Marine Natural Products from Microalgae: An -Omics Overview
Abstract
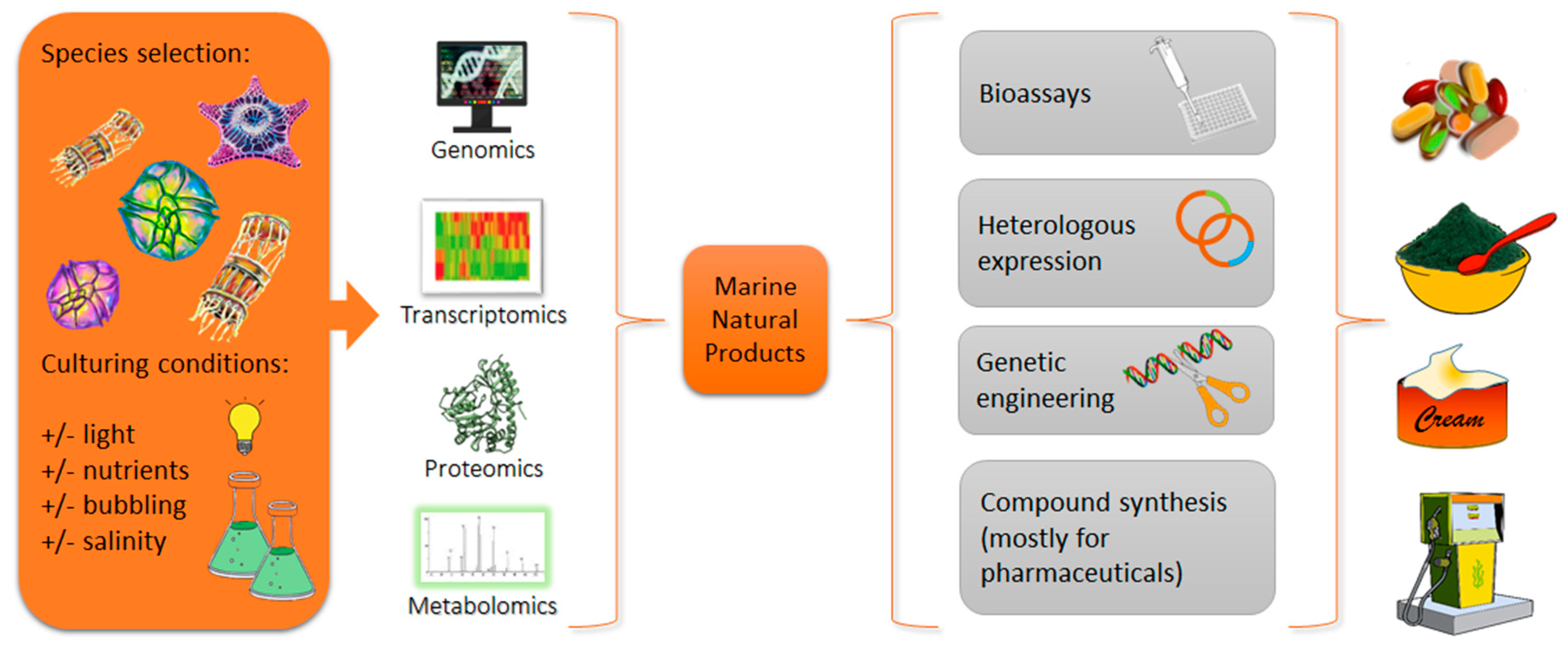
1. Introduction
2. -Omics Datasets Available for Marine Microalgae
2.1. Genomics
2.2. Transcriptomics
2.3. Proteomics
2.4. Metabolomics
3. Genetic Engineering to Increase High-Value Product Production
4. European Projects and Infrastructures
5. Conclusions and Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Abid, F.; Zahid, M.A.; Abedin, Z.U.; Nizami, S.B.; Abid, M.J.; Kazmi, S.Z.H.; Khan, S.U.; Hasan, H.; Ali, M.; Gul, A. Chapter 3—Omics Approaches in Marine Biotechnology: The Treasure of Ocean for Human Betterments. In Omics Technologies and Bio-Engineering; Barh, D., Azevedo, V., Eds.; Academic Press: New York, NY, USA, 2018; pp. 47–61. ISBN 978-0-12-804659-3. [Google Scholar]
- Romano, G.; Costantini, M.; Sansone, C.; Lauritano, C.; Ruocco, N.; Ianora, A. Marine microorganisms as a promising and sustainable source of bioactive molecules. Mar. Environ. Res. 2017, 128, 58–69. [Google Scholar] [CrossRef] [PubMed]
- Jaspars, M.; Pascale, D.D.; Andersen, J.H.; Reyes, F.; Crawford, A.D.; Ianora, A. The marine biodiscovery pipeline and ocean medicines of tomorrow. J. Mar. Biol. Assoc. UK 2016, 96, 151–158. [Google Scholar] [CrossRef]
- Rutledge, P.J.; Challis, G.L. Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat. Rev. Microbiol. 2015, 13, 509. [Google Scholar] [CrossRef] [PubMed]
- Reen, F.J.; Gutiérrez-Barranquero, J.A.; Dobson, A.D.W.; Adams, C.; O’Gara, F. Emerging concepts promising new horizons for marine biodiscovery and synthetic biology. Mar. Drugs 2015, 13, 2924–2954. [Google Scholar] [CrossRef] [PubMed]
- Romano, S.; Jackson, S.A.; Patry, S.; Dobson, A.D.W. Extending the “One Strain Many Compounds” (OSMAC) Principle to Marine Microorganisms. Mar. Drugs 2018, 16, 244. [Google Scholar] [CrossRef]
- Blunt, J.W.; Copp, B.R.; Keyzers, R.A.; Munro, M.H.G.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2015, 32, 116–211. [Google Scholar] [CrossRef]
- Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2019, 36, 122–173. [Google Scholar] [CrossRef]
- Mimouni, V.; Ulmann, L.; Pasquet, V.; Mathieu, M.; Picot, L.; Bougaran, G.; Cadoret, J.-P.; Morant-Manceau, A.; Schoefs, B. The potential of microalgae for the production of bioactive molecules of pharmaceutical interest. Curr. Pharm. Biotechnol. 2012, 13, 2733–2750. [Google Scholar] [CrossRef]
- Brillatz, T.; Lauritano, C.; Jacmin, M.; Khamma, S.; Marcourt, L.; Righi, D.; Romano, G.; Esposito, F.; Ianora, A.; Queiroz, E.F.; et al. Zebrafish-based identification of the antiseizure nucleoside inosine from the marine diatom Skeletonema marinoi. PLoS ONE 2018, 13, e0196195. [Google Scholar] [CrossRef]
- Martínez Andrade, K.A.; Lauritano, C.; Romano, G.; Ianora, A. Marine Microalgae with Anti-Cancer Properties. Mar. Drugs 2018, 16, 165. [Google Scholar] [CrossRef]
- Lauritano, C.; Martín, J.; de la Cruz, M.; Reyes, F.; Romano, G.; Ianora, A. First identification of marine diatoms with anti-tuberculosis activity. Sci. Rep. 2018, 8, 2284. [Google Scholar] [CrossRef] [PubMed]
- Yao, H.; Liu, J.; Xu, S.; Zhu, Z.; Xu, J. The structural modification of natural products for novel drug discovery. Expert Opin. Drug Discov. 2017, 12, 121–140. [Google Scholar] [CrossRef]
- Lauritano, C.; Andersen, J.H.; Hansen, E.; Albrigtsen, M.; Escalera, L.; Esposito, F.; Helland, K.; Hanssen, K.Ø.; Romano, G.; Ianora, A. Bioactivity Screening of Microalgae for Antioxidant, Anti-Inflammatory, Anticancer, Anti-Diabetes, and Antibacterial Activities. Front. Mar. Sci. 2016, 3. [Google Scholar] [CrossRef]
- Ingebrigtsen, R.A.; Hansen, E.; Andersen, J.H.; Eilertsen, H.C. Light and temperature effects on bioactivity in diatoms. J. Appl. Phycol. 2016, 28, 939–950. [Google Scholar] [CrossRef] [PubMed]
- Bode, H.B.; Bethe, B.; Höfs, R.; Zeeck, A. Big Effects from Small Changes: Possible Ways to Explore Nature’s Chemical Diversity. ChemBioChem 2002, 3, 619–627. [Google Scholar] [CrossRef]
- Nordberg, H.; Cantor, M.; Dusheyko, S.; Hua, S.; Poliakov, A.; Shabalov, I.; Smirnova, T.; Grigoriev, I.V.; Dubchak, I. The genome portal of the Department of Energy Joint Genome Institute: 2014 updates. Nucleic Acids Res. 2014, 42, D26–D31. [Google Scholar] [CrossRef]
- Vandepoele, K.; Van Bel, M.; Richard, G.; Van Landeghem, S.; Verhelst, B.; Moreau, H.; Van de Peer, Y.; Grimsley, N.; Piganeau, G. pico-PLAZA, a genome database of microbial photosynthetic eukaryotes. Environ. Microbiol. 2013, 15, 2147–2153. [Google Scholar] [CrossRef]
- Karp, P.D.; Billington, R.; Caspi, R.; Fulcher, C.A.; Latendresse, M.; Kothari, A.; Keseler, I.M.; Krummenacker, M.; Midford, P.E.; Ong, Q.; et al. The BioCyc collection of microbial genomes and metabolic pathways. Brief. Bioinform. 2017. [Google Scholar] [CrossRef]
- Kersey, P.J.; Allen, J.E.; Allot, A.; Barba, M.; Boddu, S.; Bolt, B.J.; Carvalho-Silva, D.; Christensen, M.; Davis, P.; Grabmueller, C.; et al. Ensembl Genomes 2018: An integrated omics infrastructure for non-vertebrate species. Nucleic Acids Res. 2018, 46, D802–D808. [Google Scholar] [CrossRef]
- Tanaka, T.; Maeda, Y.; Veluchamy, A.; Tanaka, M.; Abida, H.; Maréchal, E.; Bowler, C.; Muto, M.; Sunaga, Y.; Tanaka, M.; et al. Oil accumulation by the oleaginous diatom Fistulifera solaris as revealed by the genome and transcriptome. Plant Cell 2015, 27, 162–176. [Google Scholar] [CrossRef]
- Traller, J.C.; Cokus, S.J.; Lopez, D.A.; Gaidarenko, O.; Smith, S.R.; McCrow, J.P.; Gallaher, S.D.; Podell, S.; Thompson, M.; Cook, O.; et al. Genome and methylome of the oleaginous diatom Cyclotella cryptica reveal genetic flexibility toward a high lipid phenotype. Biotechnol. Biofuels 2016, 9, 258. [Google Scholar] [CrossRef]
- Basu, S.; Patil, S.; Mapleson, D.; Russo, M.T.; Vitale, L.; Fevola, C.; Maumus, F.; Casotti, R.; Mock, T.; Caccamo, M.; et al. Finding a partner in the ocean: Molecular and evolutionary bases of the response to sexual cues in a planktonic diatom. New Phytol. 2017, 215, 140–156. [Google Scholar] [CrossRef]
- Ogura, A.; Akizuki, Y.; Imoda, H.; Mineta, K.; Gojobori, T.; Nagai, S. Comparative genome and transcriptome analysis of diatom, Skeletonema costatum, reveals evolution of genes for harmful algal bloom. BMC Genom. 2018, 19, 765. [Google Scholar] [CrossRef]
- Ruan, J.; Li, H.; Chen, Z.; Coghlan, A.; Coin, L.J.; Guo, Y.; Hériché, J.K.; Hu, Y.; Kristiansen, K.; Li, R.; et al. TreeFam: 2008 Update. Nucleic Acids Res. 2008, 36, D735–D740. [Google Scholar] [CrossRef]
- Weber, T.; Blin, K.; Duddela, S.; Krug, D.; Kim, H.U.; Bruccoleri, R.; Lee, S.Y.; Fischbach, M.A.; Müller, R.; Wohlleben, W.; et al. antiSMASH 3.0—A comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015, 43, W237–W243. [Google Scholar] [CrossRef]
- Kita, M.; Ohno, O.; Han, C.; Uemura, D. Bioactive secondary metabolites from symbiotic marine dinoflagellates: Symbiodinolide and durinskiols. Chem. Rec. 2010, 10, 57–69. [Google Scholar] [CrossRef]
- Beedessee, G.; Hisata, K.; Roy, M.C.; Dolah, F.V.; Satoh, N.; Shoguchi, E. Comparative genomics-first approach to understand diversification of secondary metabolite biosynthetic pathways in symbiotic dinoflagellates. bioRxiv 2018, 376251. [Google Scholar] [CrossRef]
- Ziemert, N.; Alanjary, M.; Weber, T. The evolution of genome mining in microbes—A review. Nat. Prod. Rep. 2016, 33, 988–1005. [Google Scholar] [CrossRef]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A better web interface. Nucleic Acids Res. 2008, 36, W5–W9. [Google Scholar] [CrossRef]
- Wheeler, T.J.; Eddy, S.R. nhmmer: DNA homology search with profile HMMs. Bioinformatics 2013, 29, 2487–2489. [Google Scholar] [CrossRef]
- Liu, X. Deep Recurrent Neural Network for Protein Function Prediction from Sequence. Available online: https://arxiv.org/abs/1701.08318v1 (accessed on 29 April 2019).
- Brunson, J.K.; McKinnie, S.M.K.; Chekan, J.R.; McCrow, J.P.; Miles, Z.D.; Bertrand, E.M.; Bielinski, V.A.; Luhavaya, H.; Oborník, M.; Smith, G.J.; et al. Biosynthesis of the neurotoxin domoic acid in a bloom-forming diatom. Science 2018, 361, 1356–1358. [Google Scholar] [CrossRef]
- Mukherjee, S.; Stamatis, D.; Bertsch, J.; Ovchinnikova, G.; Katta, H.Y.; Mojica, A.; Chen, I.-M.A.; Kyrpides, N.C.; Reddy, T.B.K. Genomes OnLine database (GOLD) v.7: Updates and new features. Nucleic Acids Res. 2019, 47, D649–D659. [Google Scholar] [CrossRef]
- Chen, I.-M.A.; Chu, K.; Palaniappan, K.; Pillay, M.; Ratner, A.; Huang, J.; Huntemann, M.; Varghese, N.; White, J.R.; Seshadri, R.; et al. IMG/M v.5.0: An integrated data management and comparative analysis system for microbial genomes and microbiomes. Nucleic Acids Res. 2019, 47, D666–D677. [Google Scholar] [CrossRef]
- Villar, E.; Vannier, T.; Vernette, C.; Lescot, M.; Cuenca, M.; Alexandre, A.; Bachelerie, P.; Rosnet, T.; Pelletier, E.; Sunagawa, S.; et al. The Ocean Gene Atlas: Exploring the biogeography of plankton genes online. Nucleic Acids Res. 2018, 46, W289–W295. [Google Scholar] [CrossRef]
- Gobler, C.J.; Berry, D.L.; Dyhrman, S.T.; Wilhelm, S.W.; Salamov, A.; Lobanov, A.V.; Zhang, Y.; Collier, J.L.; Wurch, L.L.; Kustka, A.B.; et al. Niche of harmful alga Aureococcus anophagefferens revealed through ecogenomics. Proc. Natl. Acad. Sci. USA 2011, 108, 4352–4357. [Google Scholar] [CrossRef]
- Moreau, H.; Verhelst, B.; Couloux, A.; Derelle, E.; Rombauts, S.; Grimsley, N.; Van Bel, M.; Poulain, J.; Katinka, M.; Hohmann-Marriott, M.F.; et al. Gene functionalities and genome structure in Bathycoccus prasinos reflect cellular specializations at the base of the green lineage. Genome Biol. 2012, 13, R74. [Google Scholar] [CrossRef]
- Guarnieri, M.T.; Levering, J.; Henard, C.A.; Boore, J.L.; Betenbaugh, M.J.; Zengler, K.; Knoshaug, E.P. Genome Sequence of the Oleaginous Green Alga, Chlorella vulgaris UTEX 395. Front. Bioeng. Biotechnol. 2018, 6, 37. [Google Scholar] [CrossRef]
- Blanc, G.; Agarkova, I.; Grimwood, J.; Kuo, A.; Brueggeman, A.; Dunigan, D.D.; Gurnon, J.; Ladunga, I.; Lindquist, E.; Lucas, S.; et al. The genome of the polar eukaryotic microalga Coccomyxa subellipsoidea reveals traits of cold adaptation. Genome Biol. 2012, 13, R39. [Google Scholar] [CrossRef]
- Polle, J.E.W.; Barry, K.; Cushman, J.; Schmutz, J.; Tran, D.; Hathwaik, L.T.; Yim, W.C.; Jenkins, J.; McKie-Krisberg, Z.; Prochnik, S.; et al. Draft Nuclear Genome Sequence of the Halophilic and Beta-Carotene-Accumulating Green Alga Dunaliella salina Strain CCAP19/18. Genome Announc. 2017, 5, e01105-17. [Google Scholar] [CrossRef]
- Read, B.A.; Kegel, J.; Klute, M.J.; Kuo, A.; Lefebvre, S.C.; Maumus, F.; Mayer, C.; Miller, J.; Monier, A.; Salamov, A.; et al. Pan genome of the phytoplankton Emiliania underpins its global distribution. Nature 2013, 499, 209–213. [Google Scholar] [CrossRef]
- Mock, T.; Otillar, R.P.; Strauss, J.; McMullan, M.; Paajanen, P.; Schmutz, J.; Salamov, A.; Sanges, R.; Toseland, A.; Ward, B.J.; et al. Evolutionary genomics of the cold-adapted diatom Fragilariopsis cylindrus. Nature 2017, 541, 536–540. [Google Scholar] [CrossRef]
- Curtis, B.A.; Tanifuji, G.; Burki, F.; Gruber, A.; Irimia, M.; Maruyama, S.; Arias, M.C.; Ball, S.G.; Gile, G.H.; Hirakawa, Y.; et al. Algal genomes reveal evolutionary mosaicism and the fate of nucleomorphs. Nature 2012, 492, 59–65. [Google Scholar] [CrossRef]
- Worden, A.Z.; Lee, J.-H.; Mock, T.; Rouzé, P.; Simmons, M.P.; Aerts, A.L.; Allen, A.E.; Cuvelier, M.L.; Derelle, E.; Everett, M.V.; et al. Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science 2009, 324, 268–272. [Google Scholar] [CrossRef]
- Schwartz, A.S.; Brown, R.; Ajjawi, I.; McCarren, J.; Atilla, S.; Bauman, N.; Richardson, T.H. Complete Genome Sequence of the Model Oleaginous Alga Nannochloropsis gaditana CCMP1894. Genome Announc. 2018, 6, e01448-17. [Google Scholar] [CrossRef]
- Corteggiani Carpinelli, E.; Telatin, A.; Vitulo, N.; Forcato, C.; D’Angelo, M.; Schiavon, R.; Vezzi, A.; Giacometti, G.M.; Morosinotto, T.; Valle, G. Chromosome scale genome assembly and transcriptome profiling of Nannochloropsis gaditana in nitrogen depletion. Mol. Plant 2014, 7, 323–335. [Google Scholar] [CrossRef]
- Palenik, B.; Grimwood, J.; Aerts, A.; Rouzé, P.; Salamov, A.; Putnam, N.; Dupont, C.; Jorgensen, R.; Derelle, E.; Rombauts, S.; et al. The tiny eukaryote Ostreococcus provides genomic insights into the paradox of plankton speciation. Proc. Natl. Acad. Sci. USA 2007, 104, 7705–7710. [Google Scholar] [CrossRef]
- Blanc-Mathieu, R.; Verhelst, B.; Derelle, E.; Rombauts, S.; Bouget, F.-Y.; Carré, I.; Château, A.; Eyre-Walker, A.; Grimsley, N.; Moreau, H.; et al. An improved genome of the model marine alga Ostreococcus tauri unfolds by assessing Illumina de novo assemblies. BMC Genom. 2014, 15, 1103. [Google Scholar] [CrossRef]
- Blanc-Mathieu, R.; Krasovec, M.; Hebrard, M.; Yau, S.; Desgranges, E.; Martin, J.; Schackwitz, W.; Kuo, A.; Salin, G.; Donnadieu, C.; et al. Population genomics of picophytoplankton unveils novel chromosome hypervariability. Sci. Adv. 2017, 3, e1700239. [Google Scholar] [CrossRef]
- Derelle, E.; Ferraz, C.; Rombauts, S.; Rouzé, P.; Worden, A.Z.; Robbens, S.; Partensky, F.; Degroeve, S.; Echeynié, S.; Cooke, R.; et al. Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proc. Natl. Acad. Sci. USA 2006, 103, 11647–11652. [Google Scholar] [CrossRef]
- Bowler, C.; Allen, A.E.; Badger, J.H.; Grimwood, J.; Jabbari, K.; Kuo, A.; Maheswari, U.; Martens, C.; Maumus, F.; Otillar, R.P.; et al. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 2008, 456, 239–244. [Google Scholar] [CrossRef]
- Krasovec, M.; Vancaester, E.; Rombauts, S.; Bucchini, F.; Yau, S.; Hemon, C.; Lebredonchel, H.; Grimsley, N.; Moreau, H.; Sanchez-Brosseau, S.; et al. Genome Analyses of the Microalga Picochlorum Provide Insights into the Evolution of Thermotolerance in the Green Lineage. Genome Biol. Evol. 2018, 10, 2347–2365. [Google Scholar] [CrossRef]
- Lin, S.; Cheng, S.; Song, B.; Zhong, X.; Lin, X.; Li, W.; Li, L.; Zhang, Y.; Zhang, H.; Ji, Z.; et al. The Symbiodinium kawagutii genome illuminates dinoflagellate gene expression and coral symbiosis. Science 2015, 350, 691–694. [Google Scholar] [CrossRef]
- Aranda, M.; Li, Y.; Liew, Y.J.; Baumgarten, S.; Simakov, O.; Wilson, M.C.; Piel, J.; Ashoor, H.; Bougouffa, S.; Bajic, V.B.; et al. Genomes of coral dinoflagellate symbionts highlight evolutionary adaptations conducive to a symbiotic lifestyle. Sci. Rep. 2016, 6, 39734. [Google Scholar] [CrossRef] [PubMed]
- Lommer, M.; Specht, M.; Roy, A.-S.; Kraemer, L.; Andreson, R.; Gutowska, M.A.; Wolf, J.; Bergner, S.V.; Schilhabel, M.B.; Klostermeier, U.C.; et al. Genome and low-iron response of an oceanic diatom adapted to chronic iron limitation. Genome Biol. 2012, 13, R66. [Google Scholar] [CrossRef]
- Armbrust, E.V.; Berges, J.A.; Bowler, C.; Green, B.R.; Martinez, D.; Putnam, N.H.; Zhou, S.; Allen, A.E.; Apt, K.E.; Bechner, M.; et al. The genome of the diatom Thalassiosira pseudonana: Ecology, evolution, and metabolism. Science 2004, 306, 79–86. [Google Scholar] [CrossRef]
- Zheng, H.-Q.; Chiang-Hsieh, Y.-F.; Chien, C.-H.; Hsu, B.-K.J.; Liu, T.-L.; Chen, C.-N.N.; Chang, W.-C. AlgaePath: Comprehensive analysis of metabolic pathways using transcript abundance data from next-generation sequencing in green algae. BMC Genom. 2014, 15, 196. [Google Scholar] [CrossRef]
- Hockin, N.L.; Mock, T.; Mulholland, F.; Kopriva, S.; Malin, G. The Response of Diatom Central Carbon Metabolism to Nitrogen Starvation Is Different from That of Green Algae and Higher Plants. Plant Physiol. 2012, 158, 299–312. [Google Scholar] [CrossRef]
- Simionato, D.; Block, M.A.; La Rocca, N.; Jouhet, J.; Maréchal, E.; Finazzi, G.; Morosinotto, T. The response of Nannochloropsis gaditana to nitrogen starvation includes de novo biosynthesis of triacylglycerols, a decrease of chloroplast galactolipids, and reorganization of the photosynthetic apparatus. Eukaryot. Cell 2013, 12, 665–676. [Google Scholar] [CrossRef]
- Lauritano, C.; De Luca, D.; Ferrarini, A.; Avanzato, C.; Minio, A.; Esposito, F.; Ianora, A. De novo transcriptome of the cosmopolitan dinoflagellate Amphidinium carterae to identify enzymes with biotechnological potential. Sci. Rep. 2017, 7, 11701. [Google Scholar] [CrossRef]
- Lim, D.K.Y.; Schuhmann, H.; Thomas-Hall, S.R.; Chan, K.C.K.; Wass, T.J.; Aguilera, F.; Adarme-Vega, T.C.; Dal’Molin, C.G.O.; Thorpe, G.J.; Batley, J.; et al. RNA-Seq and metabolic flux analysis of Tetraselmis sp. M8 during nitrogen starvation reveals a two-stage lipid accumulation mechanism. Bioresour. Technol. 2017, 244, 1281–1293. [Google Scholar] [CrossRef]
- Lauritano, C.; De Luca, D.; Amoroso, M.; Benfatto, S.; Maestri, S.; Racioppi, C.; Esposito, F.; Ianora, A. New molecular insights on the response of the green alga Tetraselmis suecica to nitrogen starvation. Sci. Rep. 2019, 9, 3336. [Google Scholar] [CrossRef]
- Keeling, P.J.; Burki, F.; Wilcox, H.M.; Allam, B.; Allen, E.E.; Amaral-Zettler, L.A.; Armbrust, E.V.; Archibald, J.M.; Bharti, A.K.; Bell, C.J.; et al. The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): Illuminating the Functional Diversity of Eukaryotic Life in the Oceans through Transcriptome Sequencing. PLoS Biol. 2014, 12, e1001889. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, S.-F.; Lin, L.; Wang, D.-Z. Whole Transcriptomic Analysis Provides Insights into Molecular Mechanisms for Toxin Biosynthesis in a Toxic Dinoflagellate Alexandrium catenella (ACHK-T). Toxins 2017, 9, 213. [Google Scholar] [CrossRef]
- Meyer, J.M.; Rödelsperger, C.; Eichholz, K.; Tillmann, U.; Cembella, A.; McGaughran, A.; John, U. Transcriptomic characterisation and genomic glimps into the toxigenic dinoflagellate Azadinium spinosum, with emphasis on polykeitde synthase genes. BMC Genom. 2015, 16, 27. [Google Scholar] [CrossRef]
- Morey, J.S.; Monroe, E.A.; Kinney, A.L.; Beal, M.; Johnson, J.G.; Hitchcock, G.L.; Van Dolah, F.M. Transcriptomic response of the red tide dinoflagellate, Karenia brevis, to nitrogen and phosphorus depletion and addition. BMC Genom. 2011, 12, 346. [Google Scholar] [CrossRef]
- Di Dato, V.; Musacchia, F.; Petrosino, G.; Patil, S.; Montresor, M.; Sanges, R.; Ferrante, M.I. Transcriptome sequencing of three Pseudo-nitzschia species reveals comparable gene sets and the presence of Nitric Oxide Synthase genes in diatoms. Sci. Rep. 2015, 5, 12329. [Google Scholar] [CrossRef]
- Adarme-Vega, T.C.; Thomas-Hall, S.R.; Lim, D.K.Y.; Schenk, P.M. Effects of long chain fatty acid synthesis and associated gene expression in microalga Tetraselmis sp. Mar. Drugs 2014, 12, 3381–3398. [Google Scholar] [CrossRef]
- MacKinnon, S.L.; Cembella, A.D.; Burton, I.W.; Lewis, N.; LeBlanc, P.; Walter, J.A. Biosynthesis of 13-desmethyl spirolide C by the dinoflagellate Alexandrium ostenfeldii. J. Org. Chem. 2006, 71, 8724–8731. [Google Scholar] [CrossRef]
- Kobayashi, J. Amphidinolides and Its Related Macrolides from Marine Dinoflagellates. J. Antibiot. 2008, 61, 271–284. [Google Scholar] [CrossRef]
- Kellmann, R.; Stüken, A.; Orr, R.J.S.; Svendsen, H.M.; Jakobsen, K.S. Biosynthesis and molecular genetics of polyketides in marine dinoflagellates. Mar. Drugs 2010, 8, 1011–1048. [Google Scholar] [CrossRef]
- Van Dolah, F.M.; Zippay, M.L.; Pezzolesi, L.; Rein, K.S.; Johnson, J.G.; Morey, J.S.; Wang, Z.; Pistocchi, R. Subcellular localization of dinoflagellate polyketide synthases and fatty acid synthase activity. J. Phycol. 2013, 49, 1118–1127. [Google Scholar] [CrossRef]
- Kohli, G.S.; John, U.; Van Dolah, F.M.; Murray, S.A. Evolutionary distinctiveness of fatty acid and polyketide synthesis in eukaryotes. ISME J. 2016, 10, 1877–1890. [Google Scholar] [CrossRef]
- Lin, I.-P.; Jiang, P.-L.; Chen, C.-S.; Tzen, J.T.C. A unique caleosin serving as the major integral protein in oil bodies isolated from Chlorella sp. cells cultured with limited nitrogen. Plant Physiol. Biochem. PPB 2012, 61, 80–87. [Google Scholar] [CrossRef]
- Nojima, D.; Yoshino, T.; Maeda, Y.; Tanaka, M.; Nemoto, M.; Tanaka, T. Proteomics analysis of oil body-associated proteins in the oleaginous diatom. J. Proteome Res. 2013, 12, 5293–5301. [Google Scholar] [CrossRef]
- Rai, V.; Muthuraj, M.; Gandhi, M.N.; Das, D.; Srivastava, S. Real-time iTRAQ-based proteome profiling revealed the central metabolism involved in nitrogen starvation induced lipid accumulation in microalgae. Sci. Rep. 2017, 7, 45732. [Google Scholar] [CrossRef]
- Maeda, Y.; Yoshino, T.; Matsunaga, T.; Matsumoto, M.; Tanaka, T. Marine microalgae for production of biofuels and chemicals. Curr. Opin. Biotechnol. 2017, 50, 111–120. [Google Scholar] [CrossRef]
- Siegler, H.; Valerius, O.; Ischebeck, T.; Popko, J.; Tourasse, N.J.; Vallon, O.; Khozin-Goldberg, I.; Braus, G.H.; Feussner, I. Analysis of the lipid body proteome of the oleaginous alga Lobosphaera incisa. BMC Plant Biol. 2017, 17, 98. [Google Scholar] [CrossRef]
- Davidi, L.; Levin, Y.; Ben-Dor, S.; Pick, U. Proteome analysis of cytoplasmatic and plastidic β-carotene lipid droplets in Dunaliella bardawil. Plant Physiol. 2015, 167, 60–79. [Google Scholar] [CrossRef]
- Longworth, J.; Wu, D.; Huete-Ortega, M.; Wright, P.C.; Vaidyanathan, S. Proteome response of Phaeodactylum tricornutum, during lipid accumulation induced by nitrogen depletion. Algal Res. 2016, 18, 213–224. [Google Scholar] [CrossRef]
- Garibay-Hernández, A.; Barkla, B.J.; Vera-Estrella, R.; Martinez, A.; Pantoja, O. Membrane Proteomic Insights into the Physiology and Taxonomy of an Oleaginous Green Microalga. Plant Physiol. 2017, 173, 390–416. [Google Scholar] [CrossRef]
- Hurkman, W.J.; Tanaka, C.K. High-Resolution Two-Dimensional Gel Electrophoresis: A Cornerstone of Plant Proteomics. In Plant Proteomics; Šamaj, J., Thelen, J.J., Eds.; Springer: Berlin/Heidelberg, Germany, 2007; pp. 14–28. ISBN 978-3-540-72617-3. [Google Scholar]
- Fernández-Acero, F.J.; Amil-Ruiz, F.; Durán-Peña, M.J.; Carrasco, R.; Fajardo, C.; Guarnizo, P.; Fuentes-Almagro, C.; Vallejo, R.A. Valorisation of the microalgae Nannochloropsis gaditana biomass by proteomic approach in the context of circular economy. J. Proteom. 2019, 193, 239–242. [Google Scholar] [CrossRef] [PubMed]
- Burley, S.K.; Berman, H.M.; Bhikadiya, C.; Bi, C.; Chen, L.; Di Costanzo, L.; Christie, C.; Dalenberg, K.; Duarte, J.M.; Dutta, S.; et al. RCSB Protein Data Bank: Biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy. Nucleic Acids Res. 2019, 47, D464–D474. [Google Scholar] [CrossRef]
- The Uniprot Consortium. UniProt: A hub for protein information. Nucleic Acids Res. 2015, 43, D204–D212. [Google Scholar] [CrossRef]
- Kurotani, A.; Yamada, Y.; Sakurai, T. Alga-PrAS (Algal Protein Annotation Suite): A Database of Comprehensive Annotation in Algal Proteomes. Plant Cell Physiol. 2017, 58, e6. [Google Scholar] [CrossRef]
- Bule, M.H.; Ahmed, I.; Maqbool, F.; Bilal, M.; Iqbal, H.M.N. Microalgae as a source of high-value bioactive compounds. Front. Biosci. Sch. Ed. 2018, 10, 197–216. [Google Scholar]
- Giordano, D.; Costantini, M.; Coppola, D.; Lauritano, C.; Núñez Pons, L.; Ruocco, N.; di Prisco, G.; Ianora, A.; Verde, C. Biotechnological Applications of Bioactive Peptides from Marine Sources. Adv. Microb. Physiol. 2018, 73, 171–220. [Google Scholar] [PubMed]
- Guisande, C.; Frangópulos, M.; Maneiro, I.; Vergara, A.R.; Riveiro, I. Ecological Advantages of Toxin Production by the Dinoflagellate Alexandrium Minutum under Phosphorus Limitation. Mar. Ecol. Prog. Ser. 2002, 225, 169–176. [Google Scholar] [CrossRef]
- Ribalet, F.; Berges, J.A.; Ianora, A.; Casotti, R. Growth inhibition of cultured marine phytoplankton by toxic algal-derived polyunsaturated aldehydes. Aquat. Toxicol. Amst. Neth. 2007, 85, 219–227. [Google Scholar] [CrossRef]
- de Morais, M.G.; da Silva Vaz, B.; de Morais, E.G.; Costa, J.A.V. Biologically Active Metabolites Synthesized by Microalgae. BioMed Res. Int. 2015, 2015, 835761. [Google Scholar] [CrossRef]
- Gowda, G.A.N.; Djukovic, D. Overview of mass spectrometry-based metabolomics: Opportunities and challenges. Methods Mol. Biol. 2014, 1198, 3–12. [Google Scholar]
- Willette, S.; Gill, S.S.; Dungan, B.; Schaub, T.M.; Jarvis, J.M.; St. Hilaire, R.; Omar Holguin, F. Alterations in lipidome and metabolome profiles of Nannochloropsis salina in response to reduced culture temperature during sinusoidal temperature and light. Algal Res. 2018, 32, 79–92. [Google Scholar] [CrossRef]
- Sun, M.; Yang, Z.; Wawrik, B. Metabolomic Fingerprints of Individual Algal Cells Using the Single-Probe Mass Spectrometry Technique. Front. Plant Sci. 2018, 9, 571. [Google Scholar] [CrossRef] [PubMed]
- Johnson, S.R.; Lange, B.M. Open-Access Metabolomics Databases for Natural Product Research: Present Capabilities and Future Potential. Front. Bioeng. Biotechnol. 2015, 3, 22. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2019 update: Improved access to chemical data. Nucleic Acids Res. 2019, 47, D1102–D1109. [Google Scholar] [CrossRef] [PubMed]
- Caspi, R.; Billington, R.; Fulcher, C.A.; Keseler, I.M.; Kothari, A.; Krummenacker, M.; Latendresse, M.; Midford, P.E.; Ong, Q.; Ong, W.K.; et al. The MetaCyc database of metabolic pathways and enzymes. Nucleic Acids Res. 2018, 46, D633–D639. [Google Scholar] [CrossRef] [PubMed]
- Bashir, K.M.I.; Kim, M.-S.; Stahl, U.; Cho, M.-G. Microalgae engineering toolbox: Selectable and screenable markers. Biotechnol. Bioprocess Eng. 2016, 21, 224–235. [Google Scholar] [CrossRef]
- Buck, J.M.; Río Bártulos, C.; Gruber, A.; Kroth, P.G. Blasticidin-S deaminase, a new selection marker for genetic transformation of the diatom Phaeodactylum tricornutum. PeerJ 2018, 6, e5884. [Google Scholar] [CrossRef] [PubMed]
- Kilian, O.; Benemann, C.S.E.; Niyogi, K.K.; Vick, B. High-efficiency homologous recombination in the oil-producing alga Nannochloropsis sp. Proc. Natl. Acad. Sci. USA 2011, 108, 21265–21269. [Google Scholar] [CrossRef]
- Lozano, J.-C.; Schatt, P.; Botebol, H.; Vergé, V.; Lesuisse, E.; Blain, S.; Carré, I.A.; Bouget, F.-Y. Efficient gene targeting and removal of foreign DNA by homologous recombination in the picoeukaryote Ostreococcus. Plant J. Cell Mol. Biol. 2014, 78, 1073–1083. [Google Scholar] [CrossRef]
- Doudna, J.A.; Charpentier, E. The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef]
- Huang, W.; Daboussi, F. Genetic and metabolic engineering in diatoms. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2017, 372, 20160411. [Google Scholar] [CrossRef] [PubMed]
- Kroth, P.G.; Bones, A.M.; Daboussi, F.; Ferrante, M.I.; Jaubert, M.; Kolot, M.; Nymark, M.; Río Bártulos, C.; Ritter, A.; Russo, M.T.; et al. Genome editing in diatoms: Achievements and goals. Plant Cell Rep. 2018, 37, 1401–1408. [Google Scholar] [CrossRef]
- Wijffels, R.H.; Barbosa, M.J. An outlook on microalgal biofuels. Science 2010, 329, 796–799. [Google Scholar] [CrossRef]
- Hamilton, M.L.; Haslam, R.P.; Napier, J.A.; Sayanova, O. Metabolic engineering of Phaeodactylum tricornutum for the enhanced accumulation of omega-3 long chain polyunsaturated fatty acids. Metab. Eng. 2014, 22, 3–9. [Google Scholar] [CrossRef]
- Hamilton, M.L.; Warwick, J.; Terry, A.; Allen, M.J.; Napier, J.A.; Sayanova, O. Towards the Industrial Production of Omega-3 Long Chain Polyunsaturated Fatty Acids from a Genetically Modified Diatom Phaeodactylum tricornutum. PLoS ONE 2015, 10, e0144054. [Google Scholar] [CrossRef]
- Zulu, N.N.; Popko, J.; Zienkiewicz, K.; Tarazona, P.; Herrfurth, C.; Feussner, I. Heterologous co-expression of a yeast diacylglycerol acyltransferase (ScDGA1) and a plant oleosin (AtOLEO3) as an efficient tool for enhancing triacylglycerol accumulation in the marine diatom Phaeodactylum tricornutum. Biotechnol. Biofuels 2017, 10, 187. [Google Scholar] [CrossRef]
- Kadono, T.; Kira, N.; Suzuki, K.; Iwata, O.; Ohama, T.; Okada, S.; Nishimura, T.; Akakabe, M.; Tsuda, M.; Adachi, M. Effect of an Introduced Phytoene Synthase Gene Expression on Carotenoid Biosynthesis in the Marine Diatom Phaeodactylum tricornutum. Mar. Drugs 2015, 13, 5334–5357. [Google Scholar] [CrossRef]
- Eilers, U.; Bikoulis, A.; Breitenbach, J.; Büchel, C.; Sandmann, G. Limitations in the biosynthesis of fucoxanthin as targets for genetic engineering in Phaeodactylum tricornutum. J. Appl. Phycol. 2016, 28, 123–129. [Google Scholar] [CrossRef]
- Daboussi, F.; Leduc, S.; Maréchal, A.; Dubois, G.; Guyot, V.; Perez-Michaut, C.; Amato, A.; Falciatore, A.; Juillerat, A.; Beurdeley, M.; et al. Genome engineering empowers the diatom Phaeodactylum tricornutum for biotechnology. Nat. Commun. 2014, 5, 3831. [Google Scholar] [CrossRef]
- Ajjawi, I.; Verruto, J.; Aqui, M.; Soriaga, L.B.; Coppersmith, J.; Kwok, K.; Peach, L.; Orchard, E.; Kalb, R.; Xu, W.; et al. Lipid production in Nannochloropsis gaditana is doubled by decreasing expression of a single transcriptional regulator. Nat. Biotechnol. 2017, 35, 647–652. [Google Scholar] [CrossRef] [PubMed]
- Li, D.-W.; Balamurugan, S.; Yang, Y.-F.; Zheng, J.-W.; Huang, D.; Zou, L.-G.; Yang, W.-D.; Liu, J.-S.; Guan, Y.; Li, H.-Y. Transcriptional regulation of microalgae for concurrent lipid overproduction and secretion. Sci. Adv. 2019, 5, eaau3795. [Google Scholar] [CrossRef]
- Hempel, F.; Lau, J.; Klingl, A.; Maier, U.G. Algae as Protein Factories: Expression of a Human Antibody and the Respective Antigen in the Diatom Phaeodactylum tricornutum. PLoS ONE 2011, 6, e28424. [Google Scholar] [CrossRef] [PubMed]
- Hempel, F.; Bozarth, A.S.; Lindenkamp, N.; Klingl, A.; Zauner, S.; Linne, U.; Steinbüchel, A.; Maier, U.G. Microalgae as bioreactors for bioplastic production. Microb. Cell Factories 2011, 10, 81. [Google Scholar] [CrossRef]
- D’Adamo, S.; Schiano di Visconte, G.; Lowe, G.; Szaub-Newton, J.; Beacham, T.; Landels, A.; Allen, M.J.; Spicer, A.; Matthijs, M. Engineering the unicellular alga Phaeodactylum tricornutum for high-value plant triterpenoid production. Plant Biotechnol. J. 2019, 17, 75–87. [Google Scholar] [CrossRef]
- Szyjka, S.J.; Mandal, S.; Schoepp, N.G.; Tyler, B.M.; Yohn, C.B.; Poon, Y.S.; Villareal, S.; Burkart, M.D.; Shurin, J.B.; Mayfield, S.P. Evaluation of phenotype stability and ecological risk of a genetically engineered alga in open pond production. Algal Res. 2017, 24, 378–386. [Google Scholar] [CrossRef]
- Lauritano, C.; Ianora, A. Grand Challenges in Marine Biotechnology: Overview of Recent EU-Funded Projects. In Grand Challenges in Marine Biotechnology; Rampelotto, P.H., Trincone, A., Eds.; Grand Challenges in Biology and Biotechnology; Springer International Publishing: Cham, Germany, 2018; pp. 425–449. ISBN 978-3-319-69075-9. [Google Scholar]
- Brennecke, P.; Ferrante, M.I.; Johnston, I.A.; Smith, D. A Collaborative European Approach to Accelerating Translational Marine Science. J. Mar. Sci. Eng. 2018, 6, 81. [Google Scholar] [CrossRef]
- Hasin, Y.; Seldin, M.; Lusis, A. Multi-omics approaches to disease. Genome Biol. 2017, 18, 83. [Google Scholar] [CrossRef]
- Großkinsky, D.K.; Syaifullah, S.J.; Roitsch, T. Integration of multi-omics techniques and physiological phenotyping within a holistic phenomics approach to study senescence in model and crop plants. J. Exp. Bot. 2018, 69, 825–844. [Google Scholar] [CrossRef]
- Maes, A.; Martinez, X.; Druart, K.; Laurent, B.; Guégan, S.; Marchand, C.H.; Lemaire, S.D.; Baaden, M. MinOmics, an Integrative and Immersive Tool for Multi-Omics Analysis. J. Integr. Bioinform. 2018, 15. [Google Scholar] [CrossRef]
- Lin, E.; Lane, H.-Y. Machine learning and systems genomics approaches for multi-omics data. Biomark. Res. 2017, 5, 2. [Google Scholar] [CrossRef] [PubMed]
- Grapov, D.; Fahrmann, J.; Wanichthanarak, K.; Khoomrung, S. Rise of Deep Learning for Genomic, Proteomic, and Metabolomic Data Integration in Precision Medicine. OMICS J. Integr. Biol. 2018, 22, 630–636. [Google Scholar] [CrossRef] [PubMed]
| Available Genomes | Description | References |
|---|---|---|
| Aureococcus anophagefferens | Ochrophyta | [37] |
| Bathycoccus prasinos | Chlorophyta | [38] |
| Chlorella vulgaris UTEX395 | Chlorophyta | [39] |
| Coccomyxa sp. C-169 | Chlorophyta | [40] |
| Dunaliella salina | Chlorophyta | [41] |
| Emiliania huxleyi | Haptophyta | [42] |
| Fistulifera solaris | Bacillariophyta | [21] |
| Fragilariopsis cylindrus | Bacillariophyta | [43] |
| Guillardia theta | Cryptophyta | [44] |
| Micromonas pusilla (CCMP1545) | Chlorophyta | [45] |
| Micromonas pusilla (NOUM17) | Chlorophyta | [45] |
| Micromonas sp. RCC299 | Chlorophyta | [45] |
| Nannochloropsis gaditana CCMP1894 | Eustigmatophyte | [46] |
| Nannochloropsis gaditana | Eustigmatophyte | [47] |
| Ostreococcus lucimarinus | Chlorophyta | [48] |
| Ostreococcus sp. RCC809 | Chlorophyta | https://genome.jgi.doe.gov/OstRCC809_1/OstRCC809_1.home.html |
| Ostreococcus tauri | Chlorophyta | [49,50,51] |
| Phaeodactylum tricornutum | Bacillariophyta | [52] |
| Picochlorum costavermella | Chlorophyta | [53] |
| Pseudo-nitzschia multiseries | Bacillariophyta | https://genome.jgi.doe.gov/Psemu1/Psemu1.home.html |
| Pseudo-nitzschia multistriata | Bacillariophyta | [23] |
| Skeletonema costatum | Bacillariophyta | [24] |
| Symbiodinium kawagutii | Dinoflagellata | [54] |
| Symbiodinium microadriaticum | Dinoflagellata | [55] |
| Thalassiosira oceanica | Bacillariophyta | [56] |
| Thalassiosira pseudonana | Bacillariophyta | [57] |
| Project Name | Starting Date | Ending Date | Website |
|---|---|---|---|
| BAMMBO | 01/03/2011 | 01/03/2014 | www.bammbo.eu, http://cordis.europa.eu/project/rcn/97837_en.html |
| GIAVAP | 01/01/2011 | 31/12/2013 | http://cordis.europa.eu/project/rcn/97420_en.html |
| PharmaSea | October 2012 | March 2017 | www.pharma-sea.eu |
| SUNBIOPATH | 01/01/2010 | 28/02/2013 | http://cordis.europa.eu/project/rcn/92954_en.html |
| EMBRIC | 01/06/2015 | 31/05/2019 | www.embric.eu |
| NoMorFilm | 01/04/2015 | 31/03/2019 | www.nomorfilm.eu |
| TriForC | 01/10/2013 | 30/09/2017 | https://cordis.europa.eu/project/rcn/110952/factsheet/en |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lauritano, C.; Ferrante, M.I.; Rogato, A. Marine Natural Products from Microalgae: An -Omics Overview. Mar. Drugs 2019, 17, 269. https://doi.org/10.3390/md17050269
Lauritano C, Ferrante MI, Rogato A. Marine Natural Products from Microalgae: An -Omics Overview. Marine Drugs. 2019; 17(5):269. https://doi.org/10.3390/md17050269
Chicago/Turabian StyleLauritano, Chiara, Maria Immacolata Ferrante, and Alessandra Rogato. 2019. "Marine Natural Products from Microalgae: An -Omics Overview" Marine Drugs 17, no. 5: 269. https://doi.org/10.3390/md17050269
APA StyleLauritano, C., Ferrante, M. I., & Rogato, A. (2019). Marine Natural Products from Microalgae: An -Omics Overview. Marine Drugs, 17(5), 269. https://doi.org/10.3390/md17050269







