First Evidence of Dehydroabietic Acid Production by a Marine Phototrophic Gammaproteobacterium, the Purple Sulfur Bacterium Allochromatium vinosum MT86
Abstract
:1. Introduction
2. Results
2.1. Phylogenetic Analysis and Identification of the Isolate
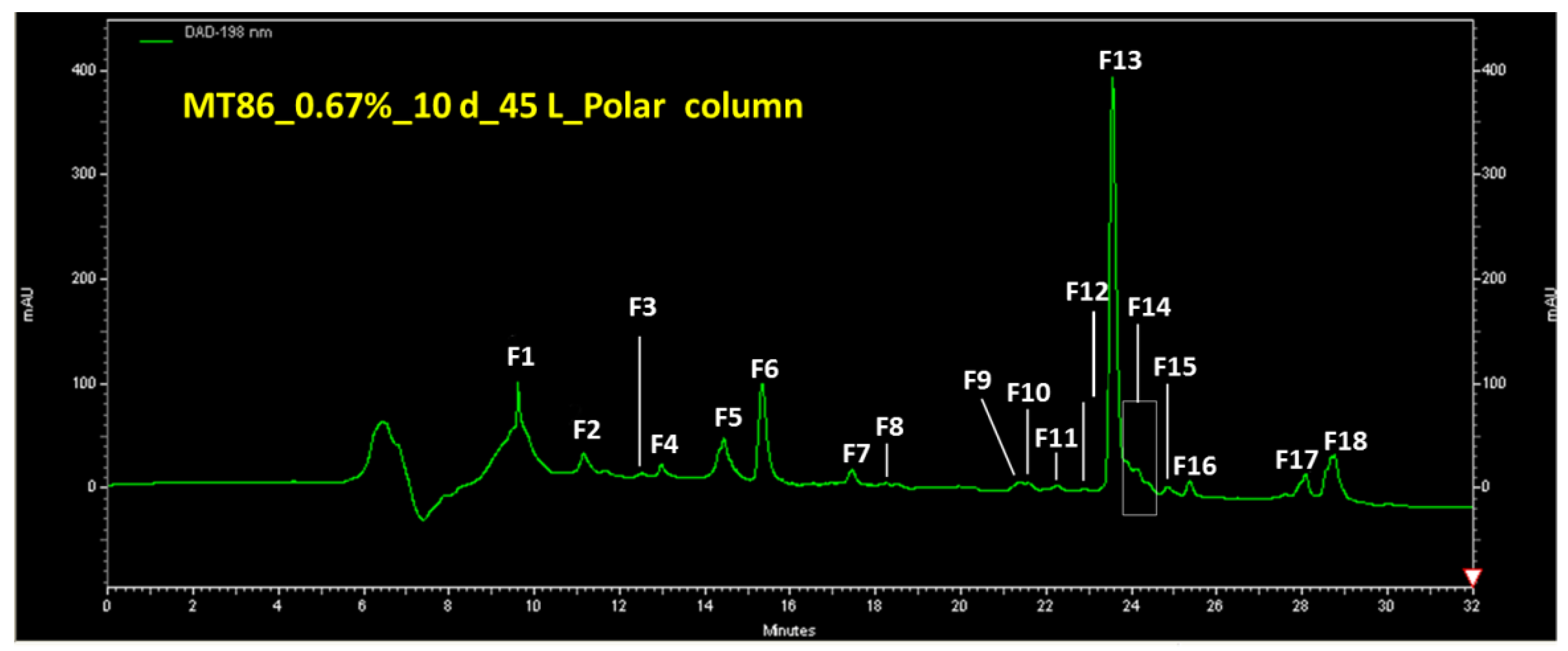
2.2. Metabolite Profiles, Fractionation of Extracts and Purification of Compound 1
2.3. Purification of Compound 1
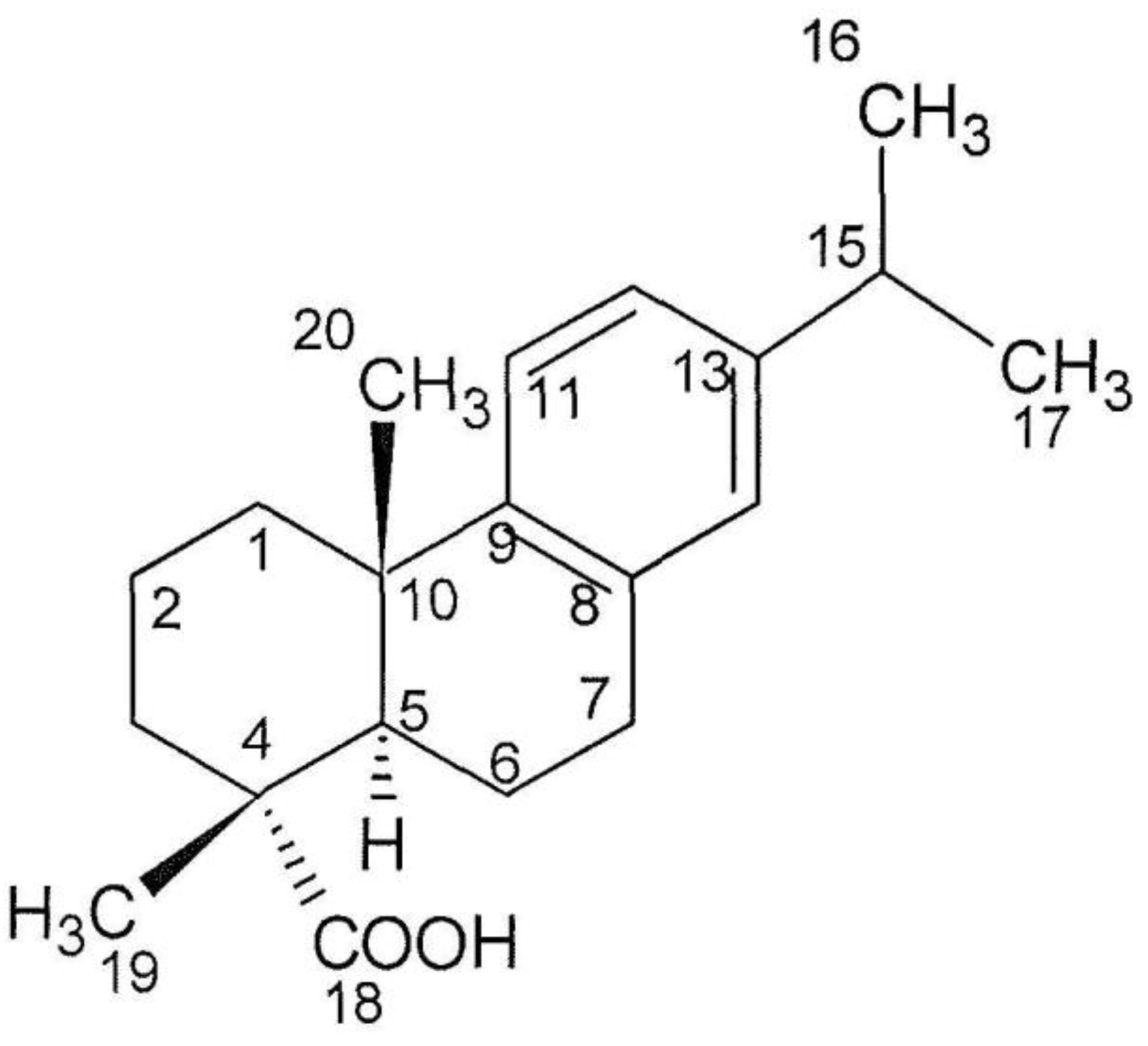
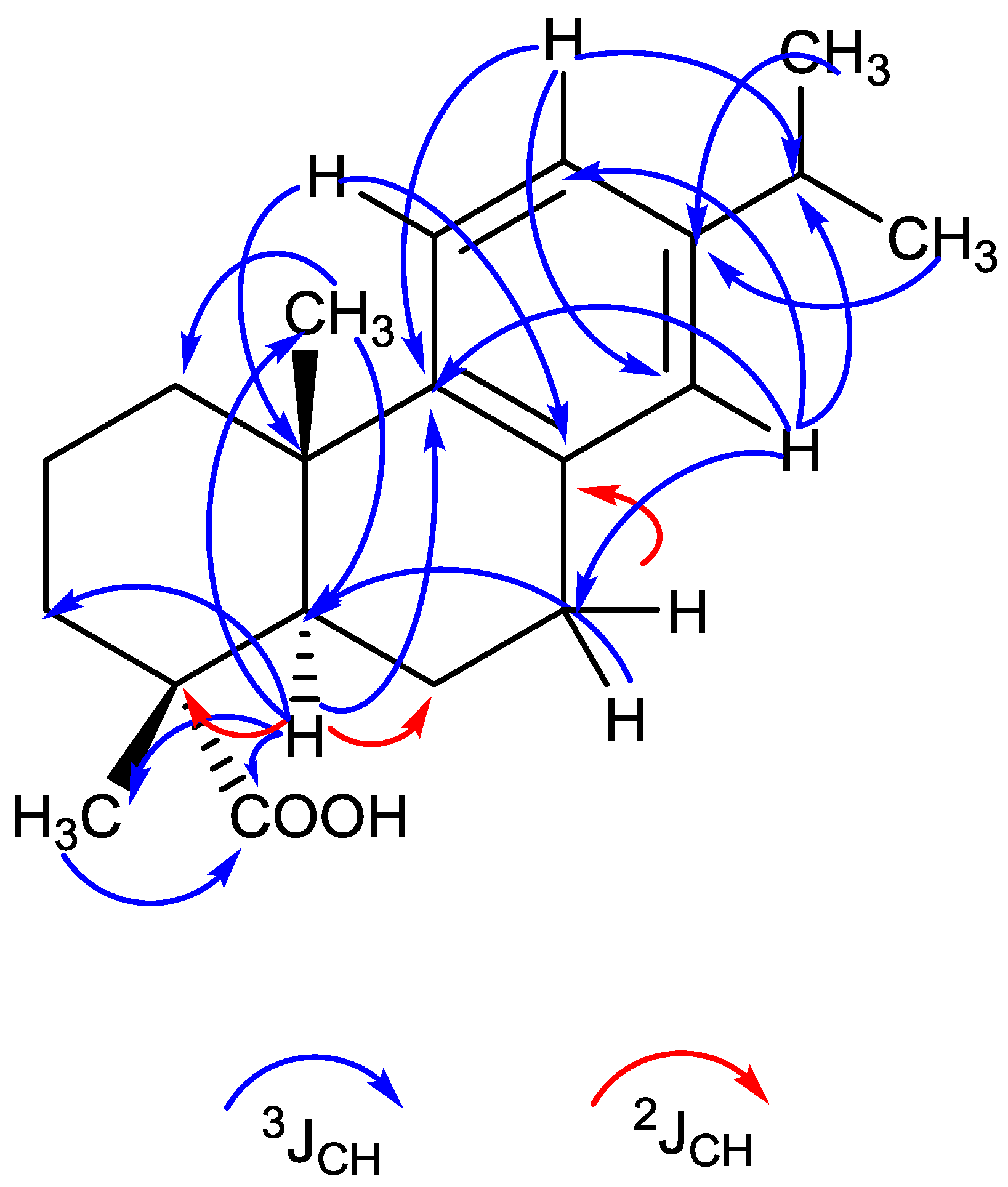
2.4. Structure Analysis of Compound 1
2.5. Biological Activities
2.6. Genetic Approval of Secondary Metabolite Biosynthetic Pathways
3. Discussion
4. Materials and Methods
4.1. Isolation and Identification of Strain MT86
4.2. Cultivation and Preparation of Cell Extracts
4.3. Analytical HPLC-DAD/MS
4.4. Semi-Preparative HPLC
4.5. NMR Analysis
4.6. ESI-MS
4.7. Antimicrobial Activity of Compound 1
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Burgess, J.G.; Miyashita, H.; Sudo, H.; Matsunaga, T. Antibiotic production by the photosynthetic bacterium Chromatium purpuratum NKPB 031704: localization of activity to the chromatophores. FEMS Microbiol. Lett. 1991, 84, 301–306. [Google Scholar] [CrossRef]
- Brinkhoff, T.; Bach, G.; Heidorn, T.; Liang, L.; Schlinghloff, A.; Simon, M. Antibiotic production by Roseobacter clade-affiliated species from the German Wadden Sea and its antagonistic effects on indigenous isolates. Appl. Environ. Microbiol. 2004, 70, 2560–2565. [Google Scholar] [CrossRef] [PubMed]
- Martens, T.; Gram, L.; Grossart, H.-P.; Kessler, D.; Müller, R.; Simon, M.; Wenzel, S.C.; Brinkhoff, T. Bacteria of the Roseobacter clade show potential for secondary metabolite production. Microbiol. Ecol. 2007, 54, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Serdyuk, O.P.; Smolygina, L.D.; Kobzar, E.F.; Gogotov, I.N. Occurrence of plant hormones in cells of the phototrophic purple bacterium Rhodospirillum rubrum 1R. FEMS Microbiol. Lett. 1993, 109, 113–116. [Google Scholar] [CrossRef]
- Ranjith, N.K.; Sasikala, Ch.; Ramana, Ch.V. Rhodethrin: A novel indole terpenoid ether produced by Rhodobacter sphaeroides has cytotoxic and phytohormonal activities. Biotechnol. Lett. 2007, 29, 1399–1402. [Google Scholar] [CrossRef] [PubMed]
- Sunayana, M.R.; Sasikala, Ch.; Ramana, Ch.V. Rhodestrin: A novel indole terpenoid phytohormone from Rhodobacter sphaeroides. Biotechnol. Lett. 2005, 27, 1897–1900. [Google Scholar] [CrossRef] [PubMed]
- Sunayana, M.R.; Sasikala, Ch.; Ramana, Ch.V. Production of a novel indole ester from 2-aminobenzoate by Rhodobacter sphaeroides OU5. J. Ind. Microbiol. Biotechnol. 2005, 32, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Smanski, M.J.; Peterson, R.M.; Huang, S.-X.; Shen, B. Bacterial diterpene synthases: new opportunities for mechanistic enzymology and engineered biosynthesis. Curr. Opin. Chem. Biol. 2012, 16, 132–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cane, D.E.; Ikeda, H. Exploration and mining of the bacterial terpenome. Acc. Chem. Res. 2012, 45, 463–472. [Google Scholar] [CrossRef] [PubMed]
- Yamada, Y.; Kuzuyama, T.; Komatsu, M.; Shin-Ya, K.; Omura, S.; Cane, D.E.; Ikeda, H. Terpene synthases are widely distributed in bacteria. PNAS 2015, 112, 857–862. [Google Scholar] [CrossRef] [PubMed]
- Hefter, J.; Richnow, H.H.; Fischer, U.; Trendel, J.M.; Michaelis, W. Verrucosan-2ß-ol from the phototrophic bacterium Chloroflexus aurantiacus: First report on a verrucosan-type diterpenoid from a prokaryote. J. Gen. Microbiol. 1993, 139, 2757–2761. [Google Scholar] [CrossRef]
- Chuck, J.; Barrow, K.D. The isolation of isoagathenediol: A new tricyclic diterpene from the lipids of Rhodospirillum rubrum. Microbiol. 1995, 141, 2659–2663. [Google Scholar] [CrossRef]
- Costa, M.S.; Rego, A.; Ramos, V.; Afonso, T.B.; Freitas, S.; Preto, M.; Lopes, V.; Vasconcelos, V.; Magalhaes, C.; Leao, P.N. The conifer biomarkers dehydroabietic acid and abietic acids are widespread in cyanobacteria. Sci. Rep. 2016, 6, 23436. [Google Scholar] [CrossRef] [PubMed]
- Chamy, M.C.; Piovano, M.; Gambaro, V.; Garbarino, J.A.; Nicoletti, M. Dehydroabietane diterpenoids from Calceolaria ascendens. Phytochemistry 1987, 26, 1763–1765. [Google Scholar] [CrossRef]
- Rao, X.-P.; Song, Z.-Q.; Shang, S.-B. Dehydroabietic acid. Acta Cryst. 2009, E65, o2402. [Google Scholar] [CrossRef] [PubMed]
- Welch, S.C.; Hagan, C.P.; Kim, J.H.; Chu, P.S. Stereoselective total syntheses of diterpene resin acids. J. Org. Chem. 1977, 42, 2879–2887. [Google Scholar] [CrossRef]
- Weber, T.; Blin, K.; Duddela, S.; Krug, D.; Kim, H.U.; Bruccoleri, R.; Lee, S.Y.; Fischbach, M.A.; Müller, R.; Wohlleben, W.; Breitling, R.; Takano, E.; Medema, M.H. AntiSMASH 3.0—A comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015, 43, W237–W243. [Google Scholar] [CrossRef] [PubMed]
- Takaishi, S. Distribution and biosynthesis of carotenoids. In The Purple Phototrophic Bacteria; Hunter, N.E., Daldal, F., Thurnauer, M.C., Beatty, J.T., Eds.; Springer: Dordrecht, The Netherlands, 2009; pp. 97–117. ISBN 978-1-4020-8814-8. [Google Scholar]
- Rohmer, M.; Bouvier-Nave, P.; Ourisson, G. Distribution of hopanoid triterpenes in prokaryotes. J. Gen. Microbiol. 1984, 130, 1137–1150. [Google Scholar] [CrossRef]
- Imhoff, J.F.; Bias-Imhoff, U. Lipids, quinones and fatty acids of anoxygenic phototrophic bacteria. In Anoxygenic Photosynthetic Bacteria; Blankenship, R.E., Madigan, M.T., Bauer, C.E., Eds.; Springer: Dordrecht, The Netherlands, 1995; pp. 179–205. ISBN 978-0-7923-3681-5. [Google Scholar]
- Luo, D.; Ni, Q.; Ji, A.; Gu, W.; Wu, J.; Jiang, C. Dehydroabietic acid derivative QC4 induces gastric cancer cell death via oncosis and apoptosis. BioMed Res. 2016, 2681061. [Google Scholar] [CrossRef] [PubMed]
- Imhoff, J.F.; Hiraishi, A.; Süling, J. Anoxygenic Phototrophic Purple Bacteria. In Bergey's Manual of Systematic Bacteriology, 2nd ed.; Garrity, G., Ed.; Springer: Dordrecht, The Netherlands, 2005; Volume 2, pp. 119–132. ISBN 978-0-387-24144-9. [Google Scholar]
- Gärtner, A.; Wiese, J.; Imhoff, J.F. Amphritea atlantica gen. nov., sp. nov., a gammaproteobacterium from the Logatchev hydrothermal vent field. Int. J. Syst. Evol. Microbiol. 2008, 58, 34–39. [Google Scholar]
- Schneemann, I.; Kajahn, I.; Ohlendorf, B.; Zinecker, H.; Erhard, A.; Nagel, K.; Wiese, J.; Imhoff, J.F. Mayamycin, a cytotoxic polyketide from a marine Streptomyces strain isolated from the marine sponge Halichondria panicea. J. Nat. Prod. 2010, 73, 1309–1312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| No. of C | δC | C-Atom | H-Atom | δH | Position |
|---|---|---|---|---|---|
| 1 | 184.52 | COOH | 1 | C-18 | |
| 2 | 147.23 | C | C-9 | ||
| 3 | 145.89 | C | C-13 | ||
| 4 | 134.99 | C | C-8 | ||
| 5 | 127.03 | CH | 1 | 6.85 | C-14 |
| 6 | 124.28 | CH | 1 | 7.13 | C-11 |
| 7 | 123.99 | CH | 1 | 6.95 | C-12 |
| 8 | 47.82 | C | C-4 | ||
| 9 | 45.22 | CH | 1 | 2.15 | C-5 |
| 10 | 38.40 | CH2 | 2 | 2.26/1.41 | C-1 |
| 11 | 37.20 | CH2 | 2 | 1.74/1.62 | C-3 |
| 12 | 37.12 | C | C-10 | ||
| 13 | 33.87 | CH | 1 | 2.79 | C-15 |
| 14 | 30.39 | CH2 | 2 | 2.85 | C-7 |
| 15 | 25.23 | CH3 | 3 | 1.19 | C-20 |
| 16 | 24.16 | CH3 | 3 | 1.20 | C-16 |
| 17 | 24.16 | CH3 | 3 | 1.20 | C-17 |
| 18 | 22.05 | CH2 | 2 | 1.80/1.50 | C-6 |
| 19 | 18.97 | CH2 | 2 | 1.74/1.66 | C-2 |
| 20 | 16.61 | CH3 | 3 | 1.22 | C-19 |
| Total | 20 C | 28 H |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Imhoff, J.F.; Sun, M.; Wiese, J.; Tank, M.; Zeeck, A. First Evidence of Dehydroabietic Acid Production by a Marine Phototrophic Gammaproteobacterium, the Purple Sulfur Bacterium Allochromatium vinosum MT86. Mar. Drugs 2018, 16, 270. https://doi.org/10.3390/md16080270
Imhoff JF, Sun M, Wiese J, Tank M, Zeeck A. First Evidence of Dehydroabietic Acid Production by a Marine Phototrophic Gammaproteobacterium, the Purple Sulfur Bacterium Allochromatium vinosum MT86. Marine Drugs. 2018; 16(8):270. https://doi.org/10.3390/md16080270
Chicago/Turabian StyleImhoff, Johannes F., Mingshuang Sun, Jutta Wiese, Marcus Tank, and Axel Zeeck. 2018. "First Evidence of Dehydroabietic Acid Production by a Marine Phototrophic Gammaproteobacterium, the Purple Sulfur Bacterium Allochromatium vinosum MT86" Marine Drugs 16, no. 8: 270. https://doi.org/10.3390/md16080270
APA StyleImhoff, J. F., Sun, M., Wiese, J., Tank, M., & Zeeck, A. (2018). First Evidence of Dehydroabietic Acid Production by a Marine Phototrophic Gammaproteobacterium, the Purple Sulfur Bacterium Allochromatium vinosum MT86. Marine Drugs, 16(8), 270. https://doi.org/10.3390/md16080270






