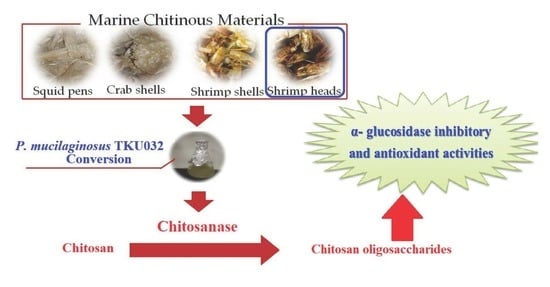
Production of a Thermostable Chitosanase from Shrimp Heads via Paenibacillus mucilaginosus TKU032 Conversion and its Application in the Preparation of Bioactive Chitosan Oligosaccharides
Abstract
:1. Introduction
2. Results and Discussion
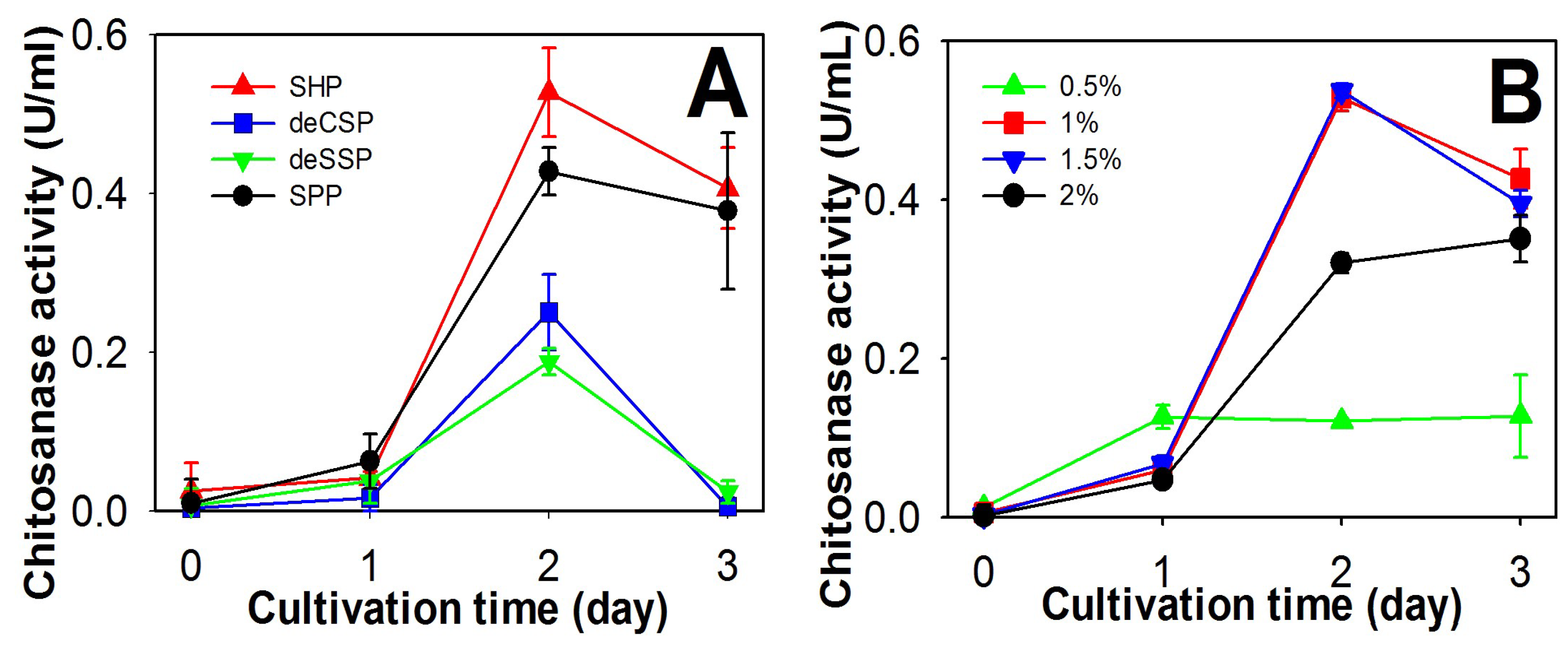
2.1. Screening of Chitinous Materials as Sole C/N Source for Chitosanase Production
2.2. Comparison of Chitosanase Production from SHP Using Different Bacteria
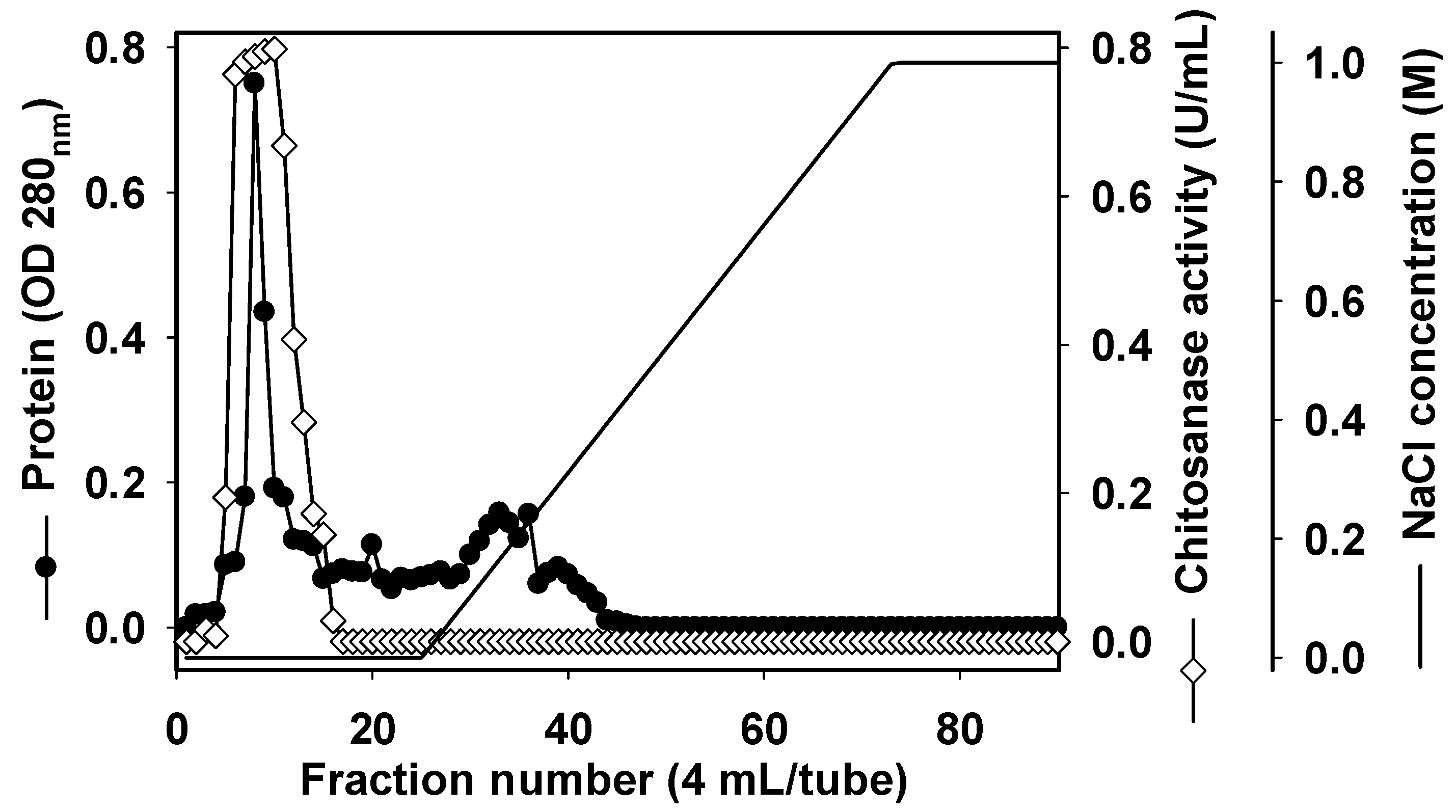
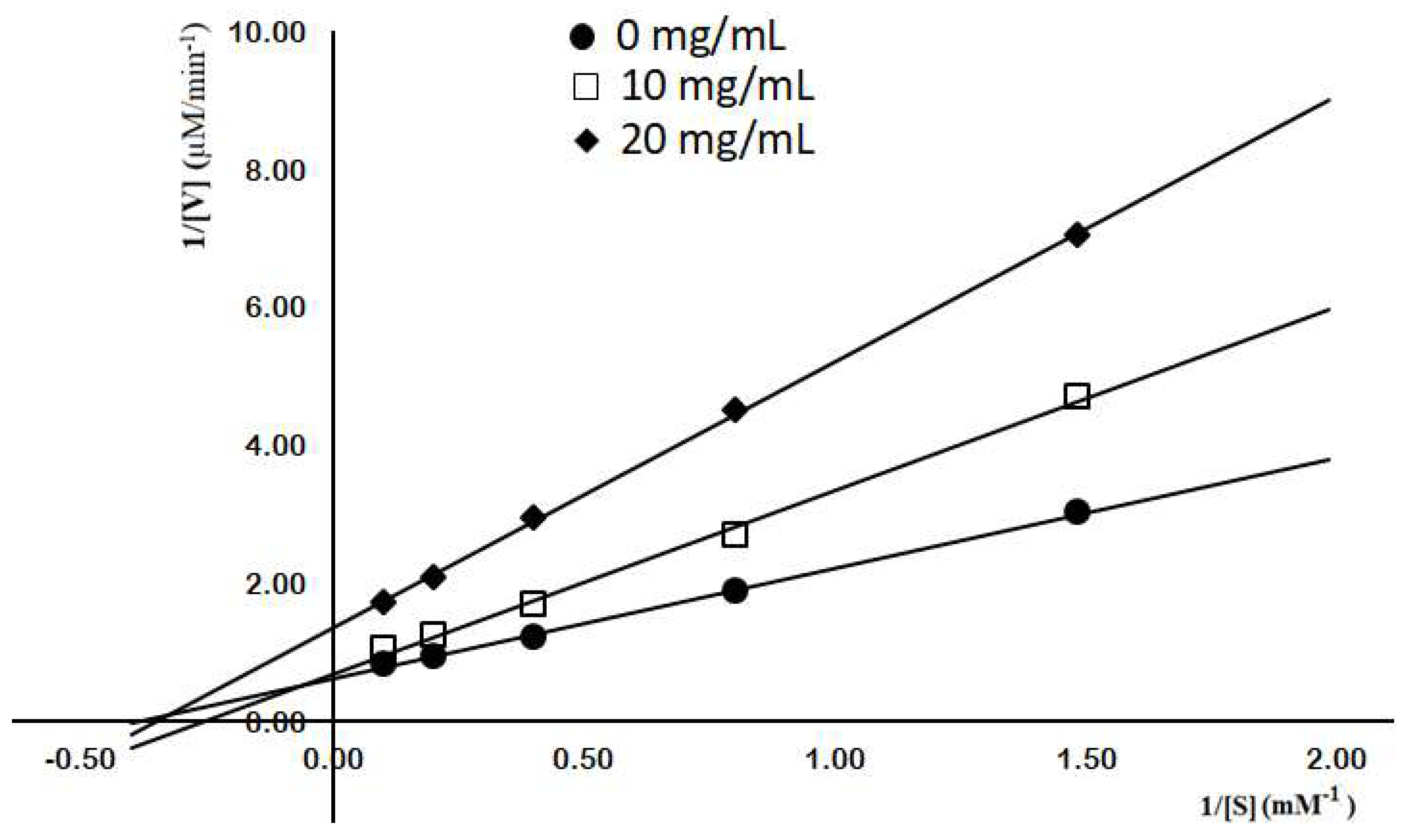
2.3. Purification and Characterization of Chitosanase
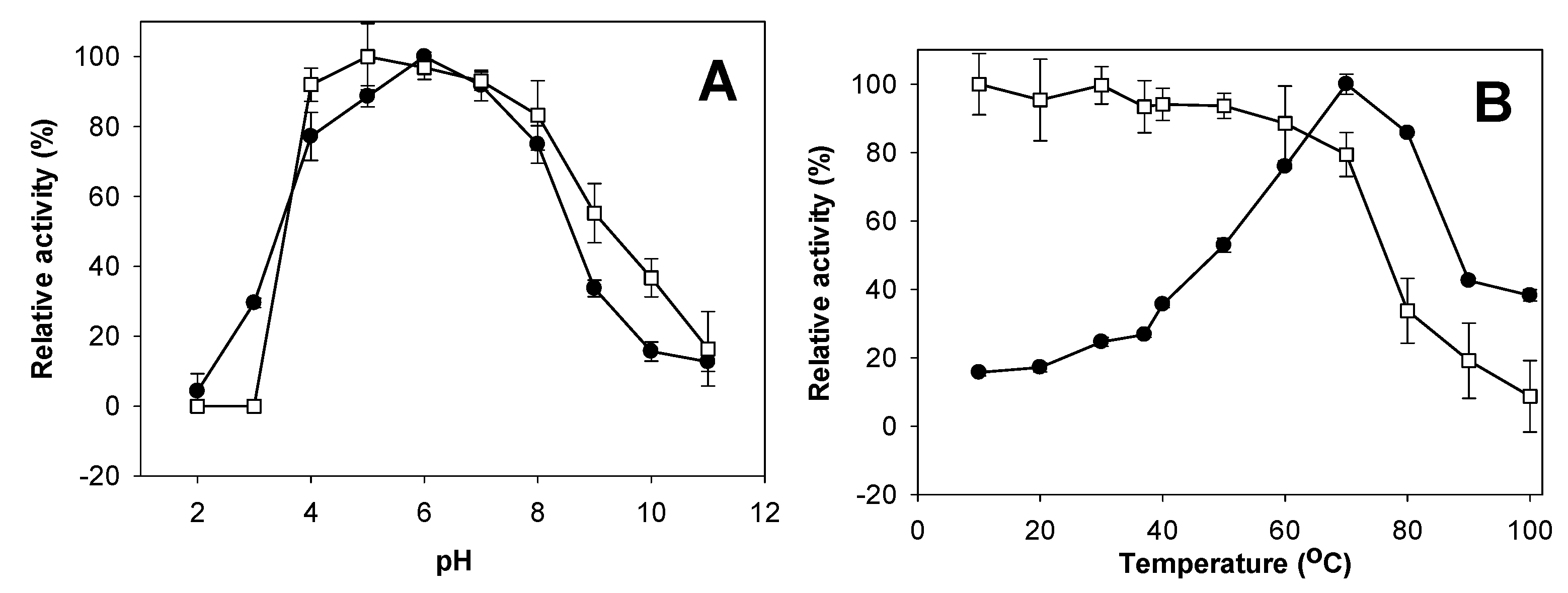
2.4. Effects of pH and Temperature on Activity and Stability of Chitosanase
2.5. Effect of Metal Ions on Activity of Chitosanase
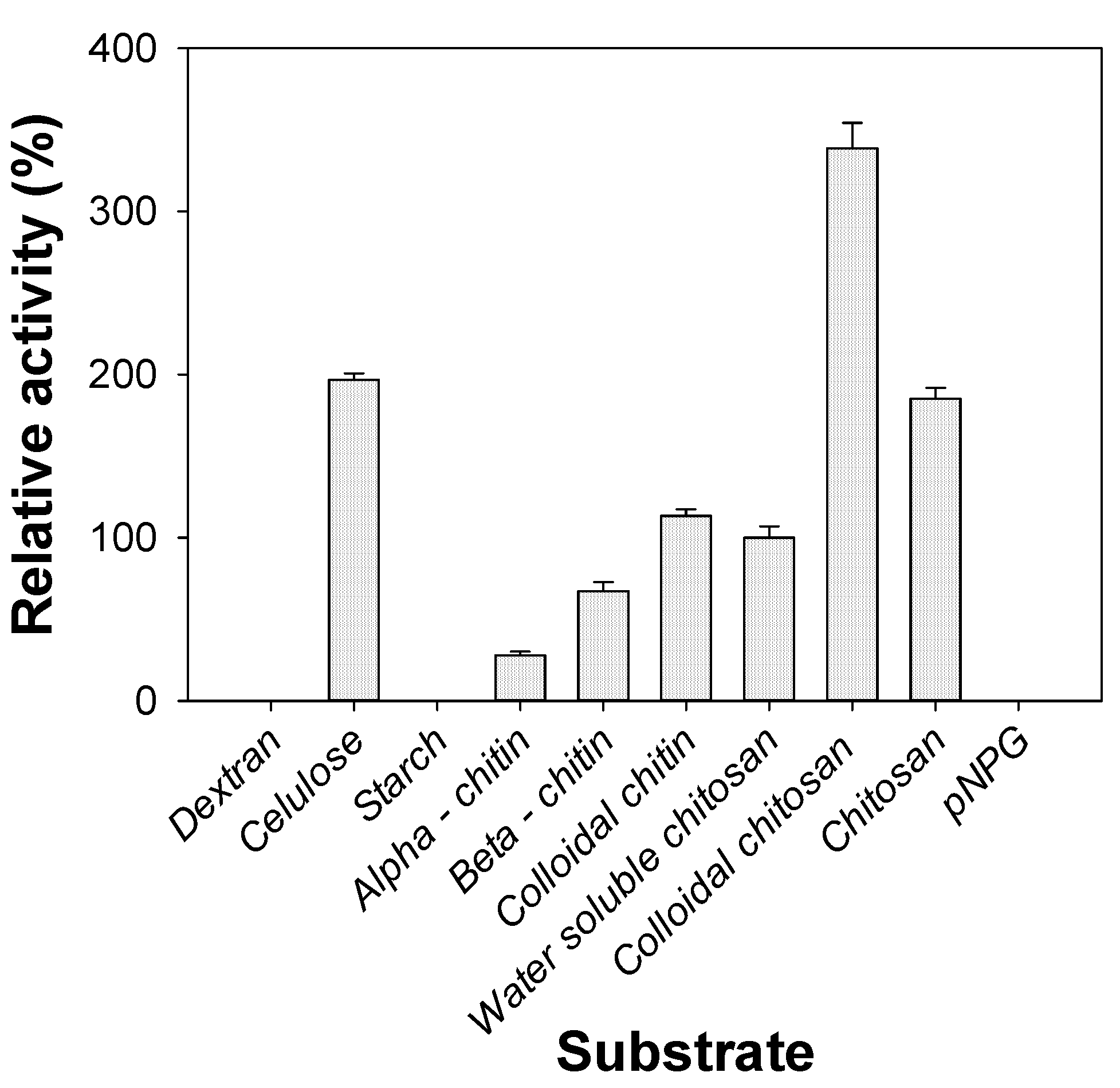
2.6. Substrate Specificity of Chitosanase
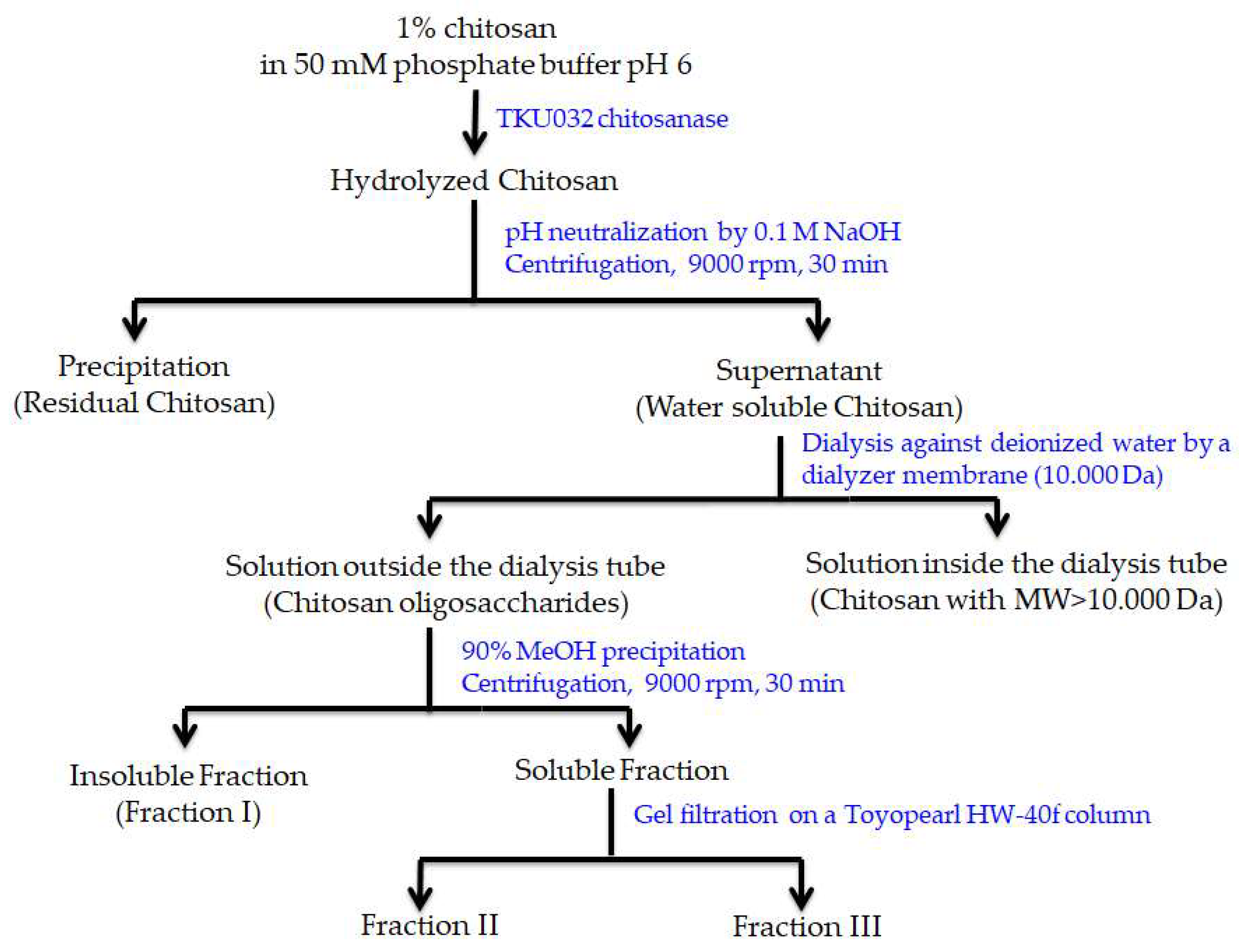
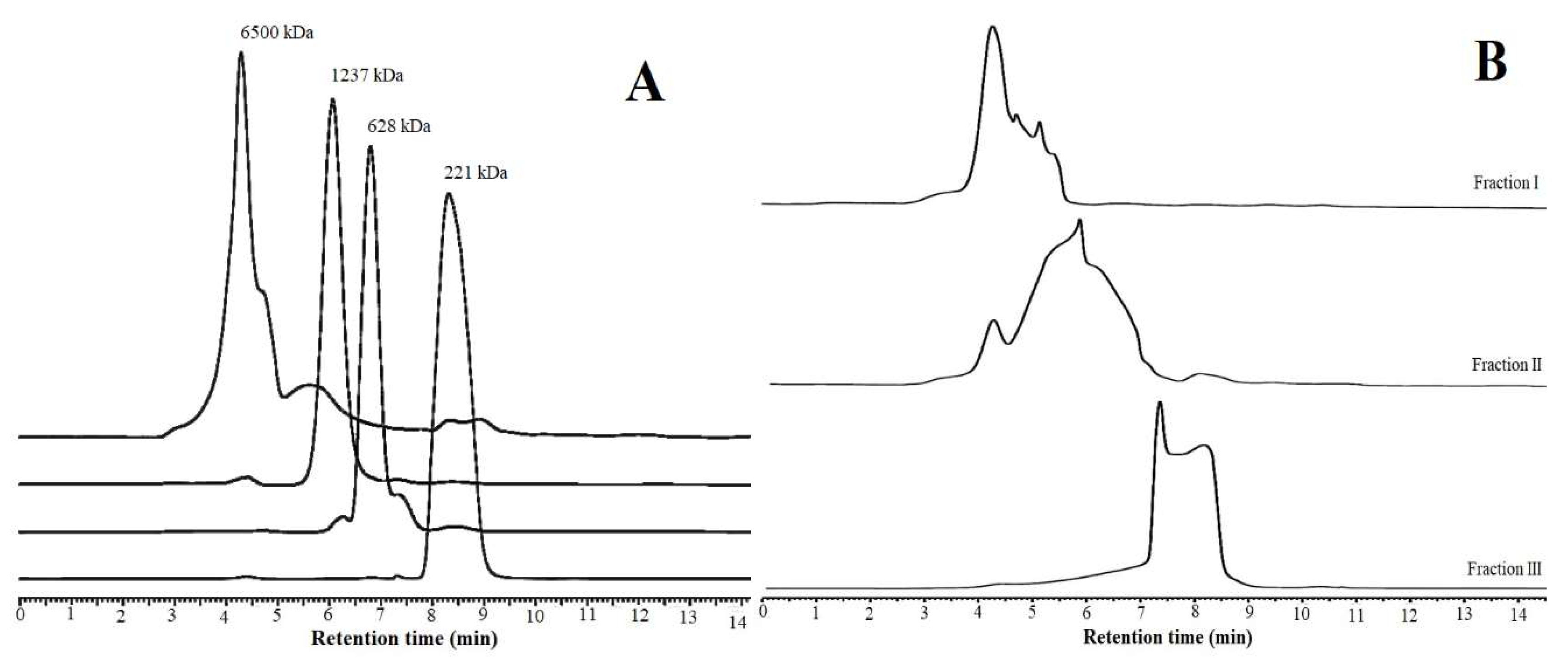
2.7. COS Production
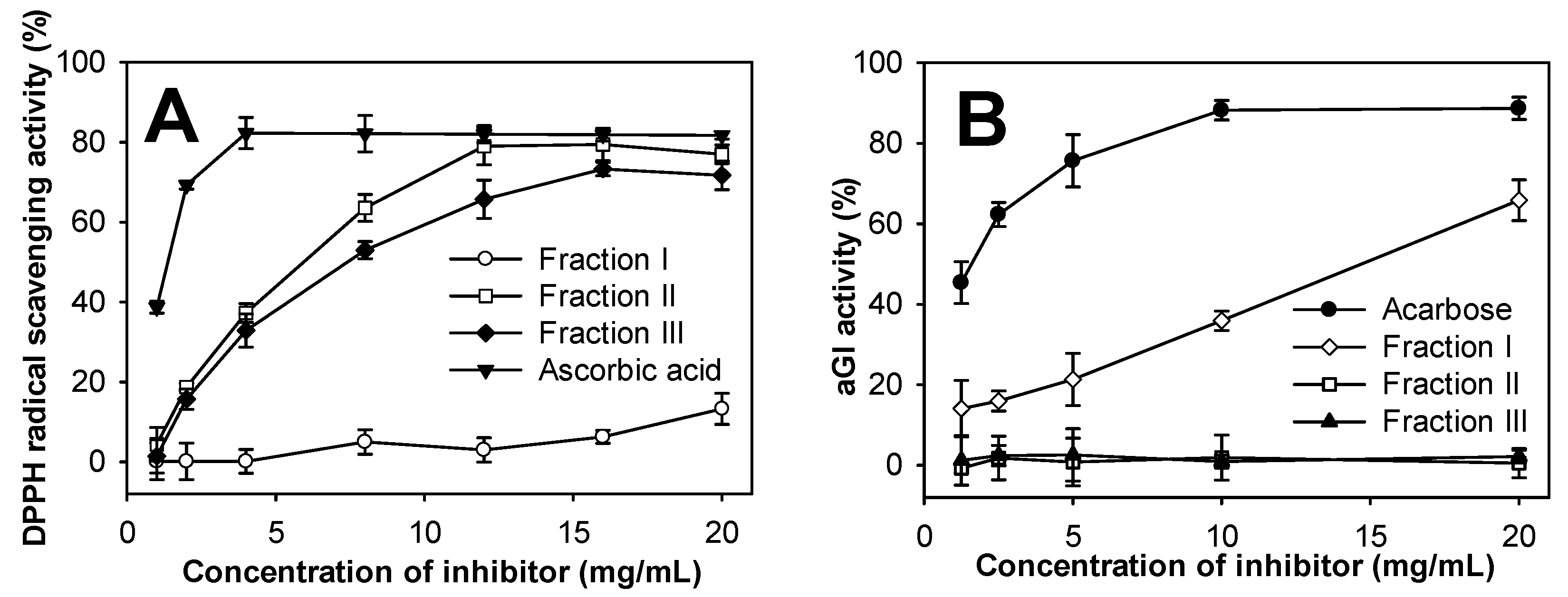
2.8. Evaluation of Antioxidant and aGI Activities of COS Fractions
3. Materials and Methods
3.1. Materials
3.2. Measurement of Chitosanase Activity
3.3. Screening of Chitinous Materials as Sole C/N Source for Chitosanase Activity
3.4. Purification of Chitosanase
3.5. Effects of pH and Temperature on Activity and Stability of Chitosanase
3.6. Effect of Metal Ions on Chitosanase Activity
3.7. Substrate Specificity of Chitosanase
3.8. Antioxidant Activity Assay
3.9. aGI Activity Assay
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Zitouni, M.; Fortin, M.; Scheerle, R.K.; Letzel, T.; Matteau, D.; Rodrigue, S.; Brzezinski, R. Biochemical and molecular characterization of a thermostable chitosanase produced by the strain Paenibacillus sp. 1794 newly isolated from compost. Appl. Microbiol. Biotechnol. 2013, 97, 5801–5813. [Google Scholar] [CrossRef]
- Kumar, M.N.V.R. A review of chitin and chitosan applications. React. Funct. Polym. 2000, 46, 1–27. [Google Scholar] [CrossRef]
- Wang, S.L.; Liang, T.W. Microbial reclamation of squid pens and shrimp shells. Res. Chem. Interm. 2017, 43, 3445. [Google Scholar] [CrossRef]
- Elieh-Ali-Komi, D.; Hamblin, M.R. Chitin and chitosan: Production and application of versatile biomedical nanomaterials. Int. J. Adv. Res. (Indore). 2016, 4, 411–427. [Google Scholar]
- Liang, T.W.; Chen, W.T.; Lin, Z.H.; Kuo, Y.H.; Nguyen, A.D.; Pan, P.S.; Wang, S.L. An amphiprotic novel chitosanase from Bacillus mycoides and its application in the production of chitooligomers with their antioxidant and anti–inflammatory evaluation. Int. J. Mol. Sci. 2017, 17, 1302. [Google Scholar] [CrossRef]
- Jo, S.H.; Ha, K.S.; Moon, K.S.; Kim, J.G.; Oh, C.G.; Kim, Y.C.; Apostolidis, E.; Kwon, Y.I. Molecular weight dependent glucose lowering effect of low molecular weight chitosan oligosaccharide (GO2KA1) on postprandial blood glucose level in SD rats model. Int. J. Mol. Sci. 2013, 14, 14214–14224. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.W.; Park, Y.S.; Choi, J.W.; Yi, S.Y.; Shin, W.S. Antidiabetic effects of chitosan oligosaccharides in neonatal streptozotocin-induced noninsulin-dependent diabetes mellitus in rats. Biol. Pharm. Bull. 2003, 26, 1100–1103. [Google Scholar] [CrossRef]
- Tran, T.N.; Doan, C.T.; Nguyen, V.B.; Nguyen, A.D.; Wang, S.L. The isolation of chitinase from Streptomyces thermocarboxydus and its application in the preparation of chitin oligomers. Res. Chem. Interm. 2019, 45, 727–742. [Google Scholar] [CrossRef]
- Doan, C.T.; Tran, T.N.; Nguyen, V.B.; Nguyen, A.D.; Wang, S.L. Reclamation of marine chitinous materials for chitosanase production via microbial conversion by Paenibacillus macerans. Mar. Drugs. 2018, 16, 429. [Google Scholar] [CrossRef]
- Azuma, K.; Osaki, T.; Minami, S.; Okamoto, Y. Anticancer and anti–inflammatory properties of chitin and chitosan oligosaccharides. J. Funct. Biomater. 2015, 6, 33–49. [Google Scholar] [CrossRef]
- Liang, T.W.; Chen, Y.J.; Yen, Y.H.; Wang, S.L. The antitumor activity of the hydrolysates of chitinous materials hydrolyzed by crude enzyme from Bacillus amyloliquefaciens V656. Process Biochem. 2007, 42, 527–534. [Google Scholar] [CrossRef]
- No, H.K.; Park, N.Y.; Lee, S.H.; Meyers, S.P. Antibacterial activity of chitosans and chitosan oligomers with different molecular weights. Int. J. Food Microbiol. 2002, 74, 65–72. [Google Scholar] [CrossRef]
- Sun, T.; Qin, Y.; Xu, H.; Xie, J.; Hu, D.; Xue, B.; Hua, X. Antibacterial activities and preservative effect of chitosan oligosaccharide Maillard reaction products on Penaeus vannamei. Int. J. Biol. Macromol. 2017, 105, 764–768. [Google Scholar] [CrossRef]
- Wang, S.L.; Liu, C.P.; Liang, T.W. Fermented and enzymatic production of chitin/chitosan oligosaccharides by extracellular chitinases from Bacillus cereus TKU027. Carbohydr. Polym. 2012, 90, 1305–1313. [Google Scholar] [CrossRef]
- Liang, T.W.; Liu, C.P.; Wu, C.; Wang, S.L. Applied development of crude enzyme from Bacillus cereus in prebiotics and microbial community changes in soil. Carbohydr. Polym. 2013, 92, 2141–2148. [Google Scholar] [CrossRef]
- Nguyen, A.D.; Huang, C.C.; Liang, T.W.; Nguyen, V.B.; Pan, P.S.; Wang, S.L. Production and purification of a fungal chitosanase and chitooligomers from Penicillium janthinellum D4 and discovery of the enzyme activators. Carbohydr. Polym. 2014, 108, 331–337. [Google Scholar] [CrossRef]
- Park, J.K.; Shimono, K.; Ochiai, N.; Shigeru, K.; Kurita, M.; Ohta, Y.; Tanaka, K.; Matsuda, H.; Kawamukai, M. Purification, characterization, and gene analysis of a chitosanase (ChoA) from Matsuebacter chitosanotabidus 3001. J. Bacteriol. 1999, 181, 6642–6649. [Google Scholar]
- Doan, C.T.; Tran, T.N.; Nguyen, V.B.; Nguyen, A.D.; Wang, S.L. Conversion of squid pens to chitosanases and proteases via Paenibacillus sp. TKU042. Mar. Drugs 2018, 16, 83. [Google Scholar] [CrossRef]
- Wang, S.L.; Yu, H.T.; Tsai, M.H.; Doan, C.T.; Nguyen, V.B.; Do, V.C.; Nguyen, A.D. Conversion of squid pens to chitosanases and dye adsorbents via Bacillus cereus. Res. Chem. Interm. 2018, 44, 4903–4911. [Google Scholar] [CrossRef]
- Wang, C.L.; Su, J.W.; Liang, T.W.; Nguyen, A.D.; Wang, S.L. Production, purification and characterization of a chitosanase from Bacillus cereus. Res. Chem. Interm. 2014, 40, 2237–2248. [Google Scholar] [CrossRef]
- Liang, T.W.; Chen, Y.Y.; Pan, P.S.; Wang, S.L. Purification of chitinase/chitosanase from Bacillus cereus and discovery of an enzyme inhibitor. Int. J. Biol. Macromol. 2014, 63, 8–14. [Google Scholar] [CrossRef]
- Liang, T.W.; Hsieh, J.L.; Wang, S.L. Production and purification of a protease, a chitosanase and chitin oligosaccharides by Bacillus cereus TKU022 fermentation. Carbohyd. Res. 2012, 362, 38–46. [Google Scholar] [CrossRef]
- Wang, S.L.; Chen, T.R.; Liang, T.W.; Wu, P.C. Conversion and degradation of shellfish wastes by Bacillus cereus TKU018 fermentation for the production of chitosanase and bioactive materials. Biochem. Eng. J. 2009, 48, 111–117. [Google Scholar] [CrossRef]
- Liang, T.W.; Kuo, Y.H.; Wu, P.C.; Wang, C.L.; Nguyen, A.D.; Wang, S.L. Purification and characterization of a chitosanase and a protease by conversion of shrimp shell wastes fermented by Serratia marcescens subsp. sakuensis TKU019. J. Chin. Chem. Soc–Taip. 2010, 57, 857–863. [Google Scholar] [CrossRef]
- Boucher, I.; Dupuy, A.; Vidal, P.; Neugebauer, W.A.; Brzezinski, R. Purification and characterization of a chitosanase from Streptomyces N174. Appl. Microbiol. Biotechnol. 1992, 38, 188–193. [Google Scholar] [CrossRef]
- Liang, T.W.; Lo, B.C.; Wang, S.L. Chitinolytic bacteria-assisted conversion of squid pen and its effect on dyes and adsorption. Mar. Drugs 2015, 13, 4576–4593. [Google Scholar] [CrossRef]
- Wang, S.L.; Wu, P.C.; Liang, T.W. Utilization of squid pen for the efficient production of chitosanase and antioxidants through prolonged autoclave treatment. Carbohydr. Res. 2009, 244, 979–984. [Google Scholar] [CrossRef]
- Wang, S.L.; Chen, S.J.; Liang, T.W.; Lin, Y.D. A novel nattokinase produced by Pseudomonas sp. TKU015 using shrimp shells as substrate. Process Biochem. 2009, 44, 70–76. [Google Scholar] [CrossRef]
- Wang, S.L.; Su, Y.C.; Nguyen, V.B.; Nguyen, A.D. Reclamation of shrimp heads for the production of α–glucosidase inhibitors by Staphylococcus sp. TKU043. Res. Chem. Intermed. 2018, 44, 4929–4937. [Google Scholar] [CrossRef]
- Wang, S.L.; Wu, Y.Y.; Liang, T.W. Purification and biochemical characterization of a nattokinase by conversion of shrimp shell with Bacillus subtilis TKU007. New Biotechnol. 2011, 28, 196–202. [Google Scholar] [CrossRef]
- Doan, C.T.; Tran, T.N.; Nguyen, M.T.; Nguyen, V.B.; Nguyen, A.D.; Wang, S.L. Anti-α-glucosidase activity by a protease from Bacillus licheniformis. Molecules 2019, 24, 691. [Google Scholar] [CrossRef]
- Nguyen, V.B.; Wang, S.L. New novel α–glucosidase inhibitors produced by microbial conversion. Process Biochem. 2018, 65, 228–232. [Google Scholar] [CrossRef]
- Nguyen, V.B.; Nguyen, T.H.; Doan, C.T.; Tran, T.N.; Nguyen, A.D.; Kuo, Y.H.; Wang, S.L. Production and bioactivity-guided isolation of antioxidants with α–glucosidase inhibitory and anti-NO properties from marine chitinous material. Molecules 2018, 23, 1124. [Google Scholar] [CrossRef]
- Nguyen, V.B.; Wang, S.L. Production of potent antidiabetic compounds from shrimp head powder via Paenibacillus conversion. Process Biochem. 2019, 76, 18–24. [Google Scholar] [CrossRef]
- Nguyen, V.B.; Nguyen, A.D.; Wang, S.L. Utilization of fishery processing byproduct squid pens for Paenibacillus sp. fermentation on producing potent α–glucosidase inhibitors. Mar. Drugs. 2017, 15, 274. [Google Scholar] [CrossRef]
- Nguyen, V.B.; Wang, S.L. Reclamation of marine chitinous materials for the production of α–glucosidase inhibitors via microbial conversion. Mar. Drugs. 2017, 15, 350. [Google Scholar] [CrossRef]
- Liang, T.W.; Tseng, S.C.; Wang, S.L. Production and characterization of antioxidant properties of exopolysaccharides from Paenibacillus mucilaginosus TKU032. Mar. Drugs. 2016, 12, 40. [Google Scholar] [CrossRef]
- Liang, T.W.; Wu, C.C.; Cheng, W.T.; Chen, Y.C.; Wang, C.L.; Wang, I.L.; Wang, S.L. Exopolysaccharides and antimicrobial biosurfactants produced by Paenibacillus macerans TKU029. Appl. Biochem. Biotech. 2014, 172, 933–950. [Google Scholar] [CrossRef]
- Doan, C.T.; Tran, T.N.; Nguyen, V.B.; Vo, T.P.K.; Nguyen, A.D.; Wang, S.L. Chitin extraction from shrimp waste by liquid fermentation using an alkaline protease–producing strain, Brevibacillus parabrevis. Int. J. Biol. Macromol. 2019, 131, 706–715. [Google Scholar] [CrossRef]
- Garcia–Gonzalez, E.; Poppinga, L.; Fünfhaus, A.; Hertlein, G.; Hedtke, K.; Jakubowska, A.; Genersch, E. Paenibacillus larvae chitin-degrading protein PlCBP49 is a key virulence factor in American foulbrood of honey bees. PLoS Pathogen. 2014, 10, e1004284. [Google Scholar] [CrossRef]
- Singh, A.K.; Chhatpar, H.S. Purification and characterization of chitinase from Paenibacillus sp. D1. Appl. Biochem. Biotechnol. 2011, 164, 77–88. [Google Scholar] [CrossRef]
- Kim, Y.H.; Park, S.K.; Hur, J.Y.; Kim, Y.C. Purification and characterization of a major extracellular chitinase from a biocontrol bacterium, Paenibacillus elgii HOA73. Plant Pathol. J. 2017, 33, 318–328. [Google Scholar] [CrossRef]
- Loni, P.P.; Patil, J.U.; Phugare, S.S.; Bajekal, S.S. Purification and characterization of alkaline chitinase from Paenibacillus pasadenensis NCIM 5434. J. Basic Microbiol. 2014, 54, 1080–1089. [Google Scholar] [CrossRef]
- Ueda, J.; Kurosawa, N. Characterization of an extracellular thermophilic chitinase from Paenibacillus thermoaerophilus strain TC22-2b isolated from compost. World J. Microbiol. Biotechnol. 2015, 31, 135–143. [Google Scholar] [CrossRef]
- Jung, W.J.; Kuk, J.K.; Kim, K.Y.; Kim, T.H.; Park, R.D. Purification and characterization of chitinase from Paenibacillus illinoisensis KJA-424. J. Microbiol. Biotechnol. 2005, 15, 274–280. [Google Scholar]
- Fu, X.; Yan, Q.; Wang, J.; Yang, S.; Jiang, Z. Purification and biochemical characterization of novel acidic chitinase from Paenibacillus barengoltzii. Int. J. Biol. Macromol. 2016, 91, 973–979. [Google Scholar] [CrossRef]
- Guo, X.; Xu, P.; Zong, M.; Lou, W. Purification and characterization of alkaline chitinase from Paenibacillus pasadenensis CS0611. Chin. J. Catalys. 2017, 38, 665–672. [Google Scholar] [CrossRef]
- Kimoto, H.; Kusaoke, H.; Yamamoto, I.; Fujii, Y.; Onodera, T.; Taketo, A. Biochemical and genetic properties of Paenibacillus glycosyl hydrolase having chitosanase activity and discoidin domain. J. Biol. Chem. 2002, 277, 14695–14702. [Google Scholar] [CrossRef]
- Itoh, T.; Sugimoto, I.; Hibi, T.; Suzuki, F.; Matsuo, K.; Fujii, Y.; Taketo, A.; Kimoto, H. Overexpression, purification, and characterization of Paenibacillus cell surface expressed chitinase ChiW with two catalytic domains. Biosci. Biotechnol. Biochem. 2014, 78, 624–634. [Google Scholar] [CrossRef]
- Meena, S.; Gothwal, R.K.; Saxena, J.; Nehra, S.; Mohan, M.K.; Ghosh, P. Effect of metal ions and chemical compounds on chitinase produced by a newly isolated thermotolerant Paenibacillus sp. BISR-047 and its shelf-life. Int. J. Curr. Microbiol. Appl. Sci. 2015, 4, 872–881. [Google Scholar]
- Nguyen, V.B.; Wang, S.L.; Nguyen, A.D.; Lin, Z.H.; Doan, C.T.; Tran, T.N.; Huang, H.T.; Kuo, Y.H. Bioactivity-Gùded purification of novel herbal antioxidant and anti-NO compounds from Euonymus laxiflorus Champ. Molecules 2019, 24, 120. [Google Scholar] [CrossRef]
- Naveed, M.; Phil, L.; Sohail, M.; Hasnat, M.; Baig, M.M.F.A.; Ihsan, A.U.; Shumzaid, M.; Kakar, M.U.; Khan, T.M.; Akabar, M.D.; Hussain, M.I.; Zhou, Q.G. Chitosan oligosaccharide (COS): An overview. Int. J. Biol. Macromol. 2019, 129, 827–843. [Google Scholar] [CrossRef]

| Bacterial Strain | Chitosanase Activity (U/mL) | Chitinase Activity (U/mL) | Exochitinase Activity (U/mL) |
|---|---|---|---|
| P. mucilaginosus TKU032 | 0.58 ± 0.10 | 0.37 ± 0.04 | - |
| P. macerans TKU029 | 0.59 ± 0.04 | 0.28 ± 0.08 | - |
| Paenibacillus sp. TKU037 | 0.05 ± 0.02 | 0.06 ± 0.05 | - |
| Paenibacillus sp. TKU042 | 0.12 ± 0.04 | 0.12 ± 0.05 | - |
| Bacillus licheniformis TKU004 | 0.01 ± 0.01 | 0.04 ± 0.01 | 10.21 ± 0.89 |
| B. subtillis TKU007 | 0.05 ± 0.02 | 0.11 ± 0.01 | - |
| Step | Total Protein (mg) | Total Activity (U) | Specific Activity (U/mg) | Recovery (%) | Purification (Fold) |
|---|---|---|---|---|---|
| Cultural supernatant | 1499.13 | 282.28 | 0.19 | 100.00 | 1.00 |
| (NH4)2SO4 precipitation | 126.97 | 89.44 | 0.70 | 31.69 | 3.74 |
| Macro-Prep High S | 15.98 | 67.92 | 4.25 | 24.06 | 22.57 |
| KW-802.5 | 5.13 | 30.89 | 6.03 | 10.94 | 32.01 |
| Metal Ion | Relative Activity (%) |
|---|---|
| Control | 100.00 ± 6.39 |
| Cu2+ | 74.37 ± 3.95 |
| Zn2+ | 76.78 ± 3.25 |
| Mg2+ | 84.39 ± 4.51 |
| Na+ | 91.71 ± 5.21 |
| Ba2+ | 62.14 ± 12.29 |
| Ca2+ | 77.07 ± 5.68 |
| Fe2+ | 65.13 ± 6.77 |
| EDTA | 84.39 ± 7.53 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Doan, C.T.; Tran, T.N.; Nguyen, V.B.; Nguyen, A.D.; Wang, S.-L. Production of a Thermostable Chitosanase from Shrimp Heads via Paenibacillus mucilaginosus TKU032 Conversion and its Application in the Preparation of Bioactive Chitosan Oligosaccharides. Mar. Drugs 2019, 17, 217. https://doi.org/10.3390/md17040217
Doan CT, Tran TN, Nguyen VB, Nguyen AD, Wang S-L. Production of a Thermostable Chitosanase from Shrimp Heads via Paenibacillus mucilaginosus TKU032 Conversion and its Application in the Preparation of Bioactive Chitosan Oligosaccharides. Marine Drugs. 2019; 17(4):217. https://doi.org/10.3390/md17040217
Chicago/Turabian StyleDoan, Chien Thang, Thi Ngoc Tran, Van Bon Nguyen, Anh Dzung Nguyen, and San-Lang Wang. 2019. "Production of a Thermostable Chitosanase from Shrimp Heads via Paenibacillus mucilaginosus TKU032 Conversion and its Application in the Preparation of Bioactive Chitosan Oligosaccharides" Marine Drugs 17, no. 4: 217. https://doi.org/10.3390/md17040217
APA StyleDoan, C. T., Tran, T. N., Nguyen, V. B., Nguyen, A. D., & Wang, S.-L. (2019). Production of a Thermostable Chitosanase from Shrimp Heads via Paenibacillus mucilaginosus TKU032 Conversion and its Application in the Preparation of Bioactive Chitosan Oligosaccharides. Marine Drugs, 17(4), 217. https://doi.org/10.3390/md17040217