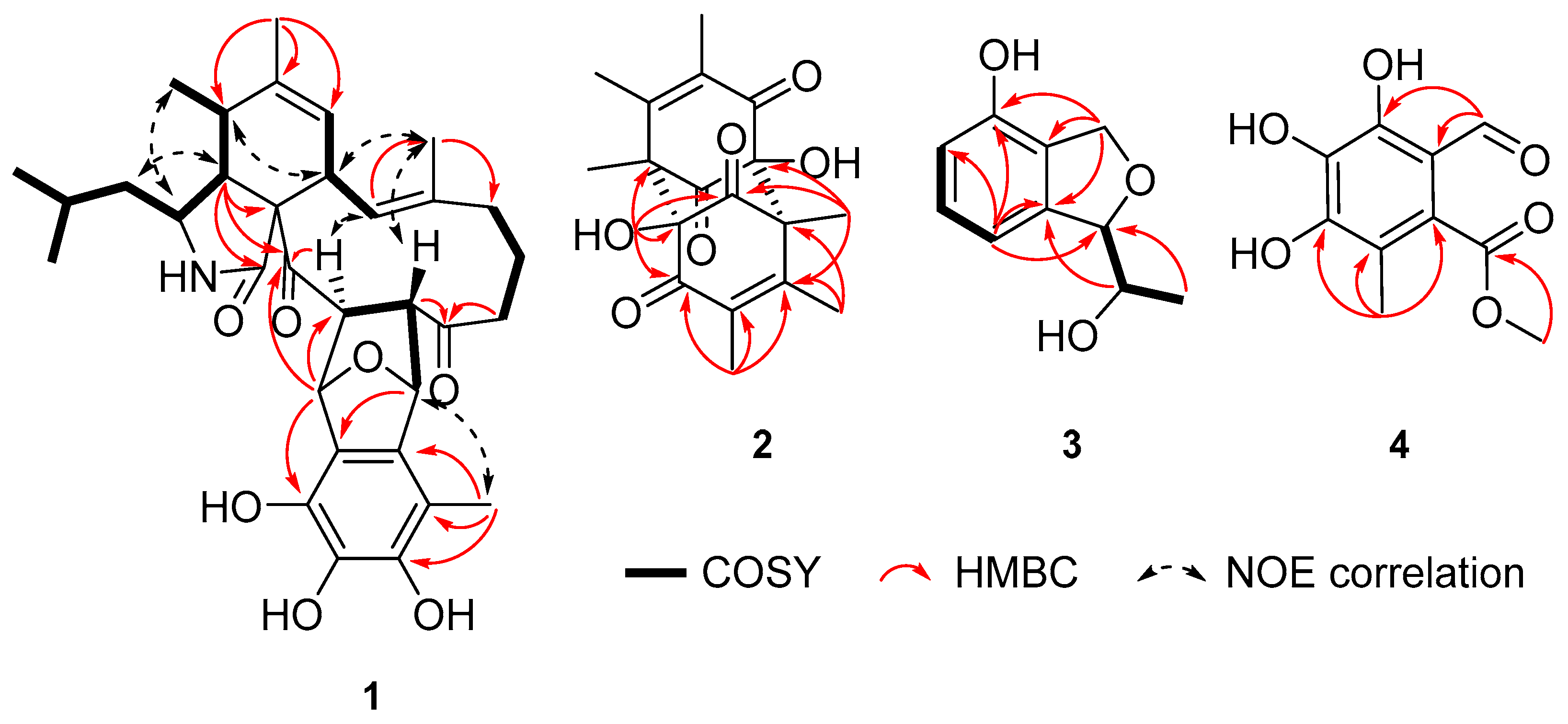
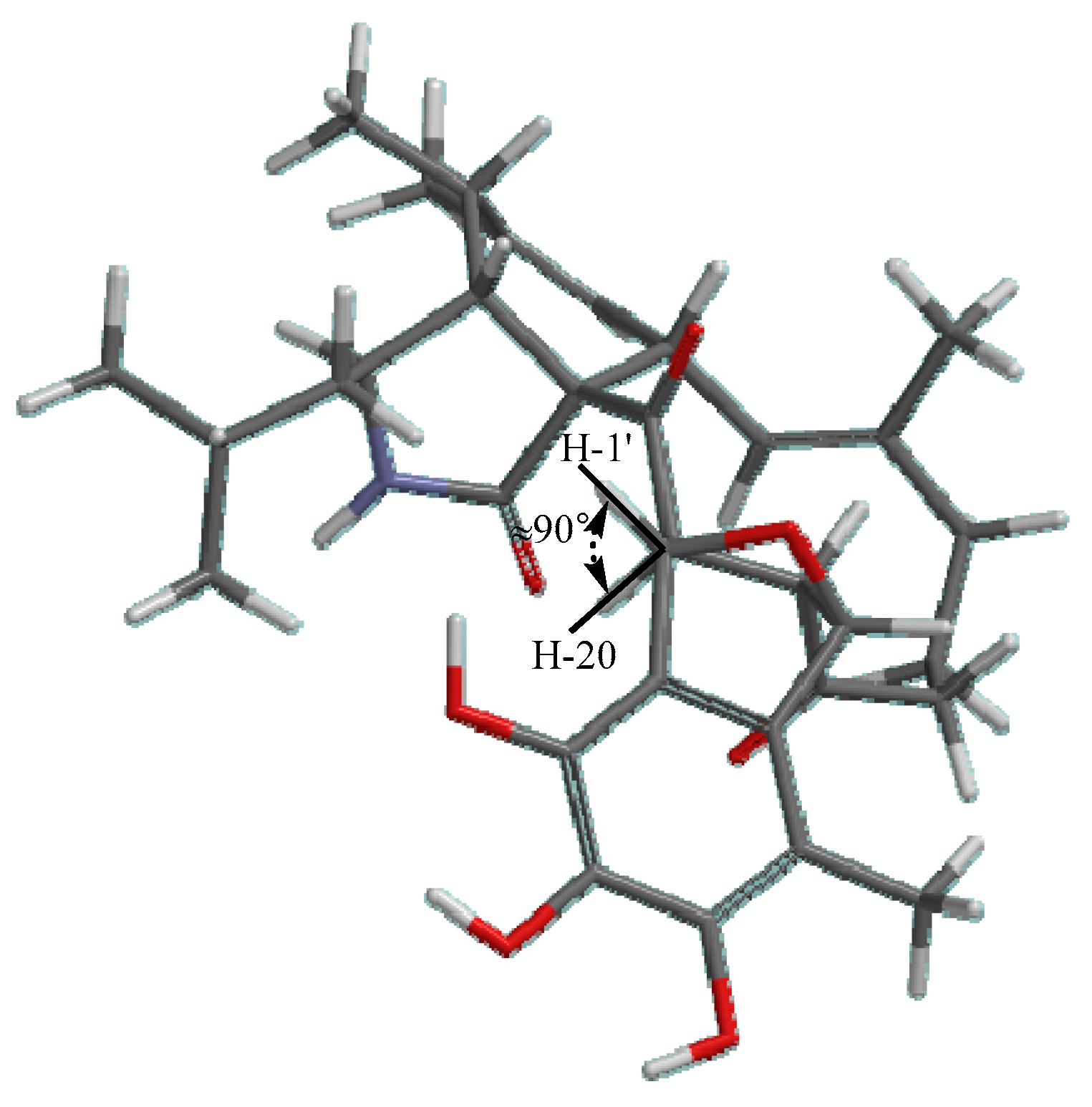
2.1. Structure Elucidation
2.2. Biological Evaluation
3. Experimental Section
3.1. General Experimental Procedures
3.2. Fungal Materials
3.3. Fermentation, Extraction and Isolation
3.4. X-Ray Crystallographic Data
3.5. Biological Assays
3.5.1. Inhibitory Activity of α-Glucosidase
3.5.2. Antioxidant Activity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wresdiyati, T.; Sa’Diah, S.; Winarto, A.; Febriyani, V. Alpha-Glucosidase Inhibition and Hypoglycemic Activities of Sweitenia mahagoni Seed Extract. HAYATI J. Biosci. 2015, 22, 73–78. [Google Scholar] [CrossRef]
- Sekar, V.; Chakraborty, S.; Mani, S.; Sali, V.K.; Vasanthi, H.R. Mangiferin from Mangifera indica fruits reduces post-prandial glucose level by inhibiting α-glucosidase and α-amylase activity. S. Afr. J. Bot. 2019, 120, 129–134. [Google Scholar] [CrossRef]
- Wang, H.; Du, Y.J.; Song, H.C. α-Glucosidase and α-amylase inhibitory activities of guava leaves. Food Chem. 2010, 123, 6–13. [Google Scholar] [CrossRef]
- Tang, H.; Ma, F.; Zhao, D.; Xue, Z. Exploring the effect of salvianolic acid C on α-glucosidase: Inhibition kinetics, interaction mechanism and molecular modelling methods. Process. Biochem. 2019, 78, 178–188. [Google Scholar] [CrossRef]
- Indrianingsih, A.W.; Tachibana, S. α-Glucosidase inhibitor produced by an endophytic fungus, Xylariaceae sp. QGS 01 from Quercus gilva Blume. Food Sci. Hum. Wellness 2017, 6, 88–95. [Google Scholar] [CrossRef]
- Yin, Z.; Zhang, W.; Feng, F.; Zhang, Y.; Kang, W. α-Glucosidase inhibitors isolated from medicinal plants. Food Sci. Hum. Wellness 2014, 3, 136–174. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, X.; Huo, D.; Cao, C.; Li, Y.; Liang, Y.; Li, B.; Li, L. Preliminary characterization, antioxidant and α-glucosidase inhibitory activities of polysaccharides from Mallotus furetianus. Carbohydr. Polym. 2019, 215, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.Z.; Chai, W.M.; Zheng, Y.L.; Huang, Q.; Ou-Yang, C. Inhibitory kinetics and mechanism of rifampicin on α-glucosidase: Insights from spectroscopic and molecular docking analyses. Int. J. Biol. Macromol. 2019, 122, 1244–1252. [Google Scholar] [CrossRef] [PubMed]
- Wei, M.; Chai, W.M.; Yang, Q.; Wang, R.; Peng, Y. Novel Insights into the Inhibitory Effect and Mechanism of Proanthocyanidins from Pyracantha fortuneana Fruit on α-Glucosidase. J. Food Sci. 2017, 82, 2260–2268. [Google Scholar] [CrossRef]
- Sun, Y.; Liu, J.; Li, L.; Gong, C.; Wang, S.; Yang, F.; Hua, H.; Lin, H. New butenolide derivatives from the marine sponge-derived fungus Aspergillus terreus. Bioorgan. Med. Chem. Lett. 2018, 28, 315–318. [Google Scholar] [CrossRef]
- Liu, H.; Chen, Z.; Zhu, G.; Wang, L.; Du, Y.; Wang, Y.; Zhu, W. Phenolic polyketides from the marine alga-derived Streptomyces sp. OUCMDZ-3434. Tetrahedron 2017, 73, 5451–5455. [Google Scholar] [CrossRef]
- Rizvi, T.S.; Hussain, I.; Ali, L.; Mabood, F.; Khan, A.L.; Shujah, S.; Rehman, N.U.; Al-Harrasi, A.; Hussain, J.; Khan, A.; et al. New gorgonane sesquiterpenoid from Teucrium mascatense Boiss, as α-glucosidase inhibitor. S. Afr. J. Bot. 2019, 124, 218–222. [Google Scholar] [CrossRef]
- Wang, C.; Guo, L.; Hao, J.; Wang, L.; Zhu, W. α-Glucosidase Inhibitors from the Marine-Derived Fungus Aspergillus flavipes HN4-13. J. Nat. Prod. 2016, 79, 2977–2981. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Lee, J.; Kim, G.J.; Yang, I.; Wang, W.; Nam, J.W.; Choi, H.; Nam, S.J.; Kang, H. Mycousfurans A and B, Antibacterial Usnic Acid Congeners from the Fungus Mycosphaerella sp., Isolated from a Marine Sediment. Mar. Drugs 2019, 17, 422. [Google Scholar] [CrossRef] [PubMed]
- Otálvaro, F.; Nanclares, J.; Vásquez, L.E.; Quiñones, W.; Echeverri, F.; Arango, R.; Schneider, B. Phenalenone-Type compounds from Musa acuminata var. “Yangambi km 5” (AAA) and Their Activity against Mycosphaerella fijiensis. J. Nat. Prod. 2007, 70, 887–890. [Google Scholar] [CrossRef] [PubMed]
- Assante, A.; Camarda, L.; Merlini, L.; Nasini, G. Secondary metabolites from Mycosphaerella ligulicola. Phytochemistry 1981, 20, 1955–1957. [Google Scholar] [CrossRef]
- Huang, H.; Feng, X.; Xiao, Z.; Liu, L.; Li, H.; Ma, L.; Lu, Y.; Ju, J.; She, Z.; Lin, Y. Azaphilones and p-Terphenyls from the Mangrove Endophytic Fungus Penicillium chermesinum (ZH4-E2) Isolated from the South China Sea. J. Nat. Prod. 2011, 74, 997–1002. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, Q.; Xia, G.; Huang, H.; Li, H.; Ma, L.; Lu, Y.; He, L.; Xia, X.; She, Z. Polyketides with α-Glucosidase Inhibitory Activity from a Mangrove Endophytic Fungus, Penicillium sp. HN29-3B1. J. Nat. Prod. 2015, 78, 1816–1822. [Google Scholar] [CrossRef]
- Liu, Z.; Chen, S.; Qiu, P.; Tan, C.; Long, Y.; Lu, Y.; She, Z. (+)- and (−)-Ascomlactone A: A pair of novel dimeric polyketides from a mangrove endophytic fungus Ascomycota sp. SK2YWS-L. Org. Biomol. Chem. 2017, 15, 10276–10280. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Liu, Y.; Nie, Y.; Liu, Z.; Chen, S.; Zhang, Z.; Lu, Y.; He, L.; Huang, X.; She, Z. Polyketides from the Mangrove-Derived Endophytic Fungus Nectria sp. HN001 and Their α–Glucosidase Inhibitory Activity. Mar. Drugs 2016, 14, 86. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Xia, G.; Li, H.; Ma, L.; Ding, B.; Lu, Y.; He, L.; Xia, X.; She, Z. Vermistatin Derivatives with α-Glucosidase Inhibitory Activity from the Mangrove Endophytic Fungus Penicillium sp. HN29-3B1. Planta Med. 2014, 80, 912–917. [Google Scholar] [CrossRef] [PubMed]
- Kemami Wangun, H.V.; Ishida, K.; Hertweck, C. Epicoccalone, a Coumarin-Type Chymotrypsin Inhibitor, and Isobenzofuran Congeners from an Epicoccum sp. Associated with a Tree Fungus. Eur. J. Org. Chem. 2008, 22, 3781–3784. [Google Scholar] [CrossRef]
- Nam, H.L.; James, B.G.; Donald, T.W. Isolation of Chromanone and Isobenzofuran Derivatives from a Fungicolous Isolate of Epicoccum purpurascens. Bull. Korean Chem. Soc. 2007, 28, 877–879. [Google Scholar]
- Ferdinand, M.T.; Timothee, J.N.K.; Birger, D.; Clovis, D.M.; Hartmut, L. Paeciloside A, a new antimicrobial and cytotoxic polyketide from Paecilomyces sp. Strain CAFT156. Planta Med. 2012, 78, 1020–1023. [Google Scholar]
- Talontsi, F.M.; Dittrich, B.; Schüffler, A.; Sun, H.; Laatsch, H. Epicoccolides: Antimicrobial and Antifungal Polyketides from an Endophytic Fungus Epicoccum sp. Associated with Theobroma cacao. Eur. J. Org. Chem. 2013, 15, 3174–3180. [Google Scholar] [CrossRef]
- Zhu, H.; Chen, C.; Xue, Y.; Tong, Q.; Li, X.; Chen, X.; Wang, J.; Yao, G.; Luo, Z.; Zhang, Y. Asperchalasine A, a Cytochalasan Dimer with an Unprecedented Decacyclic Ring System, from Aspergillus flavipes. Angew. Chem. Int. Ed. 2015, 54, 13374–13378. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.X.; Wijeratne, E.M.K.; Bigelow, D.; Pierson, L.S.; Vanetten, H.D.; Gunatilaka, A.A.L. Aspochalasins I, J, and K: Three New Cytotoxic Cytochalasans of Aspergillus flavipes from the Rhizosphere of Ericameria laricifolia of the Sonoran Desert. J. Nat. Prod. 2004, 67, 328–332. [Google Scholar] [CrossRef]
- Da Silva Araújo, F.D.; de Lima Fávaro, L.C.; Araújo, W.L.; de Oliveira, F.L.; Aparicio, R.; Marsaioli, A.J. Epicolactone—Natural Product Isolated from the Sugarcane Endophytic Fungus Epicoccum nigrum. Eur. J. Org. Chem. 2012, 27, 5225–5230. [Google Scholar] [CrossRef]
- Cai, R.; Wu, Y.; Chen, S.; Cui, H.; Liu, Z.; Li, C.; She, Z. Peniisocoumarins A–J: Isocoumarins from Penicillium commune QQF-3, an Endophytic Fungus of the Mangrove Plant Kandelia candel. J. Nat. Prod. 2018, 81, 1376–1383. [Google Scholar] [CrossRef]
- Tan, C.; Liu, Z.; Chen, S.; Huang, X.; Cui, H.; Long, Y.; Lu, Y.; She, Z. Antioxidative Polyketones from the Mangrove-Derived Fungus Ascomycota sp. SK2YWS-L. Sci. Rep. 2016, 6, 36609. [Google Scholar] [CrossRef] [PubMed]

| Position | 1 | |
|---|---|---|
| δH, mult (J in Hz) | δC, Type | |
| 1 | 176.3, C | |
| 2 | ||
| 3 | 3.21, m | 51.9, CH |
| 4 | 3.05, dd (3.9, 5.0) | 49.8, CH |
| 5 | 2.54, m, | 35.2, CH |
| 6 | 141.8, C | |
| 7 | 5.36, s | 125.1, CH |
| 8 | 2.88, d (11.1) | 43.3, CH |
| 9 | 67.3, C | |
| 10 | 1.35, t | 48.5, CH2 |
| 11 | 1.26, d (7.3) | 13.6, CH3 |
| 12 | 1.78, s | 20.1, CH3 |
| 13 | 5.93, d (11.0) | 125.8, CH |
| 14 | 137.4, C | |
| 15 | 2.09, s | 40.7, CH2 |
| 1.90, d (4.3) | ||
| 16 | 2.01, m | 22.5, CH2 |
| 1.58, m | ||
| 17 | 1.21, m | 35.1, CH2 |
| 18 | 211.6, C | |
| 19 | 3.76, t (5.3) | 56.9, CH |
| 20 | 4.46, d (5.8) | 57.3, CH |
| 21 | 203.6, C | |
| 22 | 1.62, m | 25.3, CH |
| 23 | 0.94, dd (1.2, 6.5) | 23.6, CH3 |
| 24 | 0.96, dd (1.2, 6.5) | 21.3, CH3 |
| 25 | 1.17, s | 14.2, CH3 |
| 1′ | 5.06, s | 80.7, CH |
| 2′ | 123.7, C | |
| 3′ | 132.5, C | |
| 4′ | 133.7, C | |
| 5′ | 141.3, C | |
| 6′ | 111.9, C | |
| 7′ | 132.6, C | |
| 8′ | 5.52, d (4.9) | 81.3, CH |
| 9′ | 2.05, s | 11.9, CH3 |
| Position | 2 | |
|---|---|---|
| δH, mult (J in Hz) | δC, Type | |
| 1 (1′) | 200.6, C | |
| 2 (2′) | 60.4, C | |
| 3 (3′) | 157.6, C | |
| 4 (4′) | 131.4, C | |
| 5 (5′) | 192.6, C | |
| 6 (6′) | 91.7, C | |
| 7 (7′) | 1.21, s | 12.1, CH3 |
| 8 (8′) | 2.02, s | 19.1, CH3 |
| 9 (9′) | 1.73, s | 12.2, CH3 |
| Position | 3 a | 4 b | ||
|---|---|---|---|---|
| δH, mult (J in Hz) | δC, Type | δH, mult (J in Hz) | δC, Type | |
| 1 | 152.8, C | 112.2, C | ||
| 2 | 127.3, C | 117.4, C | ||
| 3 | 142.1, C | 129.9, C | ||
| 4 | 6.82, d (7.4) | 114.3, CH | 152.5, C | |
| 5 | 7.13, t (7.7) | 129.9, CH | 134.1, C | |
| 6 | 6.69, d (7.9) | 115.0, CH | 151.2, C | |
| 7 | 5.12, dd (2.8, 11.8) | 72.4, CH2 | 9.71, s | 194.7, CH |
| 5.02, d (11.9) | ||||
| 8 | 169.9, C | |||
| 9 | 5.06, d (3.3) | 89.4, CH | 2.08, s | 12.4, CH3 |
| 10 | 3.97, qd (3.9, 6.4) | 70.6, CH | 3.90, s | 52.9, CH3 |
| 11 | 1.20, d (6.4) | 18.6, CH3 | ||
| Compounds | α-Glucosidase Inhibitory | Antioxidant | |
|---|---|---|---|
| IC50 (μM) | % Inhibition (100 μM) | EC50 (μM) | |
| 1 | 17.1 | 56 | 77.8 |
| 2 | >50 | <50 | - |
| 3 | >50 | <50 | - |
| 4 | >50 | 57 | 85.8 |
| 5 | >50 | <50 | - |
| 6 | >50 | 65 | 59.1 |
| 7 | >50 | <50 | - |
| 8 | 26.7 | 89 | 16.3 |
| 9 | 15.7 | <50 | - |
| 10 | - | - | - |
| 11 | >50 | <50 | - |
| Acarbose a | 610.2 | ||
| 1-Deoxynojirimycin a | 71.5 | ||
| Ascorbic acid a | 92 | 22.4 | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qiu, P.; Liu, Z.; Chen, Y.; Cai, R.; Chen, G.; She, Z. Secondary Metabolites with α-Glucosidase Inhibitory Activity from the Mangrove Fungus Mycosphaerella sp. SYSU-DZG01. Mar. Drugs 2019, 17, 483. https://doi.org/10.3390/md17080483
Qiu P, Liu Z, Chen Y, Cai R, Chen G, She Z. Secondary Metabolites with α-Glucosidase Inhibitory Activity from the Mangrove Fungus Mycosphaerella sp. SYSU-DZG01. Marine Drugs. 2019; 17(8):483. https://doi.org/10.3390/md17080483
Chicago/Turabian StyleQiu, Pei, Zhaoming Liu, Yan Chen, Runlin Cai, Guangying Chen, and Zhigang She. 2019. "Secondary Metabolites with α-Glucosidase Inhibitory Activity from the Mangrove Fungus Mycosphaerella sp. SYSU-DZG01" Marine Drugs 17, no. 8: 483. https://doi.org/10.3390/md17080483
APA StyleQiu, P., Liu, Z., Chen, Y., Cai, R., Chen, G., & She, Z. (2019). Secondary Metabolites with α-Glucosidase Inhibitory Activity from the Mangrove Fungus Mycosphaerella sp. SYSU-DZG01. Marine Drugs, 17(8), 483. https://doi.org/10.3390/md17080483