Culturable Diversity and Lipid Production Profile of Labyrinthulomycete Protists Isolated from Coastal Mangrove Habitats of China
Abstract
1. Introduction
2. Results
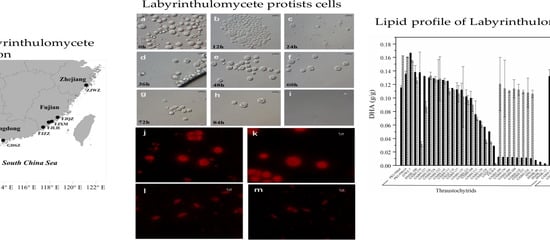
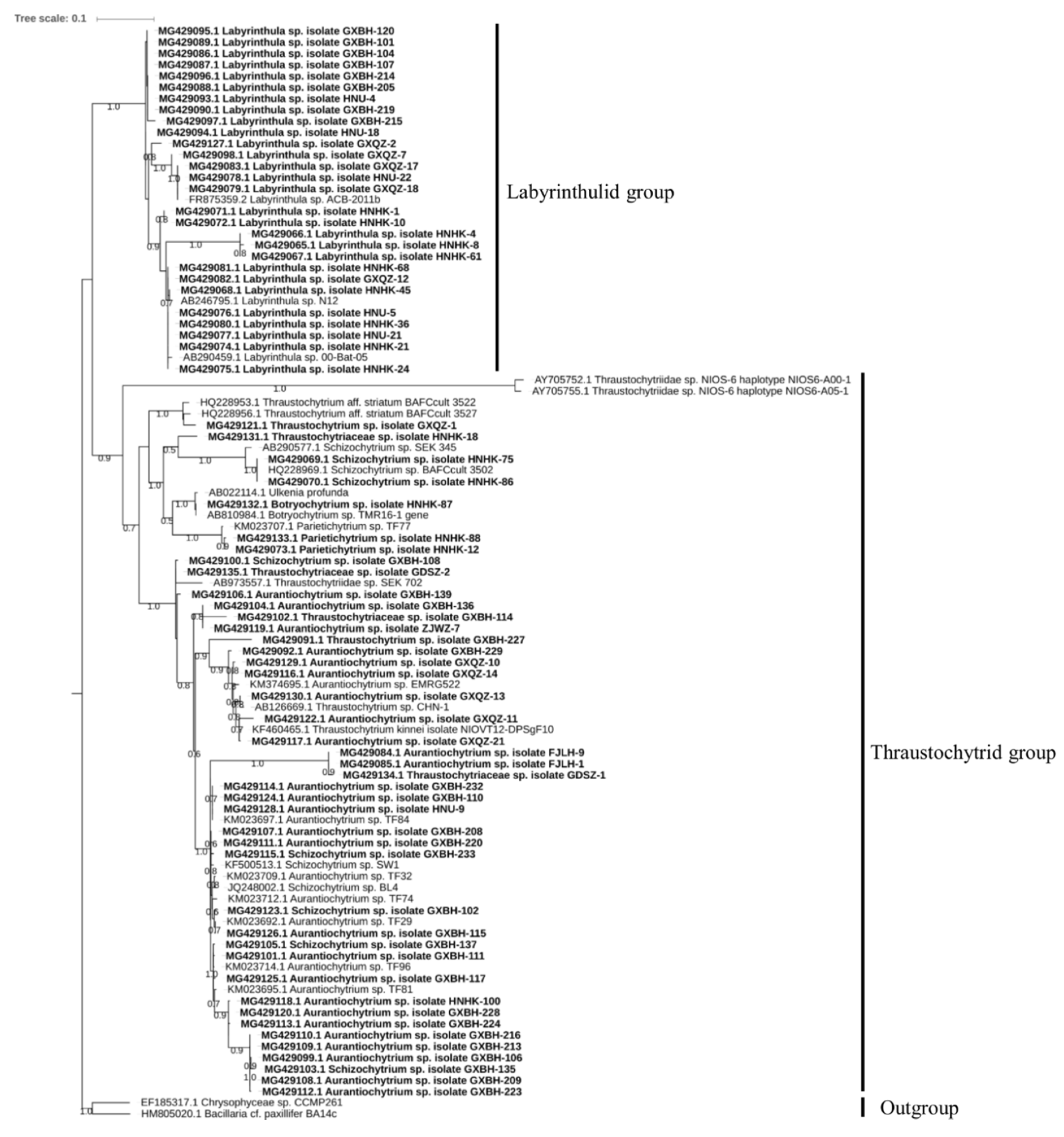
2.1. Culturable Diversity of Labyrinthulomycete Protists
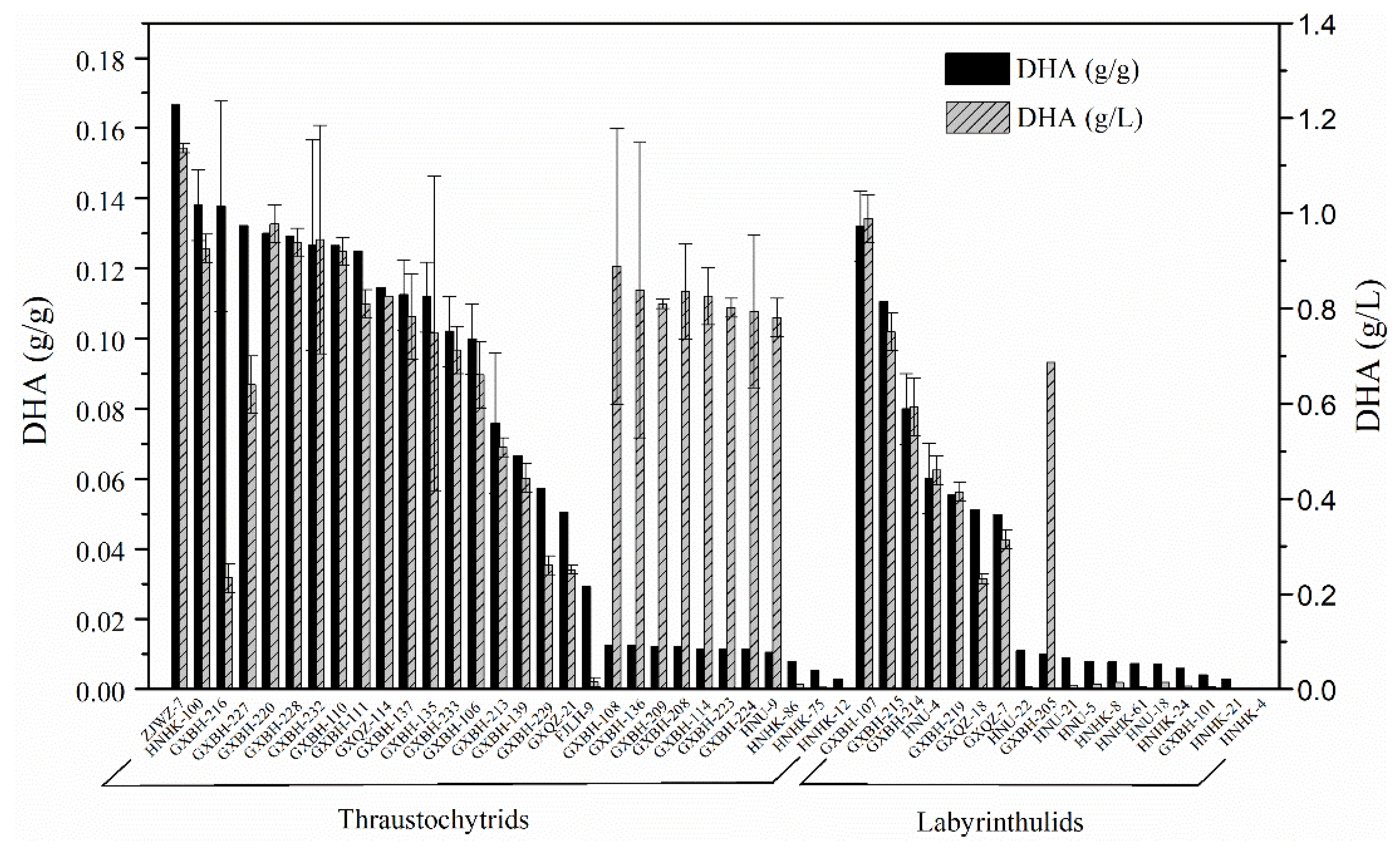
2.2. Screening for High Docosahexaenoic Acid (DHA)-Producing Strains
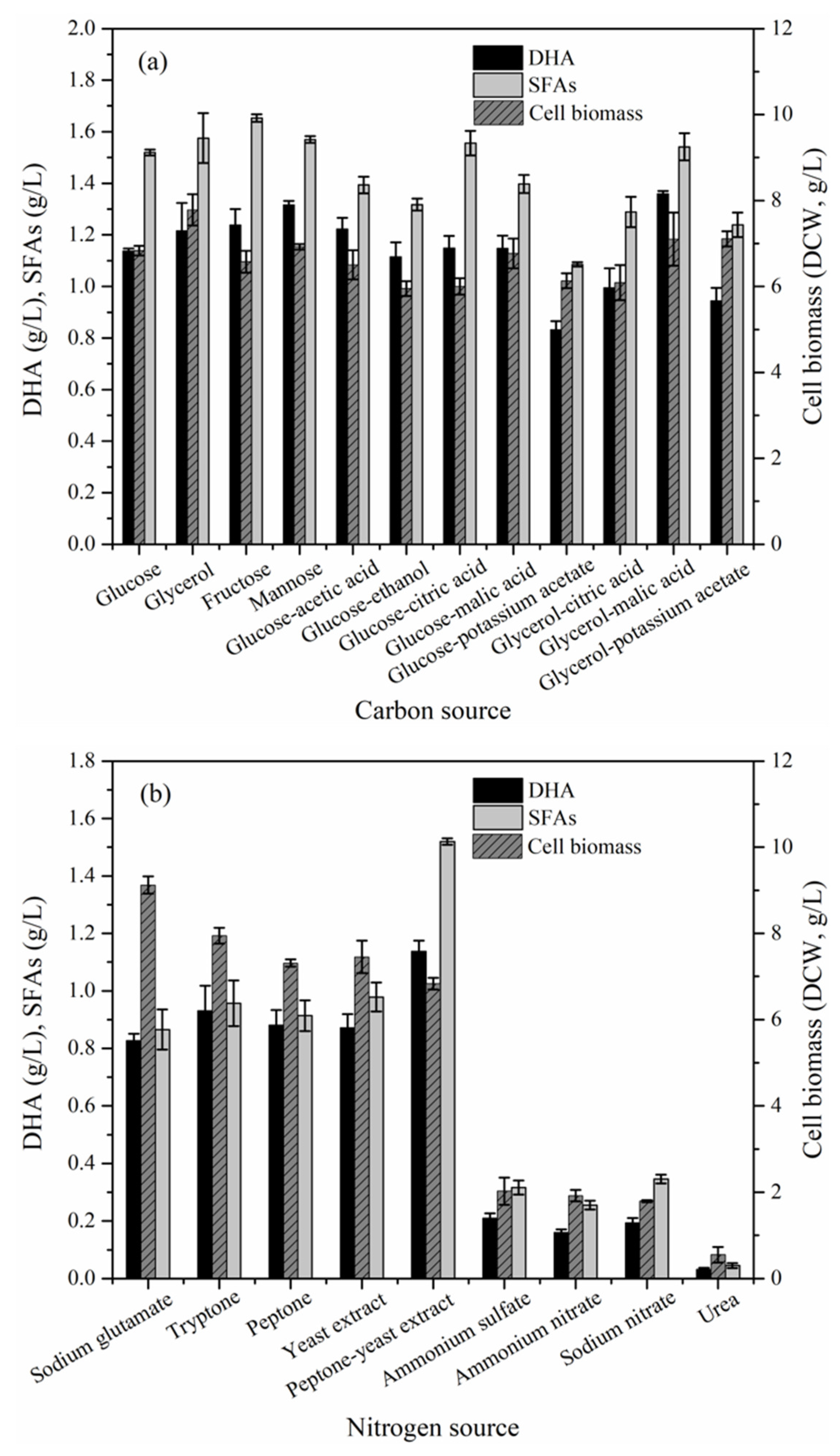
2.3. Effect of Culture Conditions on the Cell Biomass, DHA and Saturated Fatty Acids (SFAs) Production
2.4. Batch Production of DHA by Strain ZJWZ-7 under Optimal Conditions
3. Discussion
3.1. Isolation and Phylogenetic Diversity of Labyrinthulomycetes
3.2. Screening for High DHA Production Strains
3.3. Optimization and Validation of the Culture Conditions
4. Materials and Methods
4.1. Sample Collection and Strain Isolation
4.2. Sequencing and Phylogenetic Analysis
4.3. Screening for DHA-Producing Strains
4.4. Culture Optimization for the Top Strain ZJWZ-7
4.5. Analytical Methods
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bennett, R.M.; Honda, D.; Beakes, G.W.; Thines, M. Labyrinthulomycota. In Handbook of the Protists; Archibald, J.M., Simpson, A.G.B., Slamovits, C.H., Margulis, L., Melkonian, M., Chapman, D.J., Corliss, J.O., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 1–36. [Google Scholar]
- Leander, C.A.; Porter, D. Redefining the genus Aplanochytrium (phylum Labyrinthulomycota). Mycotaxon 2000, 76, 439–444. [Google Scholar]
- Leander, C.A.; Porter, D. The Labyrinthulomycota Is Comprised of Three Distinct Lineages. Mycologia 2001, 93, 459. [Google Scholar] [CrossRef]
- Leander, C.A.; Porter, D.; Leander, B.S. Comparative morphology and molecular phylogeny of aplanochytrids (Labyrinthulomycota). Eur. J. Protistology 2004, 40, 317–328. [Google Scholar] [CrossRef]
- Raghukumar, S. Ecology of the marine protists, the Labyrinthulomycetes (thraustochytrids and labyrinthulids). Eur. J. Protistology 2002, 38, 127–145. [Google Scholar] [CrossRef]
- Bremer, G.B. Lower marine fungi (Labyrinthulomycetes) and the decay of mangrove leaf litter. Hydrobiologia 1995, 295, 89–95. [Google Scholar] [CrossRef]
- Singh, P.; Liu, Y.; Li, L.; Wang, G. Ecological dynamics and biotechnological implications of thraustochytrids from marine habitats. Appl. Microbiol. Biotechnol. 2014, 98, 5789–5805. [Google Scholar] [CrossRef] [PubMed]
- Raghukumar, S. The Role of Fungi in Marine Detrital Processes. In Marine Microbiology: Facets and Opportunities; Ramaiah, N., Ed.; National Institute of Oceanography: Goa, India, 2004; pp. 91–101. [Google Scholar]
- Raghukumar, S.; Damare, V.S. Increasing evidence for the important role of Labyrinthulomycetes in marine ecosystems. Bot. Mar. 2011, 54, 3–11. [Google Scholar] [CrossRef]
- Raghukumar, S. Fungi in Coastal and Oceanic Marine Ecosystems; Springer International Publishing: Cham, Switzerland, 2017; pp. 17–38. [Google Scholar]
- Hinzpeter, I.; Quilodrán, B.; Stead, R.; Trujillo, L.; Vidal, J.; Shene, C. Isolation of thraustochytrid strains in the coastal zone of Puerto Montt, Chile and evaluation of Docosahexaenoic acid (22:6n-3, DHA) production. Afinidad 2009, 66, 482–487. [Google Scholar]
- Gupta, A.; Wilkens, S.; Adcock, J.L.; Puri, M.; Barrow, C.J. Pollen baiting facilitates the isolation of marine thraustochytrids with potential in omega-3 and biodiesel production. J. Ind. Microbiol. Biotechnol. 2013, 40, 1231–1240. [Google Scholar] [CrossRef]
- Caamaño, E.; Loperena, L.; Hinzpeter, I.; Pradel, P.; Gordillo, F.; Corsini, G.; Tello, M.; Lavín, P.; González, A.R. Isolation and molecular characterization of Thraustochytrium strain isolated from Antarctic Peninsula and its biotechnological potential in the production of fatty acids. Brazilian J. Microbiol. 2017, 48, 671–679. [Google Scholar] [CrossRef]
- Unagul, P.; Suetrong, S.; Preedanon, S.; Klaysuban, A.; Gundool, W.; Suriyachadkun, C.; Sakayaroj, J. Isolation, fatty acid profiles and cryopreservation of marine thraustochytrids from mangrove habitats in Thailand. Bot. Mar. 2017, 60, 363–379. [Google Scholar] [CrossRef]
- Sullivan, B.K.; Robinson, K.L.; Trevathan-Tackett, S.M.; Lilje, E.S.; Gleason, F.H.; Lilje, O. The First Isolation and Characterisation of the Protist Labyrinthula sp. in Southeastern Australia. J. Eukaryotic Microbiol. 2017, 64, 504–513. [Google Scholar] [CrossRef]
- Boro, M.C.; Harakava, R.; Pires-Zottarelli, C.L.A. Labyrinthulomycota from Brazilian mangrove swamps and coastal waters. Botanica Marina 2018, 61, 1. [Google Scholar] [CrossRef]
- Ueda, M.; Nomura, Y.; Doi, K.; Nakajima, M.; Honda, D. Seasonal dynamics of culturable thraustochytrids (Labyrinthulomycetes, Stramenopiles) in estuarine and coastal waters. Aquat. Microb. Ecol. 2015, 74, 187–204. [Google Scholar] [CrossRef]
- Lee Chang, K.J.; Dunstan, G.A.; Abell, G.C.J.; Clementson, L.A.; Blackburn, S.I.; Nichols, P.D.; Koutoulis, A. Biodiscovery of new Australian thraustochytrids for production of biodiesel and long-chain omega-3 oils. Appl. Microbiol. Biotechnol. 2012, 93, 2215–2231. [Google Scholar] [CrossRef]
- Burja, A.M.; Radianingtyas, H.; Windust, A.; Barrow, C.J. Isolation and characterization of polyunsaturated fatty acid producing Thraustochytrium species: Screening of strains and optimization of omega-3 production. Appl. Microbiol. Biotechnol. 2006, 72, 1161–1169. [Google Scholar] [CrossRef]
- Damare, V.S. Diversity of thraustochytrid protists isolated from brown alga, Sargassum cinereum using 18S rDNA sequencing and their morphological response to heavy metals. J. Mar. Biol. Assoc. U. K. 2015, 95, 265–276. [Google Scholar] [CrossRef]
- Jaseera, K.V.; Kaladharan, P.; Vijayan, K.K.; Sandhya, S.V.; Antony, M.L.; Pradeep, M.A. Isolation and phylogenetic identification of heterotrophic thraustochytrids from mangrove habitats along the southwest coast of India and prospecting their PUFA accumulation. J. Appl. Phycol. 2018, 1–12. [Google Scholar] [CrossRef]
- Damare, V.; Raghukumar, S. Morphology and Physiology of the Marine Straminipilan Fungi, the Aplanochytrids Isolated from the Equatorial Indian Ocean. Indian J. Mar. Sci. 2006, 35, 326–340. [Google Scholar]
- Liu, Y.; Singh, P.; Sun, Y.; Luan, S.; Wang, G. Culturable diversity and biochemical features of thraustochytrids from coastal waters of Southern China. Appl. Microbiol. Biotechnol. 2014, 98, 3241–3255. [Google Scholar] [CrossRef]
- Hong, W.K.; Rairakhwada, D.; Seo, P.S.; Park, S.Y.; Hur, B.K.; Kim, C.H.; Seo, J.W. Production of lipids containing high levels of docosahexaenoic acid by a newly isolated microalga, Aurantiochytrium sp. KRS101. Appl. Microbiol. Biotechnol. 2011, 164, 1468–1480. [Google Scholar] [CrossRef]
- Manikan, V.; Nazir, M.Y.M.; Kalil, M.S.; Isa, M.H.M.; Kader, A.J.A.; Yusoff, W.M.W.; Hamid, A.A. A new strain of docosahexaenoic acid producing microalga from Malaysian coastal waters. Algal Res. 2015, 9, 40–47. [Google Scholar] [CrossRef]
- Jaritkhuan, S.; Suanjit, S. Species diversity and polyunsaturated fatty acid content of thraustochytrids from fallen mangrove leaves in Chon Buri province, Thailand. Agrc. Nat. Resour. 2018, 52, 24–32. [Google Scholar] [CrossRef]
- Kumon, Y.; Yokoyama, R.; Haque, Z.; Yokochi, T.; Honda, D.; Nakahara, T. A New Labyrinthulid Isolate That Produces Only Docosahexaenoic Acid. Mar. Biotechnol. 2006, 8, 170–177. [Google Scholar] [CrossRef]
- Aasen, I.M.; Ertesvåg, H.; Heggeset, T.M.B.; Liu, B.; Brautaset, T.; Vadstein, O.; Ellingsen, T.E. Thraustochytrids as production organisms for docosahexaenoic acid (DHA), squalene, and carotenoids. Appl. Microbiol. Biotechnol. 2016, 100, 4309–4321. [Google Scholar] [CrossRef]
- Huang, J.; Aki, T.; Yokochi, T.; Nakahara, T.; Honda, D.; Kawamoto, S.; Shigeta, S.; Ono, K.; Suzuki, O. Grouping Newly Isolated Docosahexaenoic Acid-Producing Thraustochytrids Based on Their Polyunsaturated Fatty Acid Profiles and Comparative Analysis of 18S rRNA Genes. Mar. Biotechnol. 2003, 5, 450–457. [Google Scholar] [CrossRef] [PubMed]
- Jakobsen, A.N.; Aasen, I.M.; Josefsen, K.D.; Strøm, A.R. Accumulation of Docosahexaenoic Acid-Rich Lipid in Thraustochytrid Aurantiochytrium sp. strain T66: Effects of N and P Starvation and O2 Limitation. Appl. Microbiol. Biotechnol. 2008, 80, 297–306. [Google Scholar] [CrossRef] [PubMed]
- Chaung, K.C.; Chu, C.Y.; Su, Y.M.; Chen, Y.M. Effect of culture conditions on growth, lipid content, and fatty acid composition of Aurantiochytrium mangrovei strain BL10. AMB Express 2012, 2, 1–13. [Google Scholar] [CrossRef]
- Qu, L.; Ren, L.J.; Sun, G.N.; Ji, X.J.; Nie, Z.K.; Huang, H. Batch, fed-batch and repeated fed-batch fermentation processes of the marine thraustochytrid schizochytrium sp. For producing docosahexaenoic acid. Bioprocess. Biosyst. Eng. 2013, 36, 1905–1912. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Ye, H.; Sen, B.; Xie, Y.; He, Y.; Park, S.; Wang, G. Improved production of docosahexaenoic acid in batch fermentation by newly-isolated strains of Schizochytrium sp. and Thraustochytriidae sp. through bioprocess optimization. Synth. Syst. Biotechnol. 2018, 3, 121–129. [Google Scholar] [CrossRef]
- Wang, Q.; Sen, B.; Liu, X.; He, Y.; Xie, Y.; Wang, G. Enhanced saturated fatty acids accumulation in cultures of newly-isolated strains of Schizochytrium sp. and Thraustochytriidae sp. for large-scale biodiesel production. Sci. Total Environ. 2018, 631–632, 994–1004. [Google Scholar] [CrossRef]
- Honda, D.; Yokochi, T.; Nakahara, T.; Raghukumar, S.; Nakagiri, A.; Schaumann, K.; Higashihara, T. Molecular Phylogeny of Labyrinthulids and Thraustochytrids Based on the Sequencing of 18S Ribosomal RNA Gene. J. Eukaryotic Microbiol. 1999, 46, 637–647. [Google Scholar] [CrossRef]
- Honda, D.; Yokochi, T.; Nakahara, T.; Erata, M.; Higashihara, T. Schizochytrium limacinum sp. nov., a new thraustochytrid from a mangrove area in the west Pacific Ocean. Mycol. Res. 1998, 102, 439–448. [Google Scholar] [CrossRef]
- Fan, K.W.; Vrijmoed, L.L.P.; Jones, E.B.G. Physiological studies of subtropical mangrove thraustochytrids. Bot. Mar. 2002, 45, 50–57. [Google Scholar] [CrossRef]
- Rosa, S.M.; Galvagno, M.A.; Ve’lez, C.G. Adjusting culture conditions to isolate thraustochytrids from temperate and cold environments in southern Argentina. Mycoscience 2011, 52, 242–252. [Google Scholar] [CrossRef]
- Fuller, M.S.; Jaworski, A. Zoosporic fungi in teaching and research. Mycologia 1987, 79, 920. [Google Scholar]
- Li, Q.; Chen, G.Q.; Fan, K.W.; Lu, F.P.; Aki, T.; Jiang, Y. Screening and characterization of squalene-producing thraustochytrids from Hong Kong mangroves. J. Agric. Food. Chem. 2009, 57, 4267–4272. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Singh, D.; Byreddy, A.R.; Thyagarajan, T.; Sonkar, S.P.; Mathur, A.S.; Tuli, D.K.; Barrow, C.J.; Puri, M. Exploring omega-3 fatty acids, enzymes and biodiesel producing thraustochytrids from Australian and Indian marine biodiversity. Biotechnol. J. 2016, 11, 345–355. [Google Scholar] [CrossRef] [PubMed]
- Jain, R.; Raghukumar, S.; Sambaiah, K.; Kumon, Y.; Nakahara, T. Docosahexaenoic acid accumulation in thraustochytrids: Search for the rationale. Mar. Biol. 2007, 151, 1657–1664. [Google Scholar] [CrossRef]
- Perveen, Z.; Ando, H.; Ueno, A.; Ito, Y.; Yamamoto, Y.; Yamada, Y.; Takagi, T.; Kaneko, T.; Kogame, K.; Okuyama, H. Isolation and characterization of a novel thraustochytrid-like microorganism that efficiently produces docosahexaenoic acid. Biotechnol. Lett. 2006, 28, 197–202. [Google Scholar] [CrossRef] [PubMed]
- Marchan, L.F.; Lee Chang, K.J.; Nichols, P.D.; Polglase, J.L.; Mitchell, W.J.; Gutierrez, T. Screening of new British thraustochytrids isolates for docosahexaenoic acid (DHA) production. J. Appl. Phycol. 2017, 29, 2831–2843. [Google Scholar] [CrossRef]
- Erwin, J. Comparative biochemistry of fatty acids in eukaryotic microorganisms. Lipids Biomembr. Eukaryot. Microorg. 1973, 41–143. [Google Scholar]
- Yaguchi, T.; Tanaka, S.; Yokochi, T.; Nakahara, T.; Higashihara, T. Production of High Yields of Docosahexaenoic Acid by Schizochytrium sp. Strain SR21. J. Am. Oil Chemists’ Soc. 1997, 74, 1431–1434. [Google Scholar] [CrossRef]
- Zeng, Y.; Ji, X.J.; Lian, M.; Ren, L.J.; Jin, L.J.; Ouyang, P.K.; Huang, H. Development of a temperature shift strategy for efficient docosahexaenoic acid production by a marine fungoid protist, Schizochytrium sp. HX-308. Appl. Biochem. Biotechnol. 2011, 164, 249–255. [Google Scholar] [CrossRef]
- Huang, T.Y.; Lu, W.C.; Chu, I.M. A fermentation strategy for producing docosahexaenoic acid in Aurantiochytrium limacinum SR21 and increasing C22:6 proportions in total fatty acid. Bioresour. Technol. 2012, 123, 8–14. [Google Scholar] [CrossRef]
- Sun, X.M.; Ren, L.J.; Ji, X.J.; Chen, S.L.; Guo, D.S.; Huang, H. Adaptive evolution of Schizochytrium sp. by continuous high oxygen stimulations to enhance docosahexaenoic acid synthesis. Bioresour. Technol. 2016, 211, 374–381. [Google Scholar] [CrossRef]
- Ralph, P.J.; Short, F.T. Impact of the wasting disease pathogen, Labyrinthula zosterae, on the photobiology of eelgrass Zostera marina. Mar. Ecol. Prog. Ser. 2002, 226, 265–271. [Google Scholar] [CrossRef]
- Jakobsson-Thor, S.; Toth, G.B.; Brakel, J.; Bockelmann, A.C.; Pavia, H. Seagrass wasting disease varies with salinity and depth in natural Zostera marina populations. Mar. Ecol. Prog. Ser. 2018, 587, 105–115. [Google Scholar] [CrossRef]
- Raghukumar, S. Thraustochytrid marine protists: Production of PUFAs and other emerging technologies. Mar. Biotechnol. 2008, 10, 631–640. [Google Scholar] [CrossRef]
- Bowles, R.D.; Hunt, A.E.; Bremer, G.B.; Duchars, M.G.; Eaton, R.A. Long-chain n-3 polyunsaturated fatty acid production by members of the marine protistan group the thraustochytrids: Screening of isolates and optimization of docosahexaenoic acid production. J. Biotechnol. 1999, 70, 193–202. [Google Scholar] [CrossRef]
- Wu, S.-T.; Yu, S.-T.; Lin, L.-P. Effect of culture conditions on docosahexaenoic acid production by Schizochytrium sp. S31. Process Biochem. 2005, 40, 3103–3108. [Google Scholar] [CrossRef]
- Scott, S.D.; Armenta, R.E.; Berryman, K.T.; Norman, A.W. Use of raw glycerol to produce oil rich in polyunsaturated fatty acids by a thraustochytrid. Enzyme Microb. Technol. 2011, 48, 267–272. [Google Scholar] [CrossRef]
- Quilodrán, B.; Hinzpeter, I.; Quiroz, A.; Shene, C. Evaluation of liquid residues from beer and potato processing for the production of docosahexaenoic acid (C22:6n-3, DHA) by native thraustochytrid strains. World J. Microbiol. Biotechnol. 2009, 25, 2121. [Google Scholar] [CrossRef]
- Liang, Y.; Sarkany, N.; Cui, Y.; Yesuf, J.; Trushenski, J.; Blackburn, J.W. Use of sweet sorghum juice for lipid production by Schizochytrium limacinum SR21. Bioresour. Technol. 2010, 101, 3623–3627. [Google Scholar] [CrossRef]
- Bai, M.; Sen, B.; Wang, Q.; Xie, Y.; He, Y.; Wang, G. Molecular Detection and Spatiotemporal Characterization of Labyrinthulomycete Protist Diversity in the Coastal Waters Along the Pearl River Delta. Microb. Ecol. 2018, 77, 394–405. [Google Scholar] [CrossRef]
- Collado-Mercado, E.; Radway, J.C.; Collier, J.L. Novel uncultivated labyrinthulomycetes revealed by 18S rDNA sequences from seawater and sediment samples. Aquat. Microb. Ecol. 2010, 58, 215–228. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately Maximum-Likelihood Trees for Large Alignments. PLOS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Lepage, G.; Roy, C.C. Improved recovery of fatty acid through direct transesterification without prior extraction or purification. J. Lipid Res. 1984, 25, 1391–1396. [Google Scholar]
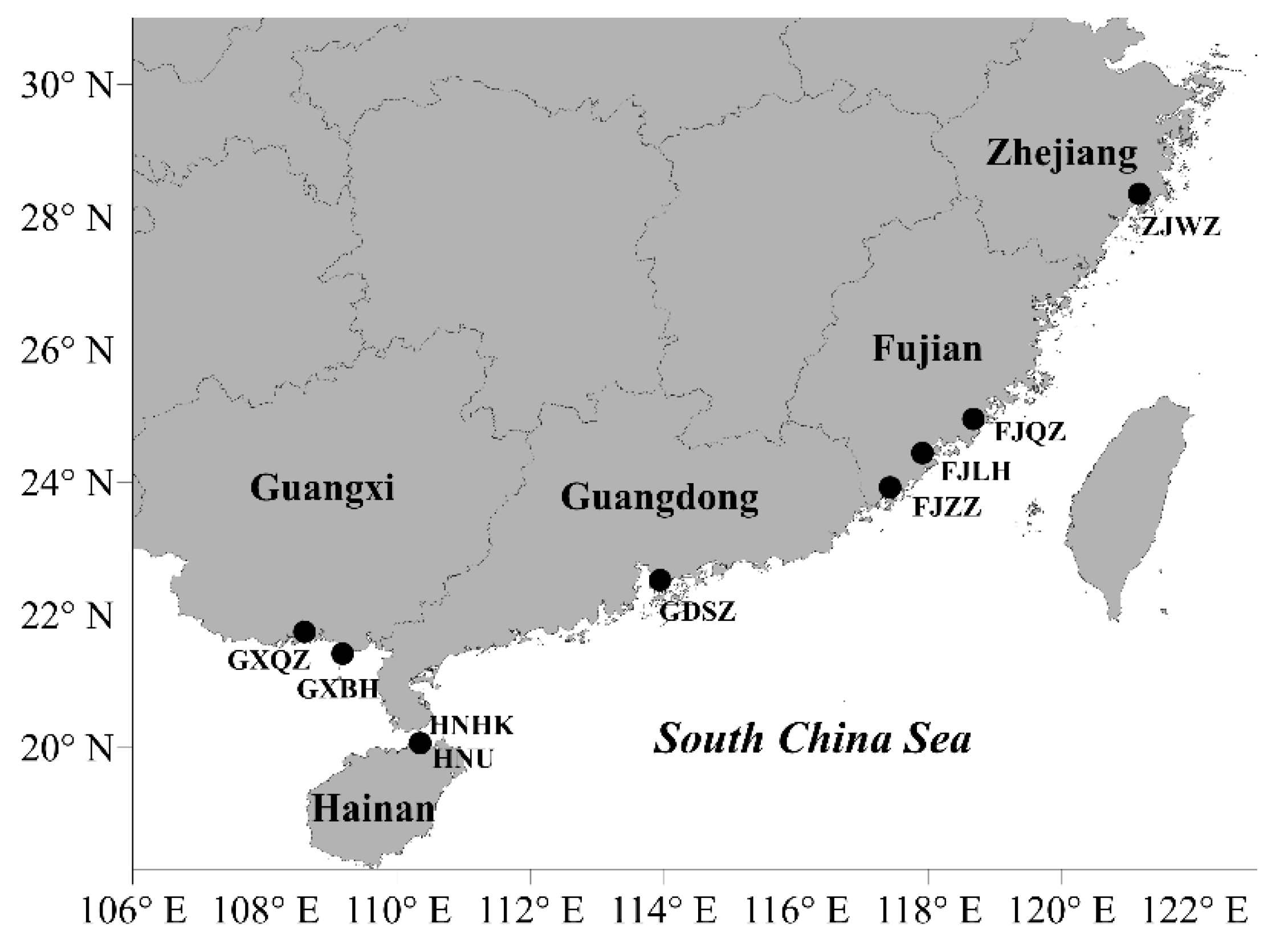
| Sampling Site | Sampling Date | Latitude (°N) | Longitude (°E) | Number of Isolates | Genus Level Classification | Number of Strains |
|---|---|---|---|---|---|---|
| Haikou, Hainan (HNHK) | 2015.07 | 20.06231 | 110.3352 | 69 | Aurantiochytrium, Schizochytrium, Thraustochytrium, Botryochytrium, Parietichytrium, Thraustochytriidae, Labyrinthula | 17 |
| Haikou, Hainan (HNU) | 2016.03 | 20.06344 | 110.3345 | 23 | Aurantiochytrium, Labyrinthula | 6 |
| Beihai, Guangxi (GXBH) | 2016.04 | 21.41175 | 109.1626 | 54 | Aurantiochytrium, Schizochytrium, Thraustochytrium, Thraustochytriidae, Labyrinthula | 32 |
| Qinzhou, Guangxi (GXQZ) | 2016.04 | 21.74423 | 108.5903 | 16 | Aurantiochytrium, Thraustochytrium, Labyrinthula | 11 |
| Shenzhen, Guangdong (GDSZ) | 2015.08 | 22.52029 | 113.9496 | 22 | Thraustochytrium, Thraustochytriidae | 2 |
| Longhai, Fujian (FJLH) | 2015.09 | 24.43823 | 117.9037 | 7 | Aurantiochytrium | 2 |
| Quanzhou, Fujian (FJQZ) | 2015.09 | 24.94337 | 118.6748 | 6 | Aurantiochytrium | 0 |
| Zhangzhou, Fujian (FJZZ) | 2015.09 | 23.92075 | 117.4167 | 10 | Aurantiochytrium, Schizochytrium, Thraustochytrium, | 0 |
| Wenzhou, Zhejiang (ZJWZ) | 2015.09 | 28.34687 | 121.1738 | 5 | Aurantiochytrium | 1 |
| Group | Genus | Number of Isolates in Each Genus | Number of Sequences Submitted |
|---|---|---|---|
| Thraustochytrids | Aurantiochytrium | 129 | 27 |
| Schizochytrium | 46 | 7 | |
| Thraustochytrium | 6 | 6 | |
| Botryochytrium | 1 | 1 | |
| Parietichytrium | 2 | 2 | |
| Labyrinthulids | Labyrinthula | 28 | 28 |
| Strain | ZJWZ-7 | HNHK-100 | GXBH-216 | GXBH-227 | GXBH-220 | GXBH-228 | GXBH-232 | GXBH-110 |
| C12:0 | 1.49 ± 0.26 | 1.45 ± 0.12 | 0.56 ± 0.08 | 0.10 ± 0.02 | 0.96 ± 0.10 | 0.43 ± 0.22 | 1.13 ± 0.10 | 0.70 ± 0.27 |
| C14:0 | 4.85 ± 0.02 | 4.35 ± 0.04 | 3.90 ± 0.06 | 4.10 ± 0.34 | 4.70 ± 0.04 | 5.01 ± 0.12 | 4.39 ± 0.09 | 5.01 ± 0.04 |
| C15:0 | 1.88 ± 0.01 | 1.74 ± 0.01 | 1.75 ± 0.18 | 4.58 ± 0.27 | 2.09 ± 0.00 | 2.01 ± 0.00 | 2.19 ± 0.04 | 1.82 ± 0.02 |
| C16:0 | 41.77 ± 0.33 | 46.08 ± 0.29 | 37.19 ± 0.63 | 46.32 ± 1.02 | 36.11 ± 0.33 | 40.09 ± 0.23 | 36.70 ± 0.37 | 42.57± 0.14 |
| C17:0 | 0.48 ± 0.00 | 0.53 ± 0.00 | 0.66 ± 0.07 | 1.40 ± 0.03 | 0.51 ± 0.01 | 0.49 ± 0.00 | 0.57 ± 0.01 | 0.44 ± 0.01 |
| C18:0 | 0.91 ± 0.02 | 1.09 ± 0.00 | 0.88 ± 0.04 | 0.95 ± 0.04 | 0.73 ± 0.04 | 0.76 ± 0.00 | 0.83 ± 0.01 | 0.80 ± 0.03 |
| C18:1n9 | 0.38 ± 0.00 | 0.32 ± 0.00 | 0.54 ± 0.10 | 0.11 ± 0.00 | 0.43 ± 0.00 | 0.49 ± 0.00 | 0.46 ± 0.00 | 0.33 ± 0.00 |
| C20:4n6 | 0.44 ± 0.00 | 0.39 ± 0.00 | 0.50 ± 0.02 | 0.02 ± 0.03 | 0.55 ± 0.00 | 0.53 ± 0.01 | 0.53 ± 0.01 | 0.49 ± 0.00 |
| C20:5n3 | 0.32 ± 0.00 | 0.24 ± 0.00 | 0.48 ± 0.06 | 0.26 ± 0.02 | 0.38 ± 0.01 | 0.37 ± 0.00 | 0.34 ± 0.01 | 0.35 ± 0.02 |
| C22:5n3 | 7.63 ± 0.01 | 7.42 ± 0.02 | 9.77 ± 0.27 | 8.23 ± 0.40 | 8.52 ± 0.06 | 7.98 ± 0.01 | 9.26 ± 0.17 | 7.48 ± 0.09 |
| C22:6n3 | 37.68 ± 0.06 | 34.31 ± 0.17 | 42.95 ± 1.02 | 28.54 ± 1.34 | 42.33 ± 0.54 | 39.60 ± 0.27 | 41.24 ± 0.35 | 37.72± 0.28 |
| Total SFAs | 51.38 | 55.25 | 44.94 | 57.45 | 45.11 | 48.79 | 45.8 | 51.42 |
| Total PUFAs | 46.07 | 42.36 | 53.69 | 37.05 | 51.77 | 48.47 | 51.38 | 46.04 |
| Biomass (g/L) | 6.83 ± 0.11 | 6.72 ± 0.22 | 1.72 ± 0.22 | 4.85 ± 0.45 | 7.52 ± 0.06 | 7.28 ± 0.23 | 7.45 ± 0.11 | 7.28 ± 0.07 |
| TFAs yield (% biomass) | 44.18 ± 1.15 | 40.2 ± 2.24 | 32.03 ± 5.88 | 47.57 ± 0.36 | 30.69 ± 0.50 | 32.57 ± 0.21 | 30.68 ± 7.09 | 33.5 ± 0.47 |
| TFAs productivity (g/L/d) | 0.74 ± 0.01 | 0.67 ± 0.02 | 0.14 ± 0.01 | 0.58 ± 0.03 | 0.57 ± 0.01 | 0.58 ± 0.02 | 0.56 ± 0.14 | 0.60 ± 0.01 |
| Strain | GXBH-111 | GXBH-114 | GXBH-137 | GXBH-135 | GXBH-233 | GXBH-107 | GXBH-215 | |
| C12:0 | 0.12 ± 0.02 | 0.66 ± 0.10 | 0.14 ± 0.03 | 0.13 ± 0.04 | 0.15 ± 0.02 | 0.62 ± 0.25 | 0.12 ± 0.01 | |
| C14:0 | 4.33 ± 0.51 | 5.70 ± 0.04 | 3.89 ± 0.16 | 4.67 ± 0.39 | 3.87 ± 0.19 | 4.56 ± 0.02 | 4.50 ± 0.22 | |
| C15:0 | 2.16 ± 0.13 | 1.72 ± 0.01 | 2.16 ± 0.09 | 1.73 ± 0.09 | 1.86 ± 0.04 | 1.54 ± 0.01 | 1.38 ± 0.02 | |
| C16:0 | 40.61 ± 0.67 | 42.16 ± 0.04 | 38.54 ± 1.71 | 46.38 ± 0.49 | 42.47 ± 0.29 | 37.03 ± 0.07 | 45.89 ± 0.51 | |
| C17:0 | 0.71 ± 0.02 | 0.44 ± 0.00 | 0.64 ± 0.11 | 0.56 ± 0.11 | 0.71 ± 0.01 | 0.38 ± 0.00 | 0.38 ± 0.01 | |
| C18:0 | 0.87 ± 0.06 | 0.77 ± 0.00 | 0.88 ± 0.03 | 0.91 ± 0.06 | 0.96 ± 0.03 | 0.71 ± 0.00 | 1.08 ± 0.09 | |
| C18:1n9 | 0.45 ± 0.09 | 0.39 ± 0.00 | 0.37 ± 0.03 | 0.38 ± 0.02 | 0.40 ± 0.01 | 0.46 ± 0.00 | 0.38 ± 0.00 | |
| C20:4n6 | 0.51 ± 0.02 | 0.48 ± 0.01 | 0.68 ± 0.29 | 0.42 ± 0.01 | 0.43 ± 0.01 | 0.58 ± 0.01 | 0.41 ± 0.01 | |
| C20:5n3 | 0.32 ± 0.03 | 0.39 ± 0.00 | 0.33 ± 0.02 | 0.45 ± 0.04 | 0.50 ± 0.02 | 0.40 ± 0.00 | 0.49 ± 0.01 | |
| C22:5n3 | 8.96 ± 0.50 | 7.31 ± 0.06 | 8.81 ± 0.42 | 7.77 ± 0.15 | 8.42 ± 0.28 | 8.36 ± 0.04 | 7.82 ± 0.04 | |
| C22:6n3 | 39.55 ± 0.89 | 37.80 ± 0.16 | 39.38 ± 2.72 | 35.54 ± 0.30 | 39.14 ± 0.30 | 42.73 ± 0.09 | 36.32 ± 0.32 | |
| Total SFAs | 48.81 | 51.44 | 46.25 | 54.38 | 50.03 | 44.85 | 53.35 | |
| Total PUFAs | 49.34 | 45.98 | 49.21 | 44.17 | 48.48 | 52.07 | 45.04 | |
| Biomass (g/L) | 6.48 ± 0.25 | 7.22 ± 0.10 | 6.97 ± 0.18 | 6.72 ± 0.18 | 7.00 ± 0.06 | 7.51 ± 0.26 | 6.8 ± 0.32 | |
| TFAs yield (% biomass) | 31.6 ± 0.98 | 30.26 ± 0.23 | 28.67 ± 4.18 | 31.48 ± 4.32 | 26.07 ± 1.90 | 30.88 ± 2.64 | 30.4 ± 0.51 | |
| TFAs productivity (g/L/d) | 0.51 ± 0.02 | 0.54 ± 0.00 | 0.50 ± 0.06 | 0.53 ± 0.24 | 0.46 ± 0.03 | 0.57 ± 0.03 | 0.52 ± 0.03 |
| Cell biomass (DCW, g/L) | DHA (g/L) | SFAs (g/L) |
|---|---|---|
| Carbon source | Carbon source | Nitrogen source |
| Nitrogen source | Nitrogen source | KH2PO4 |
| Salinity | Initial pH | Salinity |
| Initial pH | Temperature | Initial pH |
| Temperature | Rotation rate | Temperature |
| Rotation rate | Rotation rate |
| Culture Condition | Biomass (g/L) | DHA (g/L) | SFAs (g/L) | DHA (% TFAs) | SFAs (% TFAs) |
|---|---|---|---|---|---|
| KH2PO4 (g/L) | |||||
| 0 | 6.53 | 1.20 | 1.69 | 36.70 | 51.34 |
| 0.1 | 6.81 | 1.31 | 1.85 | 37.56 | 51.72 |
| 0.25 | 6.83 | 1.14 | 1.52 | 37.68 | 50.36 |
| 0.4 | 6.82 | 1.14 | 1.53 | 37.94 | 49.72 |
| 0.8 | 6.99 | 1.07 | 1.45 | 37.62 | 49.78 |
| Salinity (% seawater) | |||||
| 0 | 5.62 | 0.91 | 1.05 | 35.32 | 51.44 |
| 20 | 6.99 | 0.99 | 1.04 | 37.32 | 49.28 |
| 40 | 6.87 | 0.87 | 0.95 | 37.21 | 50.53 |
| 60 | 6.88 | 1.10 | 1.25 | 36.43 | 51.96 |
| 80 | 6.74 | 0.90 | 0.97 | 36.95 | 50.26 |
| 100 | 6.83 | 1.14 | 1.52 | 37.68 | 50.36 |
| 120 | 6.61 | 0.90 | 0.94 | 36.87 | 48.65 |
| Temperature (℃) | |||||
| 20 | 6.56 | 1.01 | 1.58 | 39.41 | 50.46 |
| 25 | 6.58 | 1.14 | 1.49 | 38.38 | 50.76 |
| 28 | 6.83 | 1.14 | 1.52 | 37.68 | 50.36 |
| 32 | 5.73 | 0.91 | 1.29 | 35.86 | 50.08 |
| 35 | 4.98 | 0.51 | 0.83 | 34.01 | 53.82 |
| Initial pH | |||||
| 4 | 7.21 | 1.21 | 1.39 | 40.62 | 46.09 |
| 5 | 6.57 | 1.04 | 1.53 | 36.33 | 52.76 |
| 6 | 6.46 | 1.07 | 1.33 | 37.87 | 50.49 |
| 6.47 | 6.83 | 1.14 | 1.52 | 37.68 | 50.36 |
| 7 | 6.54 | 1.23 | 1.41 | 40.68 | 46.51 |
| 8 | 5.49 | 0.82 | 1.23 | 35.08 | 52.80 |
| Rotation rate (rpm) | |||||
| 100 | 6.14 | 0.91 | 1.08 | 39.91 | 47.86 |
| 150 | 6.83 | 1.14 | 1.52 | 37.68 | 50.36 |
| 180 | 6.41 | 1.04 | 1.58 | 33.88 | 53.95 |
| 200 | 6.51 | 0.99 | 1.33 | 36.67 | 50.57 |
| 250 | 3.55 | 0.58 | 1.21 | 35.46 | 53.37 |
| Strains | ZJWZ-7 |
|---|---|
| C12:0 | 0.15 ± 0.00 |
| C14:0 | 3.28 ± 0.02 |
| C15:0 | 1.89 ± 0.02 |
| C16:0 | 37.63 ± 0.15 |
| C17:0 | 0.22 ± 0.00 |
| C18:0 | 0.84 ± 0.02 |
| C18:3n3 | 0.17 ± 0.00 |
| C20:0 | 0.12 ± 0.00 |
| C21:0 | 0.27 ± 0.01 |
| C20:3n6 | 0.42 ± 0.03 |
| C20:4n6 | 0.23 ± 0.00 |
| C20:5n3 | 0.06 ± 0.00 |
| C22:5n3 | 7.57 ± 0.00 |
| C22:6n3 | 44.52 ± 0.12 |
| Total PUFAs | 52.97 |
| Total SFAs | 44.40 |
| Biomass (g/L) | 9.88 |
| TFAs yield (% biomass) | 37.69 |
| DHA (g/g) | 0.17 |
| DHA (g/L) | 1.66 |
| PUFAs (g/L) | 2.00 |
| SFAs (g/g) | 0.17 |
| SFAs (g/L) | 1.68 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Q.; Ye, H.; Xie, Y.; He, Y.; Sen, B.; Wang, G. Culturable Diversity and Lipid Production Profile of Labyrinthulomycete Protists Isolated from Coastal Mangrove Habitats of China. Mar. Drugs 2019, 17, 268. https://doi.org/10.3390/md17050268
Wang Q, Ye H, Xie Y, He Y, Sen B, Wang G. Culturable Diversity and Lipid Production Profile of Labyrinthulomycete Protists Isolated from Coastal Mangrove Habitats of China. Marine Drugs. 2019; 17(5):268. https://doi.org/10.3390/md17050268
Chicago/Turabian StyleWang, Qiuzhen, Huike Ye, Yunxuan Xie, Yaodong He, Biswarup Sen, and Guangyi Wang. 2019. "Culturable Diversity and Lipid Production Profile of Labyrinthulomycete Protists Isolated from Coastal Mangrove Habitats of China" Marine Drugs 17, no. 5: 268. https://doi.org/10.3390/md17050268
APA StyleWang, Q., Ye, H., Xie, Y., He, Y., Sen, B., & Wang, G. (2019). Culturable Diversity and Lipid Production Profile of Labyrinthulomycete Protists Isolated from Coastal Mangrove Habitats of China. Marine Drugs, 17(5), 268. https://doi.org/10.3390/md17050268