Abstract
Seven new secondary metabolites classified as two perylenequinone derivatives (1 and 2), an altenusin derivative (3), two phthalide racemates (4 and 5), and two phenol derivatives (6 and 7), along with twenty-one known compounds (8–28) were isolated from cultures of the sponge-derived fungus, Alternaria sp. SCSIO41014. The structures and absolute configurations of these new compounds (1–7) were determined by spectroscopic analysis, X-ray single crystal diffraction, chiral-phase HPLC separation, and comparison of ECD spectra to calculations. Altertoxin VII (1) is the first example possessing a novel 4,8-dihydroxy-substituted perylenequinone derivative, while the phenolic hydroxy groups have commonly always substituted at C-4 and C-9. Compound 1 exhibited cytotoxic activities against human erythroleukemia (K562), human gastric carcinoma cells (SGC-7901), and hepatocellular carcinoma cells (BEL-7402) with IC50 values of 26.58 ± 0.80, 8.75 ± 0.13, and 13.11 ± 0.95 μg/mL, respectively. Compound 11 showed selectively cytotoxic activity against K562, with an IC50 value of 19.67 ± 0.19 μg/mL. Compound 25 displayed moderate inhibitory activity against Staphylococcus aureus with an MIC value of 31.25 μg/mL.
1. Introduction
Perylenequinone derivatives are secondary metabolites characterized by a conjugated aromatic pentacyclic dione, which are mainly derived from fungi [1,2]. Hypocrellins, which are the typical perylenequinone derivatives isolated from fungi Hypocrella bambusae and Shiraia bambusicola, have been studied for their light-induced antitumor and antiviral activities [3]. Furthermore, many perylenequinone derivatives isolated from Alternaria sp. showed phytotoxicity, as well as antimicrobial and anticancer activities [4,5,6,7]. Sponge-derived fungi are one of the richest sources of many structurally unique and biologically active secondary metabolites among marine sources [8]. As part of our ongoing research for bioactive natural products from sponge-derived fungi [9,10,11,12,13], the fungus Alternaria sp. SCSIO41014 was studied. Seven new (1–7) and twenty-one known (8–28) compounds (Figure 1) were isolated from the culture extract of the fungus Alternaria sp. SCSIO41014. Altertoxin VII (1) is the first example possessing a novel 4,8-dihydroxy-substituted perylenequinone derivative, while the phenolic hydroxy groups have always commonly substituted at C-4 and C-9. Herein, we describe the structure elucidation and bioactivity evaluation of new compounds 1–7.
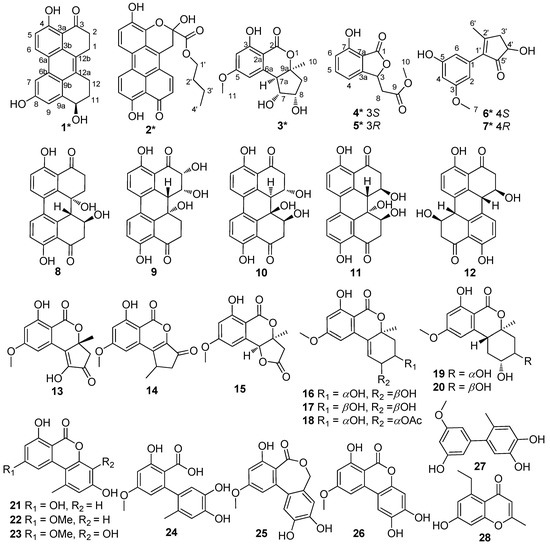
Figure 1.
Chemical structures of compounds 1–28.
2. Results and Discussion
2.1. Structural Elucidation
Compound 1 was obtained as a dark red powder. Its molecular formula was established as C20H16O4 by 13C NMR data and the high resolution electrospray ionization mass spectroscopy (HRESIMS) [M + Na]+ peak at m/z 343.0939 (calculated for C20H15O4Na, 343.0941), indicating thirteen degrees of unsaturation. Its UV spectrum showed maxima at 323 and 368 nm, which suggested that 1 featured a polycyclic aromatic hydrocarbons system. Its 1H NMR data (Table 1) showed two ortho aromatic protons (δH 8.79, d, J = 9.5 Hz, H-6 and 7.16, d, J = 9.0 Hz, H-5), two meta aromatic protons (δH 7.82, d, J = 2.0 Hz, H-7 and 7.30, d, J = 1.5 Hz, H-9), an oxygenated methine (δH 4.82, dd, J = 9.0, 3.0 Hz, H-10), and three hydroxy protons (δH 13.27, brs, OH-4; 9.87, brs, OH-8; 5.48, brs, OH-10). The striking downfield shift of OH-4 is a typical feature of a strong hydrogen bond, which is in accordance with the downfield shift of C-3 (δC 204.0) observed in the 13C NMR spectrum. The 13C NMR (DEPT) (Table 1) data also exhibited twenty carbon signals, including four sp3 methylenes, one oxygenated sp3 methine (δC 67.4, C-10), four sp2 methines (δC 133.1, C-6; 116.2, C-5; 114.4, C-9; 104.4, C-7), ten sp2 non-protonated carbons, and one carbonyl. Besides the presence of two benzene rings, a double bond, and a carbonyl, three degrees of unsaturation were left, which indicated that there were another three rings in the structure of 1. Half of its NMR data were identical to those of 4,9-dihydroxy-1,2,11,12-tetrahydroperylene-3,10-quinone [14], while compound 1 was not a symmetric structure, and the other half of the data had two obvious differences. One carbonyl and two ortho aromatic protons in 4,9-dihydroxy-1,2,11,12-tetrahydroperylene-3,10-quinone were replaced by an oxygenated methine and two meta aromatic protons, respectively. The differences were testified by the cross-peaks of H2-1 (δH 3.25)/H2-2 (δH 2.92), of H-5/H-6, and of OH-10/H-10/H2-11 (δH 2.18 and 1.88)/H2-12 (δH 3.20 and 3.00) in the 1H-1H correlation spectroscopy (COSY) spectrum (Figure 2), and the correlations from: H-7 and H-9 to C-8; from H-10 and H2-11 to C-9a; from H-7, H-9, H-10 and H2-12 to C-9b; from H-9 to C-7 and C-10; and from H2-11 and H2-12 to C-12a in the heteronuclear multiple bond correlation (HMBC) spectrum (Figure 2). Furthermore, it is allowed for the linkage of ring A (Figure 2) by the correlations from H2-1 to C-12a; from H-6 to C-6b; from H-7 to C-6a; and from H2-12 to C-12b in the HMBC spectrum. The attachment of the three hydroxy groups was further confirmed by the correlations from: OH-4 to C-3a, C-4 and C-5; from OH-8 to C-7, C-8 and C-9; and from OH-10 to C-9a (Figure 2). Therefore, the planar structure of 1 was established as 4,8,10-trihydroxy-1,2,11,12-tetrahydroperylene-3-quinone. Its nuclear Overhauser effect spectroscopy (NOESY) spectrum displayed cross-peaks of H2-1/H2-12 and of H-6/H-7, which also support the conjectural structure above. The absolute configuration of 1 was established based on comparison of its experimental electronic circular dichroism (ECD) curve with the calculated ECD curve of the 10R and the 10S model at the B3LYP/6-31G(d,p) level in Gaussian 03, and the former was in accordance with the experimental one (Figure 3). Thus, the absolute structure of 1 was defined as (10R)-4,8,10-trihydroxy-1,2,11,12-tetrahydroperylene-3-quinone, and named altertoxin VII.

Table 1.
1H NMR and 13C NMR data for compounds 1 and 2 in DMSO-d6 (500, 125 MHz).
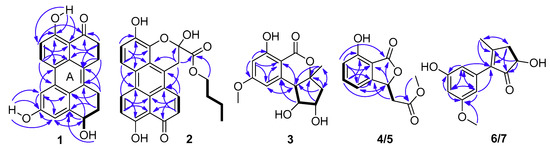
Figure 2.
COSY “H—H” and key HMBC “H→C” correlations of compounds 1–7.
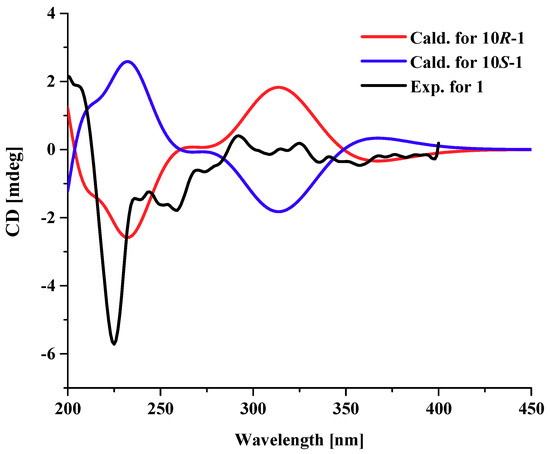
Figure 3.
Comparison between calculated and experimental electronic circular dichroism (ECD) spectra of compound 1.
Compound 2 was obtained as a dark red powder. Its molecular formula was assigned as C24H20O7 based on its [M + Na]+ ion at m/z 443.1110 and [M + H]+ ion at m/z 421.1284 in the HRESIMS spectrum, indicating fifteen degrees of unsaturation. Analysis of its 1H NMR and 13C NMR data revealed that the structural features of 2 were similar to those of xanalteric acid II [5], except for the presence of an oxygenated n-butyl group (δC/H 65.1/4.09, CH2-1′; 30.0/1.46, CH2-2′; 18.3/1.14, CH2-3′; 13.4/0.71, CH3-4′), which was confirmed by its COSY cross-peaks of H2-1′/H2-2′/H2-3′/H3-4′. The location of the oxygenated n-butyl group was ascertained by the HMBC correlation from H2-1′ to C-13. Based on the HMBC correlations (Figure 2), the planar structure of 2 was determined as butyl 2,4,9-trihydroxy-10-oxo-2,10-dihydro-1H-phenaleno[1,2,3-de] chromene-2-carboxylate and named butyl xanalterate. Due to the existence of the cyclic hemi-ketal, compound 2 was not stable under the protic solvent condition, meaning that the absolute configuration was uncertain.
Compound 3 possessed a molecular formula of C14H16O6 on the basis of its NMR data and the HRESIMS ion peak at m/z 303.0844 [M + Na]+, indicating seven indices of hydrogen deficiency. The 1H NMR spectrum (Table 2) exhibited two aromatic protons (δH 6.52, brs, H-6; 6.43, brs, H-4); three methines (δH 4.20, ddd, J = 7.0, 5.5, 2.5 Hz, H-8; 3.90, dd, J = 10.5, 5.0 Hz, H-7; 3.07, d, J = 10.0 Hz, H-7a); one methoxy group (δH 3.88, s, H3-11); and a methyl singlet (δH 1.47, s, H3-10). The 13C NMR and HSQC spectra indicated the presence of 14 carbons, including two methyls (δC 56.2, C-11; 25.7, C-10), one sp3 methylene, three sp3 methines (two oxygenated at δC 80.1, C-7; 71.2, C-8), two sp2 methines (δC 101.0, C-4; 108.9, C-6), four sp2 non-protonated carbon, and an ester carbonyl (δC 170.0, C-2). Because one benzene ring and one carbonyl accounted for five degrees of unsaturation, double ring fragment was required for the structure of 3. Its NMR data were similar to those of dihydroaltenuenes A (19) [15], the obvious differences being the disappearance of a methylene signal (δC 28.1) and the downfield shift of the oxygenated methine in 3, which indicated that a fragment of the six-membered ring in 19 was replaced by a five-membered ring in 3. The deduction was confirmed by following COSY correlations of H-7a/H-7/H-8/H2-9, as well as HMBC correlations, from H-7 and H-7a to C-6a; from H-6, H2-9 and H3-10 to C-7a; and from H-8, H2-9 and H3-10 to C-9a (Figure 2). The NOESY spectrum in DMSO-d6 (Figure 4) exhibited the cross-peaks of H-7a to H-9α, H3-10, OH-7, and OH-8, and also of H-9β to H-7, which suggested that CH3-10, H-7a, OH-7, and OH-8 were on the same side. The Cu Kα radiation for the X-ray diffraction experiment with the refined Flack parameter of −0.02(7) allowed the assignment of the absolute configuration of all the stereogenic centers in 3 as 7R, 7aR, 8S, and 9aS. Thus, the absolute structure of 3 was unambiguously elucidated and named nordihydroaltenuenes A.

Table 2.
1H NMR and 13C NMR data for compounds 3–7 in CD3OD (500, 125 MHz).
Figure 4.
Key NOESY correlations and ORTEP drawing of compound 3.
Compounds 4 and 5 were initially believed to be a single compound, which was obtained as a yellow oil. The molecular formula was established as C11H15O5 based on the HRESIMS and 13C NMR data. A literature survey suggested that the 1H NMR and 13C NMR data (Table 2) closely resembled those of isoochracinic acid [16], except for the presence of a methoxy group (δC/H 52.5/3.69, C-10) and an upfield shift of 1.3 ppm of the carbonyl (δC 171.6, C-9), which indicated the carboxyl in the isoochracinic acid was methylated. The deduction was confirmed by the HMBC correlation of H3-10 to C-9. The planar structure of 4/5 was further established by the COSY and HMBC correlations (Figure 2). The optical rotation was small ([α +1.6) and the value of the ECD spectrum close to zero, suggesting that compounds 4 and 5 belonged to a racemate. Using a chiral-phase column (Daicel Chiraloak IC-3, 250 × 4.6 mm, 5 μm), the racemate was resolved to two enantiomers, 4 and 5, whose scale was nearly 1:1 (Figure S30). The optical rotation of 4 ([α −21, c 0.1, MeOH) and 5 ([α +26, c 0.1, MeOH), as well as the ECD spectrum (Figure 5), were opposite to each other. In general, the 3R-phthalides displayed a positive optical rotation, while 3S-phthalides had a negative one [17,18,19,20]. Therefore, the absolute configuration of compound 4 was proposed as 3S and named (S)-isoochracinate A1, while compound 5 was proposed as 3R, and named (R)-isoochracinate A2.
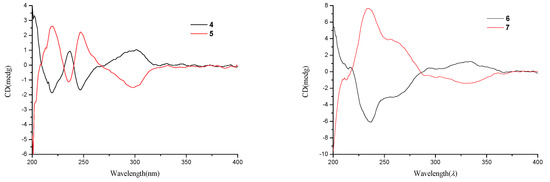
Figure 5.
Experimental ECD spectra of compounds 4–7.
Compounds 6 and 7, obtained as a yellow powder, were also believed to be a single compound at first. The HRESIMS spectrum supported a molecular formula of C13H14O4, requiring 7 degrees of unsaturation. The 1H NMR and 13C NMR data (Table 2) of 6/7 were almost identical to those of 4′-(S)-(3,5-dihydroxyphenyl)-4′-hydroxy-6′-methylcyclopent-1′-en-5′-one isolated from Penicillium sp. HN29-3B1 [21], indicating that 6/7 were a structural analog, except for the presence of a methoxy group (δC/H 55.7, C-7). The deduction was confirmed by the HMBC correlation (Table 2) from H3-7 to C-3. Analyzed by a chiral-phase column (Phenomenex Lux Cellulose-2, 250 × 4.6 mm, 5 μm), compounds 6/7 were found to be stereoisomeric mixture, whose mass ratio was about 1:3.5 (Figure S39). The single compound 6 ([α +3.1, c 0.1, MeOH) and 7 ([α −7.2, c 0.1, MeOH) were isolated, and they had opposite ECD spectra (Figure 5). The optical rotation of 6 was consistent with that of 4′-(S)-(3,5-dihydroxyphenyl)-4′-hydroxy-6′-methylcyclopent-1′-en-5′-one [21], indicating that they had the same S configuration at C-4′, opposite to that of 7. Thus, the absolute structure of 6 was determined as 4′-(S)-(3-Methoxy-5-hydroxyphenyl)-4′-hydroxy-6′-methylcyclopent-1′-en-5′-one and named (S)-alternariphent A1, while the absolute structure of 7 was determined as 4′-(R)-(3-Methoxy-5-hydroxyphenyl)-4′-hydroxy-6′-methylcyclopent-1′-en-5′-one and named (R)-alternariphent A2.
By comparing NMR data with the values in literature, the structures of the twenty-one known compounds were identified as altertoxin I (8) [22], 7-epi-8-hydroxyaltertoxin (9) [6], stemphytriol (10) [23], 6-epi-stemphytriol (11) [6], stemphyperylenol (12) [23,24], (R)-1,6-dihydroxy-8-methoxy-3a-methyl-3,3a-dihydrocyclopenta[c]iso- chromene-2,5-dione (13) [25], 1-deoxyrubralactone (14) [25], 6-hydroxy-8-methoxy-3a-methyl-3a,9b-dihydro-3H-furo[3,2-c]isochromene-2,5-dione (15) [26], altenuene (16) [27], 4′-epialtenuene (17) [28], (−)-(2R,3R,4aR)-altenuene-3-acetoxy ester (18) [29], dihydroaltenuenes A (19) [15], 3-epi-dihydroaltenuene A (20) [30], alternariol (21) [31], alternariol monomethyl ether (22) [32], 3′-hydroxyalternariol-5-O-methyl ether (23) [28], altenusin (24) [33], alterlactone (25) [28], altenuisol (26) [34], 5′-methoxy-6-methyl-biphenyl-3,4,3′-triol (27) [26], and 2,5-dimethyl-7-hydroxychromone (28) [35], respectively.
2.2. Biological Activity
The cytotoxic activities of compounds 1, 2, and 8–12 against human erythroleukemia (K562), human gastric carcinoma cells (SGC-7901), and hepatocellular carcinoma cells (BEL-7402) were evaluated using the CCK-8 method, and paclitaxel was used as a positive control with IC50 values of 0.18 ± 0.20, 0.89 ± 0.15, and 0.54 ± 0.20 μg/mL, respectively. Among the tested compounds, compound 1 exhibited cytotoxic activity against the K562, SGC-7901, and BEL-7402 cell lines with IC50 values of 26.58 ± 0.80, 8.75 ± 0.13, and 13.11±0.95 μg/mL, respectively. Compound 11 showed selectively cytotoxic activity against K562 with an IC50 value of 19.67 ± 0.19 μg/mL. All compounds were tested for their antibacterial activities against Staphylococcus aureus. Compounds 10 and 25 with 50 μg/disc displayed an inhibition zone with a diameter of about 21 and 15 mm, respectively (Figure S1). Furthermore, their minimum inhibitory concentrations (MIC) were tested, and the MIC value of compound 25 was 31.25 μg/mL, while compound 10 showed more than 500 μg/mL—perhaps due to its poor solubility. Ampicillin was used as a positive control with an MIC value of 6.25 μg/mL.
3. Materials and Methods
3.1. General Experimental Procedures
HRESIMS data were recorded on a maXis Q-TOF mass spectrometer in a positive ion mode (Bruker, Fällanden, Switzerland). 1D and 2D NMR spectra were measured on an AV 500 MHz NMR spectrometer (Bruker, Fällanden, Switzerland) with TMS as an internal standard. Chemical shifts were given as δ values, with J values reported in Hz. Optical rotations were measured using a MCP-500 polarimeter (Anton, Austria). UV spectra were recorded on a UV-2600 UV-Vis spectrophotometer (Shimadzu, Japan). X-ray diffraction intensity data were collected on an XtalLAB PRO single-crystal diffractometer using Cu Kα radiation (Rigaku, Japan). ECD spectrum was measured with a Chirascan circular dichroism spectrometer (Applied Photophysics, Surrey, UK). HPLC was performed on a Hitachi Primaide with the YMC ODS SERIES column (YMC-Pack ODS-A, YMC Co. Ltd., Kyoto, Japan, 250 × 10 mm I.D., S-5 μm, 12 nm) and chiral-phase column (Phenomenex Lux Cellulose-2 column, 4.6 mm × 25 mm and Daicel Chiraloak IC-3 column, 4.6 mm × 25 mm). Column chromatography (CC) was carried out on silica gel (200–300 mesh, Jiangyou Silica Gel Development Co., Yantai, China), Sephadex LH-20 (40–70 μm, Amersham Pharmacia Biotech AB, Uppsala, Sweden), and YMC Gel ODS-A (12 nm, S-50 μm YMC Co. Ltd., Kyoto, Japan). Spots were detected on TLC under UV light or by heating after spraying with the mixed solvent of saturated vanillin and 5% H2SO4 in EtOH. The TLC plates with silica gel GF254 (0.4–0.5 mm, Qingdao Marine Chemical Factory, Qingdao, China) were used for analysis and for the preparatives.
3.2. Fungal Material
The fungal strain SCSIO41014 was obtained from a Callyspongia sp. sponge, which was collected from the sea area near Xuwen County, Guangdong Province, China. The producing strain was stored on MB agar (malt extract 15 g, agar 16 g, sea salt 10 g, water 1 L, pH 7.4–7.8) slants at 4 °C and deposited at the Key Laboratory of Tropical Marine Bio-resources and Ecology, Chinese Academy of Science. The ITS1-5.8S-ITS2 sequence region (508 base pairs, GenBank accession No. MH444654) of strain SCSIO41014 was amplified by PCR, and DNA sequencing showed it shared a significant homology to several species of Setosphaeria. The 508 base pairs of the ITS sequence had a 99% sequence identity to that of the Alternaria alternata strain SCAU091 (GenBank accession No. MF061753.1). It was thus designated as a member of Alternaria sp. and named Alternaria sp. SCSIO41014.
3.3. Fermentation and Extraction
The strain SCSIO41014 was cultured in 100 mL flasks (×59) each containing 10 mL seed medium (malt extract: 15 g, sea salt: 2.5 g, distilled water: 1 L, pH 7.4–7.8) at 27 °C on a rotary shaker (172 rpm) for 48 h. The mass fermentation of this fungus was carried out at 25 °C for 32 days using a rice medium (rice: 200 g/flask, sea salt: 2.5 g/flask, tap water: 200 mL/flask) in the 1 L flasks (×59). The flasks were incubated statically at 25 °C under the normal day and night cycle. After 32 days, cultures were soaked in acetone (400 mL/flask), mashed into small pieces, and vibrated with ultrasound for 20 min. Then the acetone solution was evaporated under reduced pressure to afford an aqueous solution, which was extracted with ethyl acetate (EtOAc) three times. Concurrently, the rice residue was extracted with EtOAc in order to make another EtOAc solution. Both of the EtOAc solutions were combined and concentrated under reduced pressure to produce a crude extract. The extract was then suspended in MeOH and partitioned with equal volumes of petroleum ether (PE). Finally, the MeOH solution was concentrated under reduced pressure to obtain a reddish-brown extract (78.0 g).
3.4. Isolation and Purification
The reddish-brown extract was subjected to silica gel CC, which was eluted with a CH2Cl2 and MeOH mixed solvent in a step gradient (100:0–3:1, v/v) and separated into seven fractions (Fr-1–Fr-7). Fr-1 (3.4 g) was subjected to reversed-phase C-18 MPLC with MeOH/H2O (35:65–100:0, v/v) to get four fractions (Fr-1-1–Fr-1-4). Fr-1-1 was directly separated by semi-preparative HPLC (75% MeOH/H2O, 1.6 mL/min) to produce 22 (5.4 mg, tR = 25 min). Fr-1-2 was purified by semi-preparative HPLC (42% CH3CN/H2O, 2 mL/min) to yield 13 (26.0 mg, tR = 22.0 min) and 14 (5.6 mg, tR = 34.0 min). Fr-1-3 was separated by semi-preparative HPLC (30% CH3CN/H2O, 2 mL/min) to yield 15 (17.8 mg, tR = 43.0 min). Fr-1-4 was purified by semi-preparative HPLC (28% CH3CN/H2O, 2 mL/min) to afford raceme 4/5 (97.3 mg, tR = 18.0 min), part of which was further separated by HPLC with a chiral-phase column (Daicel Chiraloak IC-3 column, 4.6 mm × 25 mm, eluent n-hexane/iso-propanol, 65:35 v/v, 1 mL/min) to obtain 4 (3.3 mg, tR = 15.6 min) and 5 (2.8 mg, tR = 17.0 min). Fr-3 (15.8 g) was subjected to silica gel CC eluted with a PE and acetone mixed solvent in a step gradient (10:1–0:1, v/v) to gain five fractions (Fr-3-1–Fr-3-5). Fr-3-3 (0.94 g) was subjected to a Sephadex LH-20 column, eluted with MeOH and further purified by semi-preparative HPLC, to produce 18 (7.6 mg, 40% CH3CN/H2O, 2 mL/min, tR = 30.0 min), 23 (8.0 mg, 45% CH3CN/H2O, 2 mL/min, tR = 22.0 min), and 28 (22.5 mg, 45% CH3CN/H2O, 2 mL/min, tR = 24.0 min). Fr-3-4 (7.3 g) was subjected to reversed-phase C-18 MPLC with MeOH/H2O (10:90–100:0, v/v) to get three fractions (Fr-3-4-1–Fr-3-4-3). Fr-3-4-1 was subjected to a Sephadex LH-20 column eluted with MeOH to gain four parts. One part was purified by a preparative thin-layer chromatography, using CH2Cl2/MeOH (10:1) as a developing solvent to afford 16 (42.6 mg, Rf = 0.6) and 19 (11.3 mg, Rf = 0.7). Other parts were separated by semi-preparative HPLC to yield 3 (3.5 mg, 60% MeOH/H2O, 2 mL/min, tR = 13.5 min), 9 (4.8 mg, 60% MeOH/H2O, 2 mL/min, tR = 15.8 min), 10 (10.3 mg, 60% MeOH/H2O, 2 mL/min, tR = 19.0 min), and 25 (104.0 mg, 34% CH3CN/H2O, 2 mL/min, tR = 20.0 min). Fr-3-4-2 was subjected to a Sephadex LH-20 column, eluted with MeOH and purified by semi-preparative HPLC, to yield 11 (5.2 mg, 56% MeOH/H2O, 2 mL/min, tR = 26.0 min), 12 (13.1 mg, 35% CH3CN/H2O, 2 mL/min, tR = 21.4 min), and 21 (20.6 mg, 40% CH3CN/H2O, 2 mL/min, tR = 21.0 min). Fr-3-4-3 was subjected to silica gel CC with a PE and EtOAc mixed solvent in a step gradient (5:1–1:1, v/v), and then purified by semi-preparative HPLC to yield 26 (7.0 mg, 30% CH3CN/H2O, 2 mL/min, tR = 26.4 min). Fr-3-5 (1.1 g) was subjected to a Sephadex LH-20 column eluted with MeOH, and then purified by semi-preparative HPLC to yield 2 (6.7 mg, 55% CH3CN/H2O, 2 mL/min, tR = 32.0 min) and 8 (3.9 mg, 48% CH3CN/H2O, 2 mL/min, tR = 18.0 min). Fr-4 (8.4 g) was subjected to silica gel CC eluted with a PE and acetone mixed solvent in a step gradient (10:1–0:1, v/v) to gain three fractions (Fr-4-1–Fr-4-3). Fr-4-2 (5.1 g) was subjected to a Sephadex LH-20 column eluted with MeOH, then subjected to reversed-phase C-18 MPLC with MeOH/H2O (10:90–100:0, v/v), and further purified by semi-preparative HPLC, to obtain 1 (17.4 mg, 38% CH3CN/H2O, 2 mL/min, tR = 28.2 min), 17 (34.7 mg, 56% MeOH/H2O, 2 mL/min, tR = 21.5 min), 20 (10.6 mg, 28% CH3CN/H2O, 2 mL/min, tR = 13.0 min), and the stereoisomeric mixture 6/7 (13.2 mg, 15% CH3CN/H2O, 2 mL/min, tR = 23.6 min). Part of the mixture 6/7 was separated by HPLC with a chiral-phase column (Phenomenex Lux Cellulose-2, 4.6 mm × 25 mm, eluent n-hexane/iso-propanol, 40:60 v/v, 1 mL/min) to afford 6 (1.5 mg, tR = 7.4 min) and 7 (5.2 mg, tR = 8.4 min). Fr-5 (13.1 g) was subjected to silica gel CC eluted with a PE and acetone mixed solvent in a step gradient (10:1–0:1, v/v) to gain three fractions (Fr-5-1–Fr-5-3). Fr-5-2 (3.7 g) was subjected to a Sephadex LH-20 column eluted with MeOH, which was then subjected to reversed-phase C-18 MPLC with MeOH/H2O (10:90–100:0, v/v), and further purified by semi-preparative HPLC, to yield 27 (8.2 mg, 52% MeOH/H2O, 2 mL/min, tR = 17.6 min) and 24 (21.9 mg, 52% MeOH/H2O, 2 mL/min, tR = 28.0 min).
3.5. Spectral Data
Altertoxin VII (1): dark red powder; [α −9.0 (c 0.02, MeOH); UV(MeOH) λmax(log ε) 226 (3.06), 253 (3.36), 323 (2.68), 368 (2.45) nm; ECD (0.38 mM, MeOH) λmax (Δε) 204 (+0.92), 224 (–2.77) and 253 (−0.87) nm; 1H NMR (DMSO-d6, 500 MHz) and 13C NMR (DMSO-d6, 125 MHz), Table 1; HRESIMS m/z 343.0939 [M + Na]+ (calcd for C20H16NaO4, 343.0941).
Butyl xanalterate (2): dark red powder; [α −42 (c 0.02, MeOH); 1H NMR (DMSO-d6, 500 MHz) and 13C NMR (DMSO-d6, 125 MHz), Table 1; HRESIMS m/z 443.1110 [M + Na]+ (calcd for C24H20NaO7, 443.1101) and 421.1284 [M + H]+ (calcd for C24H21O7, 421.1282).
Nordihydroaltenuenes A (3), colourless oil, [α +113 (c 0.07, MeOH); UV(MeOH) λmax(log ε) 211 (3.26), 269 (2.97), 307 (2.64) nm; ECD (0.24 mM, MeOH) λmax (Δε) 207 (–15.34), 232 (+7.75), 246 (+0.43) and 271 (+6.89) nm; 1H NMR (CD3OD, 500 MHz) and 13C NMR (CD3OD, 125 MHz), Table 2; HRESIMS m/z 303.0844 [M + Na]+ (calcd for C14H16NaO6, 303.0839).
Isoochracinate A (4 and 5): yellow oil; [α +1.6 (c 0.1, MeOH); UV (MeOH) λmax (log ε) 207 (3.34), 233 (2.59), 300 (2.38) nm; 1H NMR (CD3OD, 500 MHz) and 13C NMR (CD3OD, 125 MHz), Table 2; HRESIMS m/z 223.0606 [M + H]+ (calcd for C11H11O5, 223.0607); (S)-isoochracinate A1 (4), [α −21 (c 0.1, MeOH), ECD (0.45 mM, MeOH) λmax (Δε) 219 (–1.25), 237 (+0.64), 247 (–1.12) and 301 (+0.70) nm; (R)-isoochracinate A2 (5), [α +26 (c 0.1, MeOH), ECD (0.32 mM, MeOH) λmax (Δε) 220 (+1.77), 235 (–0.76), 247 (+1.50) and 300 (–0.99) nm.
Alternariphent A (6 and 7): yellow powder; [α −4.6 (c 0.1, MeOH); UV(MeOH) λmax(log ε) 206 (2.37) nm; 1H NMR (CD3OD, 500 MHz) and 13C NMR (CD3OD, 125 MHz), Table 2; HRESIMS m/z 257.0787 [M + Na]+ (calcd for C13H14NaO4, 257.0784). (S)-Alternariphent A1 (6), [α +3.1 (c 0.1, MeOH), ECD (0.64 mM, MeOH) λmax (Δε) 211 (+0.47), 237 (–2.90), 259 (–1.46) and 334 (+0.59) nm; (R)-Alternariphent A2 (7), [α −7.2 (c 0.1, MeOH), ECD (0.51 mM, MeOH) λmax (Δε) 213 (–0.59), 233 (+4.53), 256 (+2.34) and 328 (–0.84) nm.
3.6. X-ray Crystal Structure Analysis
Crystallographic data for compound nordihydroaltenuenes A (3) were collected on a Rigaku XtaLAB PRO single-crystal diffractometer using Cu Kα radiation. The structure of 3 was solved by direct methods (SHELXS 97), expanded using difference Fourier techniques, and refined by using the full-matrix least-squares calculation. The non-hydrogen atoms were refined anisotropically, and hydrogen atoms were fixed at calculated positions. Crystallographic data for the structure 3 has been deposited with the Cambridge Crystallographic Data Centre with the supplementary publication number CCDC-1847869. Copies of the data can be obtained free of charge from the CCDC at www.ccdc.cam.ac.uk.
Crystal data for3: Moiety formula: C14H16O6 (MW = 280.27), colourless block, crystal size = 0.2 × 0.1 × 0.1 mm3, orthorhombic, space group P212121; unit cell dimensions: a = 6.75000(10) Å, b = 8.05910(10) Å, c = 22.8555(3) Å, V = 1243.31(3) Å3, Z = 4, ρcalcd = 1.497 g cm−3, T = 100.00(10) K, μ(Cu Kα) = 0.995 mm−1. A total of 5819 reflections were measured with 2434 independent reflections (Rint = 0.0209, Rsigma = 0.0222). Final R indices (I > 2σ (I)): R1 = 0.0277, wR2 = 0.0730. Final R indexes (all date): R1 = 0.0281, wR2 = 0.0734, Flack parameter = −0.02(7). Largest diff. peak and hole = 0.15 and −0.19 eÅ−3.
3.7. ECD Calculations
The theoretical calculations of new compound 1 was performed by using the density functional theory (DFT) as carried out in the Gaussian 03 [36]. Conformational analysis was initially conducted by using SYBYL-X 2.0 software. All ground-state geometries were optimized at the B3LYP/6-31G(d) level. TDDFT at B3LYP/6-31G(d) was employed to calculate the electronic excitation energies and rotational strengths in methanol [37,38]. Solvent effects of methanol solution were evaluated at the same DFT level by using the SCRF/PCM method [39].
3.8. Biological Assays
3.8.1. Antibacterial Activity Assay
Compounds 1–28 were tested for antibacterial activities against Staphylococcus aureus using the agar filter-paper diffusion method. Then, compounds which had an inhibition zone were evaluated in 96-well plates using a modified version of the broth microdilution method, and ampicillin was used as a positive control [40].
3.8.2. Antitumor Activity Assay
The in vitro cytotoxic activities against the three tumor cell lines (K562, SGC-7901 and BEL-7402) were assessed by the CCK-8 method, and the positive control was taxol [41].
4. Conclusions
Seven new structurally diverse polyketide derivatives (1–7), along with 21 known compounds (8–28), were isolated from cultures of the sponge-derived fungus, Alternaria sp. SCSIO41014. The structures and absolute configurations of these new compounds (1–7) were determined by spectroscopic analysis, X-ray single crystal diffraction, chiral separation, and comparison of ECD spectra to the calculations. Altertoxin VII (1) was the first example to possess a novel 4,8-dihydroxy substituted perylenequinone derivative, while the phenolic hydroxy groups always commonly substituted at C-4 and C-9. Compound 1 exhibited cytotoxic activities against the K562, SGC-7901, and BEL-7402 cell lines, with IC50 values of 26.58 ± 0.80, 8.75 ± 0.13, and 13.11 ± 0.95 μg/mL, respectively. Compound 11 showed selectively cytotoxic activity against K562 with an IC50 value of 19.67 ± 0.19 μg/mL. Compound 25 displayed moderate inhibitory activity against Staphylococcus aureus, with an MIC value of 31.25 μg/mL.
Supplementary Materials
The following materials are available online at http://www.mdpi.com/1660-3397/16/8/280/s1. The 16S rRNA gene sequences data of Alternaria sp. SCSIO41014, the 1D and 2D NMR spectra, HRESIMS for the new compounds 1–7, and X-ray crystallographic files of compound 3 (in CIF format).
Author Contributions
X.P., J.W., and Y.L. contributed conception and design of the study. X.P. performed experiments, analyzed data and wrote the manuscript, X.L. did the isolation and identification of the fungus. All authors contributed to manuscript revision, read and approved the submitted version.
Funding
This research was funded by the National Natural Science Foundation of China (Nos. 21172230, 21772210, 21502204, 31270402, 41476135 and 41776169), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA11030403), the Science and Technology Project of Guangdong Province (No. 2016A020222010), and Pearl River S&T Nova Program of Guangzhou (No. 201710010136).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Chagas, F.O.; Dias, L.G.; Pupo, M.T. New perylenequinone derivatives from the endophytic fungus Alternaria tenuissima SS77. Tetrahedron Lett. 2016, 57, 3185–3189. [Google Scholar] [CrossRef]
- Chowdhury, P.K.; Das, K.; Datta, A.; Liu, W.Z.; Zhang, H.Y.; Petrich, J.W. A comparison of the excited-state processes of nearly symmetrical perylene quinones: Hypocrellin A and hypomycin B. J. Photoch. Photobiol. A 2002, 154, 107–116. [Google Scholar] [CrossRef]
- Mazzini, S.; Merlini, L.; Mondelli, R.; Scaglioni, L. Conformation and tautomerism of hypocrellins. Revised structure of shiraiachrome A. J. Chem. Soc. Perk. Trans. 2 2001, 409–416. [Google Scholar] [CrossRef]
- Zhang, N.D.; Zhang, C.Y.; Xiao, X.; Zhang, Q.Y.; Huang, B.K. New cytotoxic compounds of endophytic fungus Alternaria sp. isolated from Broussonetia papyrifera (L.) Vent. Fitoterapia 2016, 110, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Kjer, J.; Wray, V.; Edrada-Ebel, R.; Ebel, R.; Pretsch, A.; Lin, W.; Proksch, P. Xanalteric acids I and II and related phenolic compounds from an endophytic Alternaria sp. isolated from the mangrove plant Sonneratia alba. J. Nat. Prod. 2009, 72, 2053–2057. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Li, X.; Wang, B. Perylene derivatives produced by Alternaria alternata, An endophytic fungus isolated from Laurencia species. Nat. Prod. Commun. 2009, 4, 1477–1480. [Google Scholar] [PubMed]
- Hradil, C.M.; Hallock, Y.F.; Clardy, J.; Kenfield, D.; Strobel, G. Phytotoxins from Alternaria cassia. Phytochemistry 1989, 28, 73–75. [Google Scholar] [CrossRef]
- Bugni, T.S.; Ireland, C.M. Marine-derived fungi: A chemically and biologically diverse group of microorganisms. Nat. Prod. Rep. 2004, 21, 143–163. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.F.; Wei, X.Y.; Qin, X.C.; Lin, X.P.; Zhou, X.F.; Liao, S.R.; Yang, B.; Liu, J.; Tu, Z.C.; Liu, Y.H. Arthpyrones A-C, Pyridone alkaloids from a sponge-derived fungus Arthrinium arundinis ZSDS1-F3. Org. Lett. 2015, 17, 656–659. [Google Scholar] [CrossRef] [PubMed]
- Pang, X.Y.; Lin, X.P.; Wang, J.F.; Liang, R.; Tian, Y.Q.; Salendra, L.; Luo, X.W.; Zhou, X.F.; Yang, B.; Tu, Z.C.; et al. Three new highly oxygenated sterols and one new dihydroisocoumarin from the marine sponge-derived fungus Cladosporium sp. SCSIO41007. Steroids 2018, 129, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Pang, X.Y.; Lin, X.P.; Tian, Y.Q.; Liang, R.; Wang, J.F.; Yang, B.; Zhou, X.F.; Kaliyaperumal, K.; Luo, X.W.; Tu, Z.C.; et al. Three new polyketides from the marine sponge-derived fungus Trichoderma sp. SCSIO41004. Nat. Prod. Res. 2018, 32, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.Q.; Lin, X.P.; Wang, Z.; Zhou, X.F.; Qin, X.C.; Kaliyaperumal, K.; Zhang, T.Y.; Tu, Z.C.; Liu, Y. Asteltoxins with antiviral activities from the marine sponge-derived fungus Aspergillus sp. SCSIO XWS02F40. Molecules 2016, 21, 34. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.F.; Lin, X.P.; Qin, C.; Liao, S.R.; Wan, J.T.; Zhang, T.Y.; Liu, J.; Fredimoses, M.; Chen, H.; Yang, B.; et al. Antimicrobial and antiviral sesquiterpenoids from sponge-associated fungus, Aspergillus sydowii ZSDS1-F6. J. Antibiot. 2014, 67, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Jing, P.Z.; Xu, N.L.; Meng, D.L.; Sha, Y. A new perylenequinone from the fruit bodies of Bulgaria inquinans. J. Asian Nat. Prod. Res. 2006, 8, 743–746. [Google Scholar]
- Jiao, P.; Gloer, J.B.; Campbell, J.; Shearer, C.A. Altenuene derivatives from an unidentified freshwater fungus in the family Tubeufiaceae. J. Nat. Prod. 2006, 69, 612–615. [Google Scholar] [CrossRef] [PubMed]
- Phaopongthai, J.; Wiyakrutta, S.; Meksuriyen, D.; Sriubolmas, N.; Suwanborirux, K. Azole-synergistic anti-candidal activity of altenusin, A biphenyl metabolite of the endophytic fungus Alternaria alternata isolated from Terminalia chebula Retz. J. Microbiol. 2013, 51, 821–828. [Google Scholar] [CrossRef] [PubMed]
- Chou, T.H.; Chen, I.S.; Hwang, T.L.; Wang, T.C.; Lee, T.H.; Cheng, L.Y.; Chang, Y.C.; Cho, J.Y.; Chen, J.J. Phthalides from Pittosporum illicioides var. illicioides with inhibitory activity on superoxide generation and elastase release by neutrophils. J. Nat. Prod. 2008, 71, 1692–1695. [Google Scholar] [CrossRef] [PubMed]
- Tianpanich, K.; Prachya, S.; Wiyakrutta, S.; Mahidol, C.; Ruchirawat, S.; Kittakoop, P. Radical scavenging and antioxidant activities of isocoumarins and a phthalide from the endophytic fungus Colletotrichum sp. J. Nat. Prod. 2011, 74, 79–81. [Google Scholar] [CrossRef] [PubMed]
- Nakano, H.; Kumagai, N.; Matsuzaki, H.; Kabuto, C.; Hongo, H. Enantioselective addition of diethylzinc to aldehydes using 2-azanorbornylmethanols and 2-azanorbornylmethanethiol as acatalyst. Tetrahedron Lett. 1997, 8, 1391–1401. [Google Scholar] [CrossRef]
- Takahashi, H.; Tsubuki, T.; Higashiyama, K. Highly diastereoselective reaction of chiral o-[2-(1,3-oxazolidinyl)] benzaldehydes with alkylmetallic reagents: Synthesis of chiral 3-substituted phthalides. Chem. Pharm. Bull. 1991, 39, 3136–3139. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, Q.; Xia, G.; Huang, H.; Li, H.; Ma, L.; Lu, Y.; He, L.; Xia, X.; She, Z. Polyketides with alpha-glucosidase inhibitory activity from a mangrove endophytic fungus, Penicillium sp. HN29-3B1. J. Nat. Prod. 2015, 78, 1816–1822. [Google Scholar] [CrossRef] [PubMed]
- Stack, M.E.; Mazzola, E.P.; Page, S.W.; Pohlan, A.E. Mutagenic perylenequinone metabolites of Alternaria Alternata: alteroxins I, II, and III. J. Nat. Prod. 1986, 49, 866–871. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.J.; Fu, X.M.; Zhang, X.L.; Kong, W.W.; Wang, C.Y. Bioactive perylene derivatives from a soft coral-derived fungus Alternaria sp. (ZJ-2008017). Chem. Nat. Compd. 2015, 51, 766–768. [Google Scholar] [CrossRef]
- Zhao, D.L.; Wang, D.; Tian, X.Y.; Cao, F.; Li, Y.Q.; Zhang, C.S. Anti-phytopathogenic and cytotoxic activities of crude extracts and secondary metabolites of marine-derived fungi. Mar. Drugs 2018, 16, 36. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wu, Y.; Zhai, R.; Liu, Z.; Huang, X.; She, Z. Altenusin derivatives from mangrove endophytic fungus Alternaria sp. SK6YW3L. RSC Adv. 2016, 6, 72127–72132. [Google Scholar] [CrossRef]
- Wang, Q.X.; Bao, L.; Yang, X.L.; Guo, H.; Yang, R.N.; Ren, B.; Zhang, L.X.; Dai, H.Q.; Guo, L.D.; Liu, H.W. Polyketides with antimicrobial activity from the solid culture of an endolichenic fungus Ulocladium sp. Fitoterapia 2012, 83, 209–214. [Google Scholar] [CrossRef] [PubMed]
- Bradburn, N.; Coker, R.D.; Blunedn, G.; Turner, C.H.; Crabb, T.A. 5-epialtenuene and neoaltenuene, dibenzo-a-pyrones from Alternaria alternata cultured on rice. Phtochemistry 1994, 35, 665–669. [Google Scholar] [CrossRef]
- Aly, A.H.; Edrada-Ebe, R.; Indriani, I.D.; Wray, V.; Muller, W.E.G.; Totzke, F.; Zirrgiebel, U.; Schachtele, C.; Kubbutat, M.H.G.; Lin, W.H.; et al. Cytotoxic metabolites from the fungal endophyte Alternaria sp. and their subsequent detection in its host plant polygonum senegalense. J. Nat. Prod. 2008, 71, 972–980. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yang, M.H.; Wang, X.B.; Li, T.X.; Kong, L.Y. Bioactive metabolites from the endophytic fungus Alternaria alternata. Fitoterapia 2014, 99, 153–158. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.; Fu, L.; Zhang, Z.; Dong, X.; Xu, D.; Mao, Z.; Liu, Y.; Lai, D.; Zhou, L. Dibenzo-alpha-pyrones from the endophytic fungus Alternaria sp. Samif01: Isolation, Structure elucidation, And their antibacterial and antioxidant activities. Nat. Prod. Res. 2016, 31, 387–396. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, L.; Zhuang, Y.; Kong, F.; Zhang, C.; Zhu, W. Phenolic polyketides from the co-cultivation of marine-derived Penicillium sp. WC-29-5 and Streptomyces fradiae 007. Mar. Drugs 2014, 12, 2079–2088. [Google Scholar] [CrossRef] [PubMed]
- Kellogg, J.J.; Todd, D.A.; Egan, J.M.; Raja, H.A.; Oberlies, N.H.; Kvalheim, O.M.; Cech, N.B. Biochemometrics for natural products research: comparison of data analysis approaches and application to identification of bioactive compounds. J. Nat. Prod. 2016, 79, 376–386. [Google Scholar] [CrossRef] [PubMed]
- De Souza, G.D.; Mithofer, A.; Daolio, C.; Schneider, B.; Rodrigues-Filho, E. Identification of Alternaria alternata mycotoxins by LC-SPE-NMR and their cytotoxic effects to soybean (Glycine max) cell suspension culture. Molecules 2013, 18, 2528–2538. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.; Sohn, M.J.; Koshino, H.; Kim, E.H.; Kim, W.G. Verrulactone C with an unprecedented dispiro skeleton, A new inhibitor of Staphylococcus aureus enoyl-ACP reductase, from Penicillium verruculosum F375. Bioorg. Med. Chem. Lett. 2014, 24, 83–86. [Google Scholar] [CrossRef] [PubMed]
- Kashiwada, Y.; Nonaka, G.I.; Nishioka, I. Chromone glucosides from Rhubarb. Phtochemistry 1990, 29, 1007–1009. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Montgomery, J.A.; Vreven, T., Jr.; Kudin, K.N.; Burant, J.C.; et al. Gaussian 03, Revision E.01; Gaussian, Inc.: Wallingford, CT, USA, 2004; Available online: http://gaussian.com/g03citation/ (accessed on 8 August 2018).
- Tomasi, J.; Persico, M. Molecular interactions in solution: An overview of methods based on continuous distributions of the solvent. Chem. Rev. 1994, 94, 2027–2094. [Google Scholar] [CrossRef]
- Cammi, R.; Tomasi, J. Remarks on the use of the apparent surface charges (ASC) methods in solvation problems: Iterative versus Matrix-Inversion procedures and the renormalization of the apparent charges. J. Comput. Chem. 1995, 16, 1449–1458. [Google Scholar] [CrossRef]
- Gross, E.K.U.; Dobson, J.F.; Petersilka, M. Density functional theory of time-dependent phenomena. Top. Curr. Chem. 1996, 181, 81–172. [Google Scholar]
- Wang, J.F.; Cong, Z.W.; Huang, X.L.; Hou, C.X.; Chen, W.B.; Tu, Z.C.; Huang, D.Y.; Liu, Y.H. Soliseptide A, A cyclic hexapeptide possessing piperazic acid groups from Streptomyces solisilvae HNM30702. Org. Lett. 2018, 20, 1371–1374. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.F.; Wei, X.Y.; Qin, X.C.; Tian, X.P.; Liao, L.; Li, K.M.; Zhou, X.F.; Yang, X.W.; Wang, F.Z.; Zhang, T.Y.; et al. Antiviral merosesquiterpenoids produced by the antarctic fungus Aspergillus ochraceopetaliformis SCSIO 05702. J. Nat. Prod. 2016, 79, 59–65. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).