Characterization of the Noncanonical Regulatory and Transporter Genes in Atratumycin Biosynthesis and Production in a Heterologous Host
Abstract
1. Introduction
2. Results
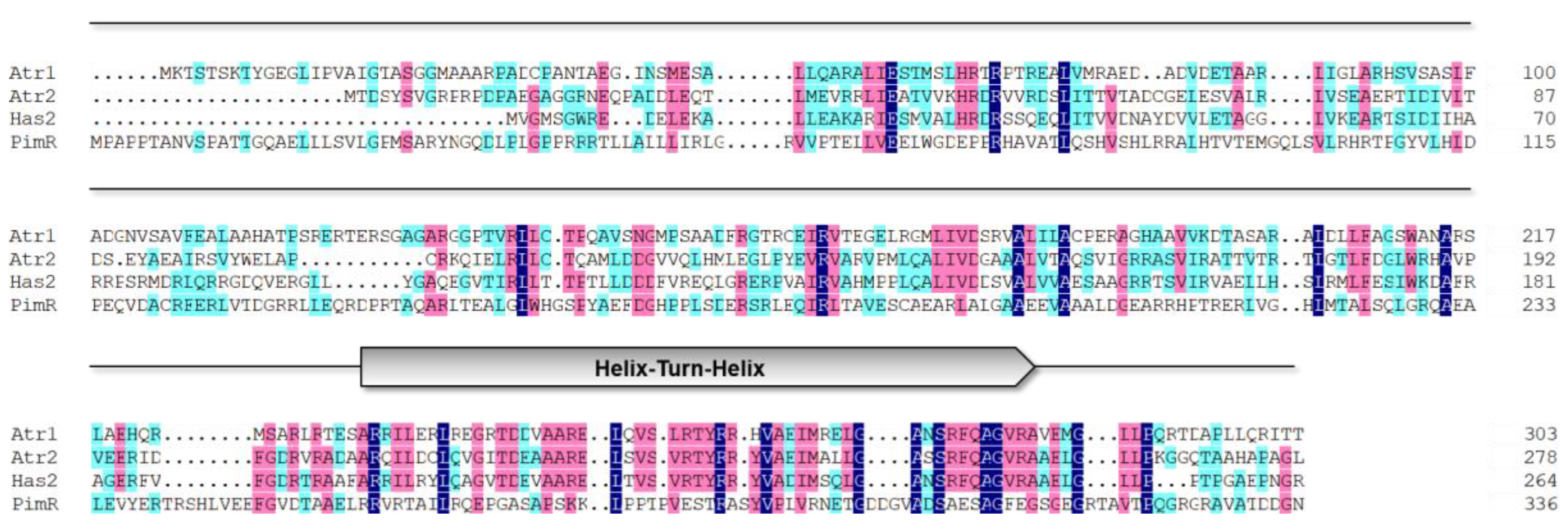
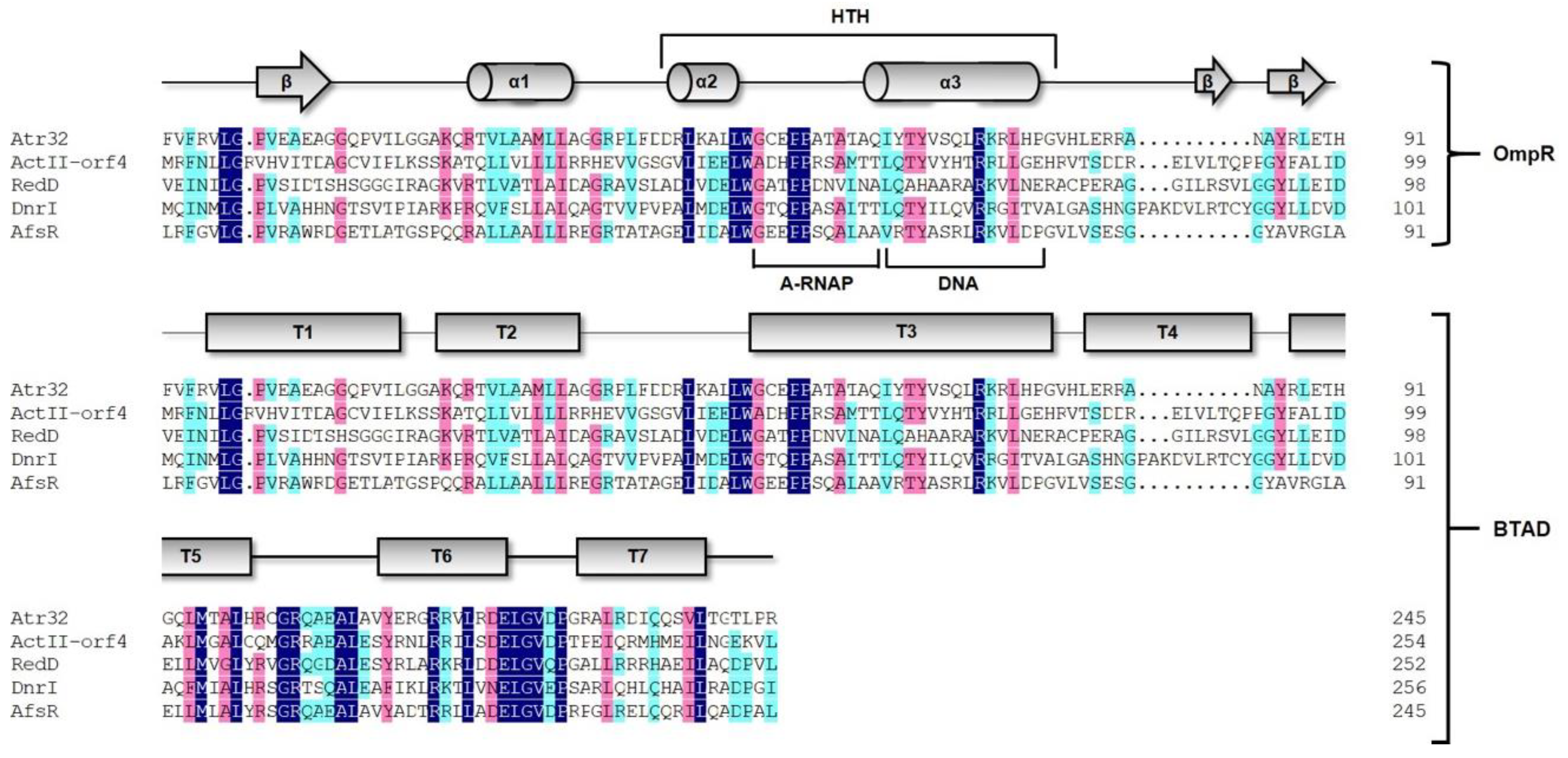
2.1. In Silico Analysis of Regulatory Genes in the Atr BGC
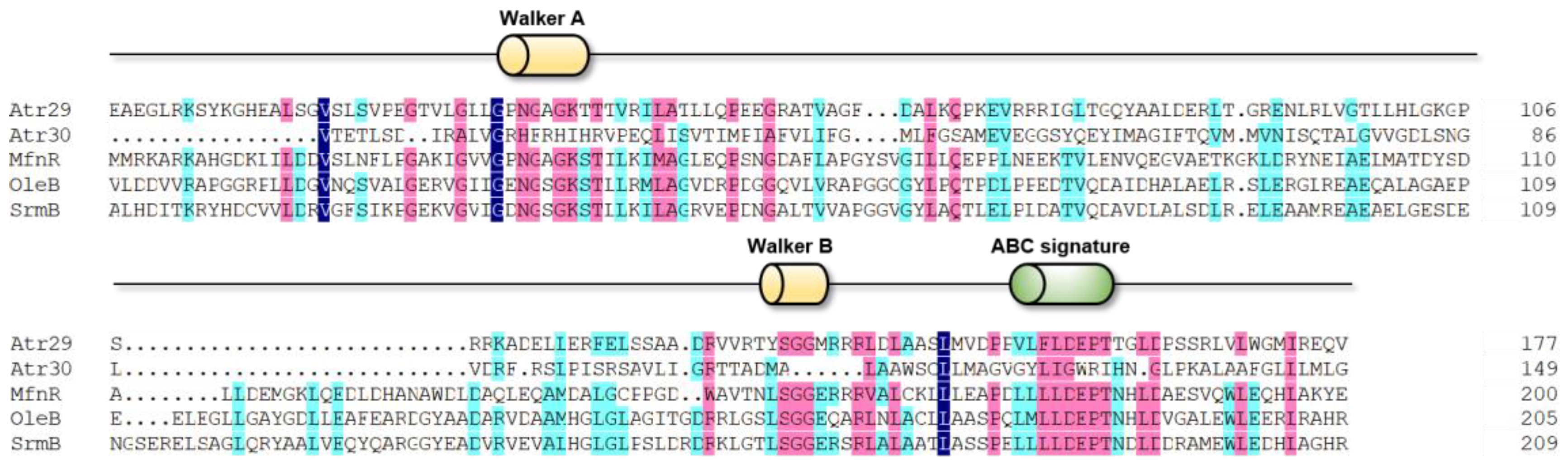
2.2. In Silico Analysis of Transporter Genes in the Atr BGC
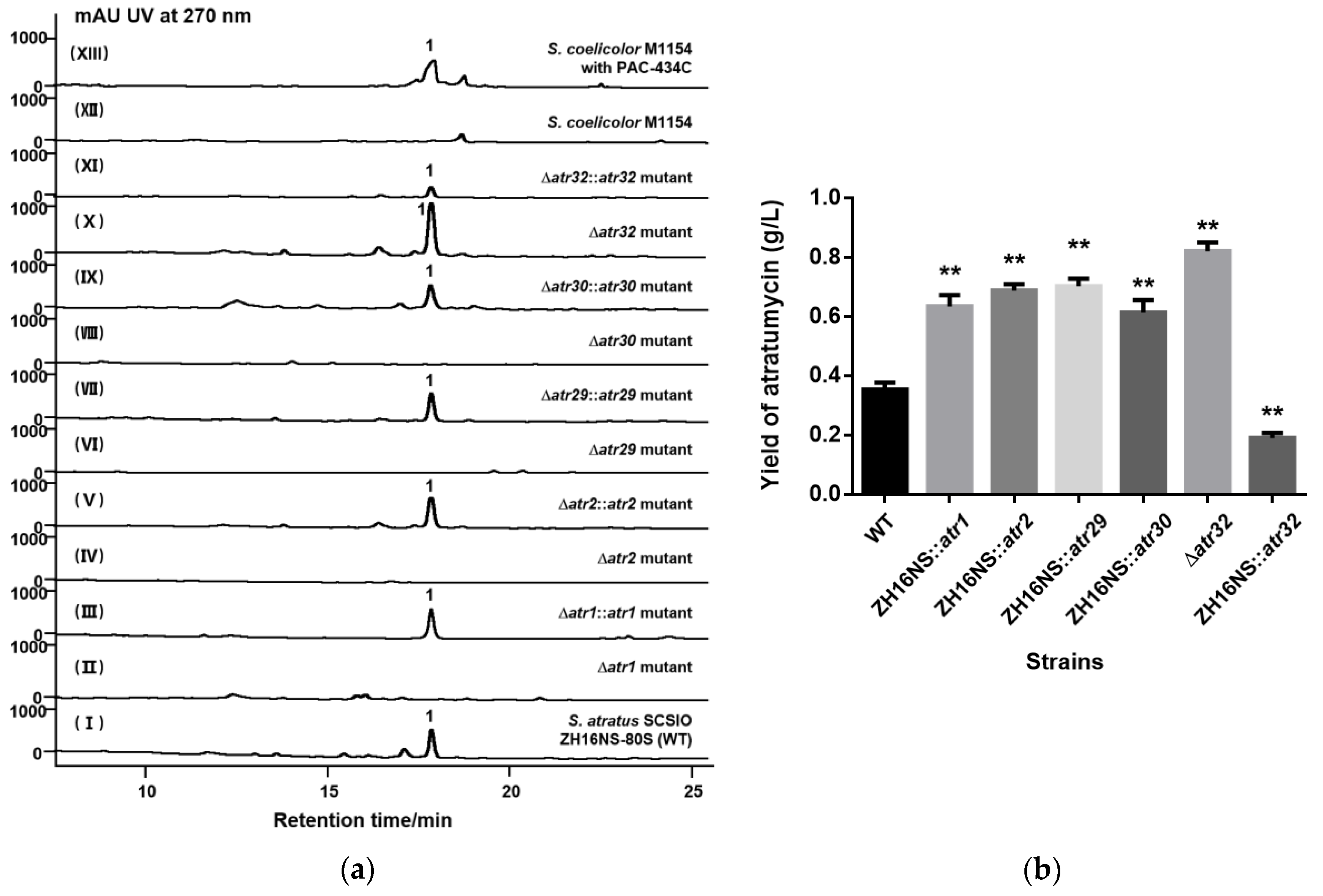
2.3. Gene Inactivation and Trans-Complementation of Regulatory and Transporter Genes
2.4. Construction of the High Producing Strain by Over-Expression of Regulatory and Transporter Genes
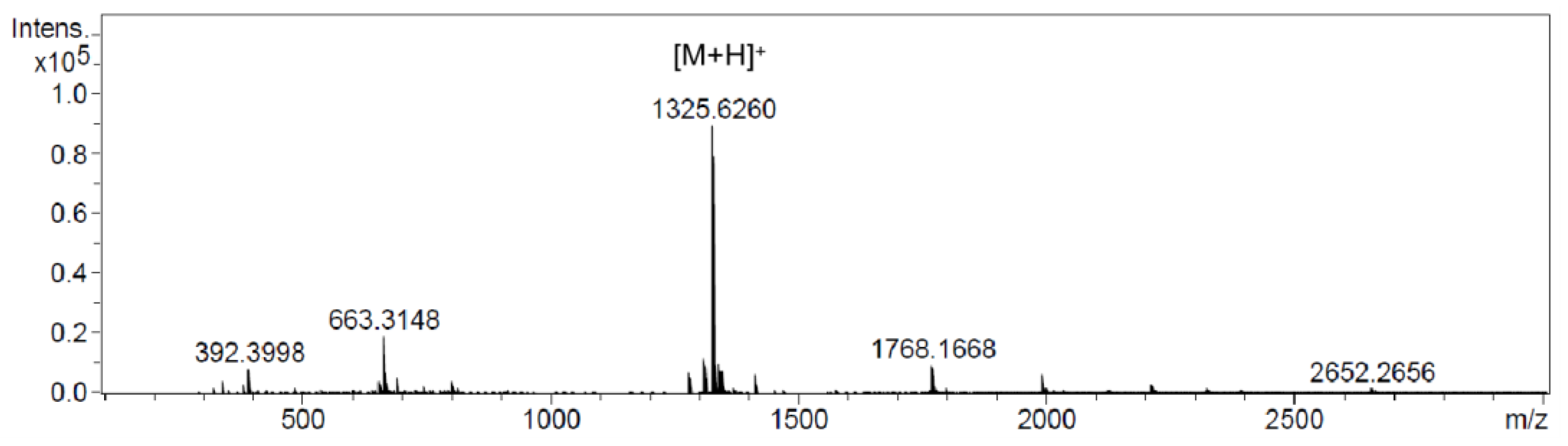
2.5. Heterologous Expression of Atratumycin
3. Discussion
4. Materials and Methods
4.1. General Materials and Experimental Procedures
4.2. Bacterial Strains, Plasmids and Culture Conditions
4.3. In Silico Analysis of Atratumycin Gene Cluster and Sequence Alignment of Proteins
4.4. Construction of Gene-Inactivated Mutants
4.5. Fermentation, Extraction and Quantitative Analysis
4.6. Complementation and Over-Expression in Mutants or the WT Producer Strain
4.7. Construction of Genomic PAC Library and Heterologous Expression of the Atr Gene Cluster
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Abdelmohsen, U.R.; Balasubramanian, S.; Oelschlaeger, T.A.; Grkovic, T.; Ngoc, B.P.; Quinn, R.J.; Hentschel, U. Potential of marine natural products against drug-resistant fungal, viral, and parasitic infections. Lancet Infect. Dis. 2017, 17, E30–E41. [Google Scholar] [CrossRef]
- Skropeta, D.; Wei, L. Recent advances in deep-sea natural products. Nat. Prod. Rep. 2014, 31, 999–1025. [Google Scholar] [CrossRef]
- Furin, J.; Cox, H.; Pai, M. Tuberculosis. Lancet 2019, 393, 1642–1656. [Google Scholar] [CrossRef]
- Ma, J.; Huang, H.; Xie, Y.; Liu, Z.; Zhao, J.; Zhang, C.; Jia, Y.; Zhang, Y.; Zhang, H.; Zhang, T.; et al. Biosynthesis of ilamycins featuring unusual building blocks and engineered production of enhanced anti-tuberculosis agents. Nat. Commun. 2017, 8, 391. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Yang, Z.; Zhang, C.; Liu, Z.; He, J.; Liu, Q.; Zhang, T.; Ju, J.; Ma, J. Genome mining of Streptomyces atratus SCSIO ZH16: Discovery of atratumycin and identification of its biosynthetic gene cluster. Org. Lett. 2019, 21, 1453–1457. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Chater, K.F.; Chandra, G.; Niu, G.; Tan, H. Molecular regulation of antibiotic biosynthesis in Streptomyces. Microbiol. Mol. Biol. Rev. 2013, 77, 112–143. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Xie, J. Role and regulation of bacterial LuxR-like regulators. J. Cell. Biochem. 2011, 112, 2694–2702. [Google Scholar] [CrossRef]
- Nasser, W.; Reverchon, S. New insights into the regulatory mechanisms of the LuxR family of quorum sensing regulators. Anal. Bioanal. Chem. 2007, 387, 381–390. [Google Scholar] [CrossRef]
- Yang, M.; Giel, J.L.; Cai, T.; Zhong, Z.; Zhu, J. The LuxR family quorum-sensing activator MrtR requires its cognate autoinducer for dimerization and activation but not for protein folding. J. Bacteriol. 2009, 191, 434–438. [Google Scholar] [CrossRef] [PubMed]
- Fuqua, C. The QscR quorum-sensing regulon of Pseudomonas aeruginosa: An orphan claims its identity. J. Bacteriol. 2006, 188, 3169–3171. [Google Scholar] [CrossRef]
- Patankar, A.V.; González, J.E. Orphan LuxR regulators of quorum sensing. FEMS Microbiol. Rev. 2009, 33, 739–756. [Google Scholar] [CrossRef] [PubMed]
- Covaceuszach, S.; Degrassi, G.; Venturi, V.; Lamba, D. Structural insights into a novel interkingdom signaling circuit by cartography of the ligand-binding sites of the homologous quorum sensing LuxR-Family. Int. J. Mol. Sci. 2013, 14, 20578–20596. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Hackert, E.; Stock, A.M. Structural relationships in the OmpR family of winged-helix transcription factors. J. Mol. Biol. 1997, 269, 301–312. [Google Scholar] [CrossRef]
- Martinez-Hackert, E.; Stock, A.M. The DNA-binding domain of OmpR: Crystal structures of a winged helix transcription factor. Structure 1997, 5, 109–124. [Google Scholar] [CrossRef]
- Takken, F.L.; Goverse, A. How to build a pathogen detector: Structural basis of NB-LRR function. Curr. Opin. Plant Biol. 2012, 15, 375–384. [Google Scholar] [CrossRef]
- Wietzorrek, A.; Bibb, M. A novel family of proteins that regulates antibiotic production in streptomycetes appears to contain an OmpR-like DNA-binding fold. Mol. Microbiol. 1997, 25, 1181–1184. [Google Scholar] [CrossRef]
- Romero-Rodriguez, A.; Robledo-Casados, I.; Sanchez, S. An overview on transcriptional regulators in Streptomyces. Biochim. Biophys. Acta 2015, 1849, 1017–1039. [Google Scholar] [CrossRef]
- Tanaka, A.; Takano, Y.; Ohnishi, Y.; Horinouchi, S. AfsR recruits RNA polymerase to the afsS promoter: A model for transcriptional activation by SARPs. J. Mol. Biol. 2007, 369, 322–333. [Google Scholar] [CrossRef] [PubMed]
- Takano, H.; Toriumi, N.; Hirata, M.; Amano, T.; Ohya, T.; Shimada, R.; Kusada, H.; Amano, S.-I.; Matsuda, K.-I.; Beppu, T.; et al. An ABC transporter involved in the control of streptomycin production in Streptomyces griseus. FEMS Microbiol. Lett. 2016, 363. [Google Scholar] [CrossRef]
- Yu, L.; Yan, X.; Wang, L.; Chu, J.; Zhuang, Y.; Zhang, S.; Guo, M. Molecular cloning and functional characterization of an ATP-binding cassette transporter OtrC from Streptomyces rimosus. BMC Biotechnol. 2012, 12, 52. [Google Scholar] [CrossRef] [PubMed]
- Rice, A.J.; Park, A.; Pinkett, H.W. Diversity in ABC transporters: Type I, II and III importers. Crit. Rev. Biochem. Mol. Biol. 2014, 49, 426–437. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.; Deng, Z.; Bai, L. Research advances in actinomycete ATP-binding cassette transporters—A review. Acta Microbiol. Sin. 2012, 52, 801–808. [Google Scholar]
- Hong, B.; Phornphisutthimas, S.; Tilley, E.; Baumberg, S.; McDowall, K.J. Streptomycin production by Streptomyces griseus can be modulated by a mechanism not associated with change in the adpA component of the A-factor cascade. Biotechnol. Lett. 2007, 29, 57–64. [Google Scholar] [CrossRef]
- Sosio, M.; Giusino, F.; Cappellano, C.; Bossi, E.; Puglia, A.M.; Donadio, S. Artificial chromosomes for antibiotic-producing actinomycetes. Nat. Biotechnol. 2000, 18, 343–345. [Google Scholar] [CrossRef]
- Arias, P.; Fernández-Moreno, M.A.; Malpartida, F. Characterization of the pathway-specific positive transcriptional regulator for Actinorhodin biosynthesis in Streptomyces coelicolor A3(2) as a DNA-Binding Protein. J. Bacteriol. 1999, 181, 6958–6968. [Google Scholar] [PubMed]
- Suzuki, T.; Mochizuki, S.; Yamamoto, S.; Arakawa, K.; Kinashi, H. Regulation of Lankamycin biosynthesis in Streptomyces rochei by two SARP genes, srrY and srrZ. Biosci. Biotechnol. Biochem. 2010, 74, 819–827. [Google Scholar] [CrossRef] [PubMed]
- Nováková, R.; Rehakova, A.; Kutas, P.; Fecková, L.; Kormanec, J. The role of two SARP family transcriptional regulators in regulation of the auricin gene cluster in Streptomyces aureofaciens CCM 3239. Microbiology 2011, 157, 1629–1639. [Google Scholar] [CrossRef]
- Tameling, W.I.; Vossen, J.H.; Albrecht, M.; Lengauer, T.; Berden, J.A.; Haring, M.A.; Cornelissen, B.J.; Takken, F.L. Mutations in the NB-ARC domain of I-2 that impair ATP hydrolysis cause autoactivation. Plant Physiol. 2006, 140, 1233–1245. [Google Scholar] [CrossRef]
- Van Der Biezen, E.A.; Jones, J.D. The NB-ARC domain: a novel signalling motif shared by plant resistance gene products and regulators of cell death in animals. Curr. Biol. 1998, 8, R226–R227. [Google Scholar] [CrossRef]
- Berdy, J. Bioactive microbial metabolites—A personal view. J. Antibiot. 2005, 58, 1–26. [Google Scholar] [CrossRef]
- Martín, J.F.; Casqueiro, J.; Liras, P. Secretion systems for secondary metabolites: How producer cells send out messages of intercellular communication. Curr. Opin. Microbiol. 2005, 8, 282–293. [Google Scholar] [CrossRef] [PubMed]
- Widdick, D.; Royer, S.F.; Wang, H.; Vior, N.M.; Gomez-Escribano, J.P.; Davis, B.G.; Bibb, M.J. Analysis of the Tunicamycin biosynthetic gene cluster of Streptomyces chartreusis reveals new insights into Tunicamycin production and immunity. Antimicrob. Agents Chemother. 2018, 62. [Google Scholar] [CrossRef] [PubMed]
- Mo, S.; Yoo, Y.J.; Ban, Y.H.; Lee, S.K.; Kim, E.; Suh, J.W.; Yoon, Y.J. Roles of fkbN in positive regulation and tcs7 in negative regulation of FK506 Biosynthesis in Streptomyces sp. strain KCTC 11604BP. Appl. Environ. Microbiol. 2012, 78, 2249–2255. [Google Scholar] [CrossRef] [PubMed]
- Yuan, T.; Yin, C.; Zhu, C.; Zhu, B.; Hu, Y. Improvement of antibiotic productivity by knock-out of dauW in Streptomyces coeruleobidus. Microbiol. Res. 2011, 166, 539–547. [Google Scholar] [CrossRef]
- Zhao, G.; Li, S.; Guo, Z.; Sun, M.; Lu, C. Overexpression of div8 increases the production and diversity of divergolides in Streptomyces sp. W112. RSC Adv. 2015, 5, 98209–98214. [Google Scholar] [CrossRef]
- Tu, J.; Li, S.; Chen, J.; Song, Y.; Fu, S.; Ju, J.; Li, Q. Characterization and heterologous expression of the neoabyssomicin/abyssomicin biosynthetic gene cluster from Streptomyces koyangensis SCSIO 5802. Microb. Cell Fact. 2018, 17, 28. [Google Scholar] [CrossRef] [PubMed]
- Flett, F.; Mersinias, V.; Smith, C.P. High efficiency intergeneric conjugal transfer of plasmid DNA from Escherichia coli to methyl DNA-restricting streptomycetes. FEMS Microbiol. Lett. 1997, 155, 223–229. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Z.; Wei, X.; He, J.; Sun, C.; Ju, J.; Ma, J. Characterization of the Noncanonical Regulatory and Transporter Genes in Atratumycin Biosynthesis and Production in a Heterologous Host. Mar. Drugs 2019, 17, 560. https://doi.org/10.3390/md17100560
Yang Z, Wei X, He J, Sun C, Ju J, Ma J. Characterization of the Noncanonical Regulatory and Transporter Genes in Atratumycin Biosynthesis and Production in a Heterologous Host. Marine Drugs. 2019; 17(10):560. https://doi.org/10.3390/md17100560
Chicago/Turabian StyleYang, Zhijie, Xin Wei, Jianqiao He, Changli Sun, Jianhua Ju, and Junying Ma. 2019. "Characterization of the Noncanonical Regulatory and Transporter Genes in Atratumycin Biosynthesis and Production in a Heterologous Host" Marine Drugs 17, no. 10: 560. https://doi.org/10.3390/md17100560
APA StyleYang, Z., Wei, X., He, J., Sun, C., Ju, J., & Ma, J. (2019). Characterization of the Noncanonical Regulatory and Transporter Genes in Atratumycin Biosynthesis and Production in a Heterologous Host. Marine Drugs, 17(10), 560. https://doi.org/10.3390/md17100560





