Cross-Sectional Study: Associations of A20 and Cezanne with Leukocyte Accumulation in B-Cell Acute Lymphoblastic Leukemia
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Control Subjects
2.2. DNA Sequencing of A20, CYLD and Cezanne Genes
2.3. Cytokine Quantification
2.4. RNA Extraction and Real-Time RT-PCR
2.5. Immunostaining and Flow Cytometry
2.6. Data Analysis
2.7. Statistics
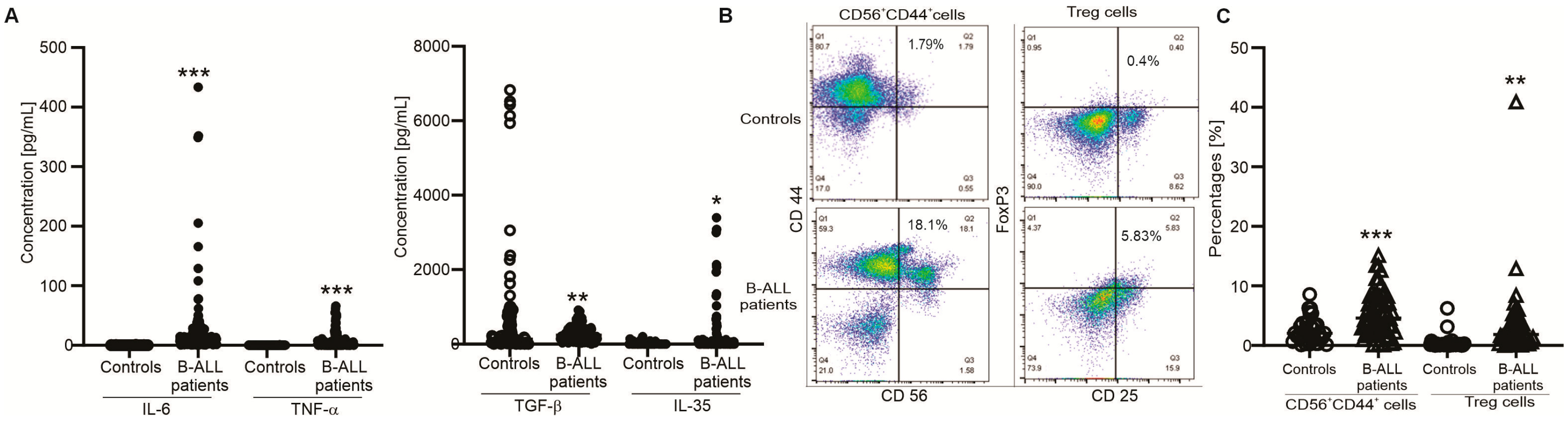
3. Results
3.1. Clinical Associations and Immunophenotype in B-ALL Patients
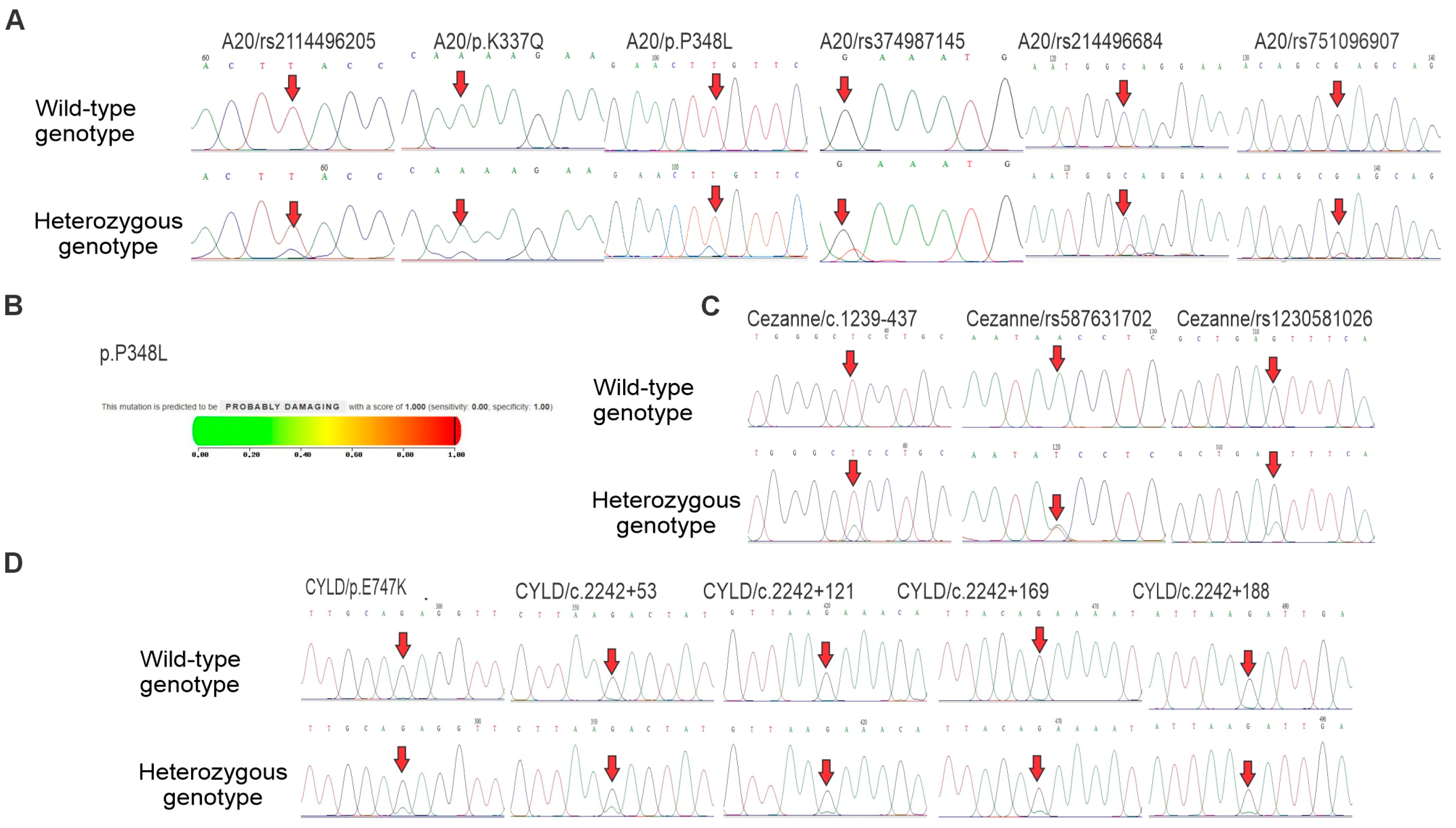
3.2. DNA Sequencing of A20, Cezanne and CYLD Genes in B-ALL Patients
3.3. Associations of the A20 Expression and Polymorphisms with Clinical Characteristics and Immunophenotype in B-ALL Patients
4. Discussion
5. Conclusions
Limitations
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ALL | Acute lymphoblastic leukemia |
| ALT | Alanine aminotransferase |
| AST | Aspartate transaminase |
| BM | Bone marrow |
| CYLD | Cylindromatosis |
| DUB | Deubiquitinases |
| GAPDH | Glyceraldehyde-3-Phosphate Dehydrogenase |
| GGT | Gamma glutamyl transferase |
| HWE | Hardy–Weinberg equilibrium |
| IL-6 | Interleukin-6 |
| NK | Natural Killer |
| LDH | Lactate dehydrogenase |
| MAF | Minor allele frequency |
| NF-κB | Nuclear Factor-kappa B |
| MAPK | Mitogen-activated protein kinase |
| PCR | Polymerase Chain Reaction |
| SNP | Single nucleotide polymorphism |
| STAT | Signal transducer and activator of transcription |
| TNFAIP3, A20 | Tumor necrosis factor-α (TNFα)-induced protein 3 |
| TGF-β | Transforming growth factor-β |
| TNF-α | Tumor necrosis factor-α |
| Treg | Regulatory T-cells |
| WBC | Ưhite blood cell |
References
- Della Starza, I.; Chiaretti, S.; De Propris, M.S.; Elia, L.; Cavalli, M.; De Novi, L.A.; Soscia, R.; Messina, M.; Vitale, A.; Guarini, A.; et al. Minimal Residual Disease in Acute Lymphoblastic Leukemia: Technical and Clinical Advances. Front. Oncol. 2019, 9, 726. [Google Scholar] [CrossRef] [PubMed]
- Rabin, K.R.; Poplack, D.G. Management strategies in acute lymphoblastic leukemia. Oncology 2011, 25, 328–335. [Google Scholar] [PubMed]
- Passaro, D.; Irigoyen, M.; Catherinet, C.; Gachet, S.; Da Costa De Jesus, C.; Lasgi, C.; Tran Quang, C.; Ghysdael, J. CXCR4 Is Required for Leukemia-Initiating Cell Activity in T Cell Acute Lymphoblastic Leukemia. Cancer Cell 2015, 27, 769–779. [Google Scholar] [CrossRef]
- Thomas, X.; Le, Q.H. Prognostic factors in adult acute lymphoblastic leukemia. Hematology 2003, 8, 233–242. [Google Scholar] [CrossRef]
- Sarmiento Palao, H.; Tarin, F.; Martirena, F.; Barragan, E.; Such, E.; Sempere, A.; Tasso, M.; Manresa, P.; Lopez, F. A reproducible strategy for analysis of minimal residual disease measured by Standardized multiparametric flow cytometry in b acute lymphoblastic leukemia. Cytom. B Clin. Cytom. 2019, 96, 12–15. [Google Scholar] [CrossRef]
- Martino, M.; Alati, C.; Canale, F.A.; Musuraca, G.; Martinelli, G.; Cerchione, C. A Review of Clinical Outcomes of CAR T-Cell Therapies for B-Acute Lymphoblastic Leukemia. Int. J. Mol. Sci. 2021, 22, 2150. [Google Scholar] [CrossRef]
- Thomas, D.A.; O’Brien, S.; Kantarjian, H.M. Monoclonal antibody therapy with rituximab for acute lymphoblastic leukemia. Hematol. Oncol. Clin. N. Am. 2009, 23, 949–971. [Google Scholar] [CrossRef][Green Version]
- Chaturvedi, A.; Shetty, D.; Ghogale, S.G.; Deshpande, N.; Badrinath, Y.; Chatterjee, G.; Girase, K.; Sriram, H.; Khanka, T.; Mishra, C.; et al. Detecting hypodiploidy with endoreduplication and masked hypodiploidy in B-cell acute lymphoblastic leukemia using multicolor flow cytometry. Cytom. B Clin. Cytom. 2022, 102, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Enesa, K.; Zakkar, M.; Chaudhury, H.; Luong le, A.; Rawlinson, L.; Mason, J.C.; Haskard, D.O.; Dean, J.L.; Evans, P.C. NF-kappaB suppression by the deubiquitinating enzyme Cezanne: A novel negative feedback loop in pro-inflammatory signaling. J. Biol. Chem. 2008, 283, 7036–7045. [Google Scholar] [CrossRef]
- Meng, Z.; Xu, R.; Xie, L.; Wu, Y.; He, Q.; Gao, P.; He, X.; Chen, Q.; Xie, Q.; Zhang, J.; et al. A20/Nrdp1 interaction alters the inflammatory signaling profile by mediating K48- and K63-linked polyubiquitination of effectors MyD88 and TBK1. J. Biol. Chem. 2021, 297, 100811. [Google Scholar] [CrossRef]
- Zhu, G.; Herlyn, M.; Yang, X. TRIM15 and CYLD regulate ERK activation via lysine-63-linked polyubiquitination. Nat. Cell Biol. 2021, 23, 978–991. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Hu, J.; Wang, X.; Xuan, L.; Lai, J.; Xu, L.; Chen, S.; Yang, L.; Luo, G.; Zhu, K.; et al. Overexpression of MALT1-A20-NF-kappaB in adult B-cell acute lymphoblastic leukemia. Cancer Cell Int. 2015, 15, 73. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Xing, H.; Li, S.; Yu, J.; Li, H.; Liu, S.; Tian, Z.; Tang, K.; Rao, Q.; Wang, M.; et al. Up-regulated A20 promotes proliferation, regulates cell cycle progression and induces chemotherapy resistance of acute lymphoblastic leukemia cells. Leuk. Res. 2015, 39, 976–983. [Google Scholar] [CrossRef]
- Espinosa, L.; Cathelin, S.; D’Altri, T.; Trimarchi, T.; Statnikov, A.; Guiu, J.; Rodilla, V.; Ingles-Esteve, J.; Nomdedeu, J.; Bellosillo, B.; et al. The Notch/Hes1 pathway sustains NF-kappaB activation through CYLD repression in T cell leukemia. Cancer Cell 2010, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Wu, S.; Fan, J.; Meng, Z.; Gao, G.; Liu, T.; Wang, Q.; Xia, H.; Wang, X.; Wu, K. CYLD regulates cell ferroptosis through Hippo/YAP signaling in prostate cancer progression. Cell Death Dis. 2024, 15, 79. [Google Scholar] [CrossRef]
- Pareja, F.; Ferraro, D.A.; Rubin, C.; Cohen-Dvashi, H.; Zhang, F.; Aulmann, S.; Ben-Chetrit, N.; Pines, G.; Navon, R.; Crosetto, N.; et al. Deubiquitination of EGFR by Cezanne-1 contributes to cancer progression. Oncogene 2012, 31, 4599–4608. [Google Scholar] [CrossRef]
- McNally, R.S.; Davis, B.K.; Clements, C.M.; Accavitti-Loper, M.A.; Mak, T.W.; Ting, J.P. DJ-1 enhances cell survival through the binding of Cezanne, a negative regulator of NF-kappaB. J. Biol. Chem. 2011, 286, 4098–4106. [Google Scholar] [CrossRef]
- Zhu, L.; Zhang, F.; Shen, Q.; Chen, S.; Wang, X.; Wang, L.; Yang, L.; Wu, X.; Huang, S.; Schmidt, C.A.; et al. Characteristics of A20 gene polymorphisms in T-cell acute lymphocytic leukemia. Hematology 2014, 19, 448–454. [Google Scholar] [CrossRef]
- Arora, M.; Kaul, D.; Varma, N. Functional nature of a novel mutant CYLD observed in pediatric lymphoblastic B-cell leukemia. Pediatr. Blood Cancer 2015, 62, 1066–1069. [Google Scholar] [CrossRef]
- Duy, P.N.; Thuy, N.T.; Trang, B.K.; Giang, N.H.; Van, N.T.H.; Xuan, N.T. Regulation of NF-kappaB- and STAT1-mediated plasmacytoid dendritic cell functions by A20. PLoS ONE 2019, 14, e0222697. [Google Scholar] [CrossRef]
- Nishanth, G.; Deckert, M.; Wex, K.; Massoumi, R.; Schweitzer, K.; Naumann, M.; Schluter, D. CYLD enhances severe listeriosis by impairing IL-6/STAT3-dependent fibrin production. PLoS Pathog. 2013, 9, e1003455. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.G.; Boone, D.L.; Chai, S.; Libby, S.L.; Chien, M.; Lodolce, J.P.; Ma, A. Failure to regulate TNF-induced NF-kappaB and cell death responses in A20-deficient mice. Science 2000, 289, 2350–2354. [Google Scholar] [CrossRef] [PubMed]
- Jin, W.; Reiley, W.R.; Lee, A.J.; Wright, A.; Wu, X.; Zhang, M.; Sun, S.C. Deubiquitinating enzyme CYLD regulates the peripheral development and naive phenotype maintenance of B cells. J. Biol. Chem. 2007, 282, 15884–15893. [Google Scholar] [CrossRef]
- Reiley, W.W.; Zhang, M.; Jin, W.; Losiewicz, M.; Donohue, K.B.; Norbury, C.C.; Sun, S.C. Regulation of T cell development by the deubiquitinating enzyme CYLD. Nat. Immunol. 2006, 7, 411–417. [Google Scholar] [CrossRef]
- Vilchis-Ordonez, A.; Contreras-Quiroz, A.; Vadillo, E.; Dorantes-Acosta, E.; Reyes-Lopez, A.; Quintela-Nunez del Prado, H.M.; Venegas-Vazquez, J.; Mayani, H.; Ortiz-Navarrete, V.; Lopez-Martinez, B.; et al. Bone Marrow Cells in Acute Lymphoblastic Leukemia Create a Proinflammatory Microenvironment Influencing Normal Hematopoietic Differentiation Fates. BioMed Res. Int. 2015, 2015, 386165. [Google Scholar] [CrossRef]
- Dai, Q.; Zhang, G.; Wang, Y.; Ye, L.; Shi, R.; Peng, L.; Guo, S.; He, J.; Yang, H.; Zhang, Y.; et al. Cytokine network imbalance in children with B-cell acute lymphoblastic leukemia at diagnosis. Cytokine 2023, 169, 156267. [Google Scholar] [CrossRef] [PubMed]
- Urwanisch, L.; Luciano, M.; Horejs-Hoeck, J. The NLRP3 Inflammasome and Its Role in the Pathogenicity of Leukemia. Int. J. Mol. Sci. 2021, 22, 1271. [Google Scholar] [CrossRef]
- Schober, S.; Rottenberger, J.M.; Hilz, J.; Schmid, E.; Ebinger, M.; Feuchtinger, T.; Handgretinger, R.; Lang, P.; Queudeville, M. Th1 cytokines in pediatric acute lymphoblastic leukemia. Cancer Immunol. Immunother. 2023, 72, 3621–3634. [Google Scholar] [CrossRef]
- Solati, H.; Zareinejad, M.; Ghavami, A.; Ghasemi, Z.; Amirghofran, Z. IL-35 and IL-18 Serum Levels in Children With Acute Lymphoblastic Leukemia: The Relationship With Prognostic Factors. J. Pediatr. Hematol. Oncol. 2020, 42, 281–286. [Google Scholar] [CrossRef]
- Turnis, M.E.; Sawant, D.V.; Szymczak-Workman, A.L.; Andrews, L.P.; Delgoffe, G.M.; Yano, H.; Beres, A.J.; Vogel, P.; Workman, C.J.; Vignali, D.A. Interleukin-35 Limits Anti-Tumor Immunity. Immunity 2016, 44, 316–329. [Google Scholar] [CrossRef]
- Harris, N.L.; Jaffe, E.S.; Diebold, J.; Flandrin, G.; Muller-Hermelink, H.K.; Vardiman, J.; Lister, T.A.; Bloomfield, C.D. The World Health Organization classification of hematological malignancies report of the Clinical Advisory Committee Meeting, Airlie House, Virginia, November 1997. Mod. Pathol. 2000, 13, 193–207. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Pui, C.H.; Dodge, R.K.; Dahl, G.V.; Rivera, G.; Look, A.T.; Kalwinsky, D.; Bowman, W.P.; Ochs, J.; Abromowitch, M.; Mirro, J.; et al. Serum lactic dehydrogenase level has prognostic value in childhood acute lymphoblastic leukemia. Blood 1985, 66, 778–782. [Google Scholar] [CrossRef]
- Esteban, R.E.; Christianne, B.; Alvaro, A.; Demichelis-Gomez, R. Prognostic Effect of CD20 Expression in Adult B-cell Acute Lymphoblastic Leukemia. Clin. Lymphoma Myeloma Leuk. 2018, 18, 361–367. [Google Scholar] [CrossRef]
- Tiftik, N.; Bolaman, Z.; Batun, S.; Ayyildiz, O.; Isikdogan, A.; Kadikoylu, G.; Muftuoglu, E. The importance of CD7 and CD56 antigens in acute leukaemias. Int. J. Clin. Pract. 2004, 58, 149–152. [Google Scholar] [CrossRef]
- Kutlu, H.; Avci, E.; Ozyurt, F. White blood cells detection and classification based on regional convolutional neural networks. Med. Hypotheses 2020, 135, 109472. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Wu, D.; Lin, S.; Li, P. CD34 and CD38 are prognostic biomarkers for acute B lymphoblastic leukemia. Biomark. Res. 2016, 4, 23. [Google Scholar] [CrossRef] [PubMed]
- Liao, C.; Shen, D.Y.; Xu, X.J.; Song, H.; Xu, W.Q.; Zhao, F.Y.; Yang, S.L.; Tang, Y.M. High CD38 expression in childhood T-cell acute lymphoblastic leukemia is not associated with prognosis. Cancer Biomark. 2020, 27, 277–284. [Google Scholar] [CrossRef]
- Chen, H.; Hu, L.; Luo, Z.; Zhang, J.; Zhang, C.; Qiu, B.; Dong, L.; Tan, Y.; Ding, J.; Tang, S.; et al. A20 suppresses hepatocellular carcinoma proliferation and metastasis through inhibition of Twist1 expression. Mol. Cancer 2015, 14, 186. [Google Scholar] [CrossRef]
- Wang, J.H.; Wei, W.; Guo, Z.X.; Shi, M.; Guo, R.P. Decreased Cezanne expression is associated with the progression and poor prognosis in hepatocellular carcinoma. J. Transl. Med. 2015, 13, 41. [Google Scholar] [CrossRef][Green Version]
- Kachuri, L.; Jeon, S.; DeWan, A.T.; Metayer, C.; Ma, X.; Witte, J.S.; Chiang, C.W.K.; Wiemels, J.L.; de Smith, A.J. Genetic determinants of blood-cell traits influence susceptibility to childhood acute lymphoblastic leukemia. Am. J. Hum. Genet. 2021, 108, 1823–1835. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Li, L.; Wu, J.; Zhang, L.; Sun, Z.; Li, X.; Wang, X.; Yu, H.; Chang, Y.; Wu, X.; et al. Complete Blood Count Score Model Integrating Reduced Lymphocyte-Monocyte Ratio, Elevated Neutrophil-Lymphocyte Ratio, and Elevated Platelet-Lymphocyte Ratio Predicts Inferior Clinical Outcomes in Adult T-Lymphoblastic Lymphoma. Oncologist 2019, 24, e1123–e1131. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xu, Y.; Liang, L.; Xu, Y.; Wang, C.; Wang, L.; Chen, S.; Yang, L.; Wu, X.; Li, B.; et al. Abnormal expression of A20 and its regulated genes in peripheral blood from patients with lymphomas. Cancer Cell Int. 2014, 14, 36. [Google Scholar] [CrossRef] [PubMed]
- Hjelmeland, A.B.; Wu, Q.; Wickman, S.; Eyler, C.; Heddleston, J.; Shi, Q.; Lathia, J.D.; Macswords, J.; Lee, J.; McLendon, R.E.; et al. Targeting A20 decreases glioma stem cell survival and tumor growth. PLoS Biol. 2010, 8, e1000319. [Google Scholar] [CrossRef]
- Wang, M.; Li, S. Bladder polypoid cystitis-derived A20 associates with tumorigenesis. Cell Biochem. Biophys. 2013, 67, 669–673. [Google Scholar] [CrossRef]

| Characteristics | Normal Range | Total (n = 147) |
|---|---|---|
| Age (years) | 30.95 ± 21.7 | |
| Sex. male (n. %) | 87 (59.18) | |
| Urea (mmol/L) | 2.5–7.5 | 6 ± 3.38 |
| Glucose (mmol/L) | 3.9–6.4 | 7.28 ± 7.47 |
| Creatinine (µmol/L) | 62–120 | 86.17 ± 56.54 |
| Uric acid (µmol/L) | 420 (M)/360 (F) | 460.48 ± 203.5 |
| Total bilirubin (µmol/L) | ≤17 | 16.53 ± 25 |
| Direct bilirunbin (µmol/L) | ≤4.3 | 5.63 ± 18.04 |
| Indirect bilirubin (µmol/L) | ≤12.7 | 11.37 ± 8.98 |
| Total protein (g/L) | 65–82 | 77.04 ± 63.06 |
| Albumin (g/L) | 35–50 | 38.72 ± 5.18 |
| Globulin (g/L) | 24–38 | 33.21 ± 6.44 |
| Ferritin (µg/L) | 30–300 | 766.93 ± 494.14 |
| AST (GOT) (U/L) | ≤37 | 63.8 ± 66.17 |
| ALT (GPT) (U/L) | ≤40 | 44.8 ± 43.84 |
| GGT (UI/L) | ≤60 | 150.22 ± 158.06 |
| LDH (IU/L) | 230–460 | 2942.7 ± 4268.1 |
| Erythrocyte count (T/L) | 3.9–5.03 | 3.89 ± 7.3 |
| Hemoglobin (g/L) | 120–155 | 92.93 ± 24.87 |
| Hematocrit (%) | 37–43 | 29 ± 7.45 |
| Nucleated erythrocyte count (G/L) | 0 | 0.23 ± 0.56 |
| Reticulocytes (%) | 0.5–1.5 | 1.45 ± 1.16 |
| Platelet count (G/L) | 150–450 | 90.29 ± 100.5 |
| WBC count (G/L) | 3.5–10.5 | 69.78 ± 105.65 |
| BM blasts (%) | 0–20 | 70.95 ± 27.83 |
| Neutrophil count (G/L) | 2.8–5.5 | 5.68 ± 17.2 |
| Eosinophil count (G/L) | 0.16–0.8 | 0.23 ± 0.8 |
| Basophil count (G/L) | 0.01–0.12 | 0.12 ± 0.39 |
| Monocyte count (G/L) | 0.05–0.3 | 1.27 ± 3.6 |
| Lymphocyte count (G/L) | 1.2–3 | 11.48 ± 34.87 |
| SNP | Gene | Test Model | Controls (n = 144) | Cases (n = 147) | p-Value |
|---|---|---|---|---|---|
| rs2114496205 | A20 | TT | 144 (100%) | 144 (97.96%) | |
| TC | 0 (0%) | 3 (2.04%) | 0.497 | ||
| p.K337Q | A20 | AA | 144 (100%) | 140 (95.24%) | |
| AC | 0 (0%) | 7 (4.76%) | 0.059 | ||
| p.P348L | A20 | TT | 144 (100%) | 137 (93.2%) | |
| TC | 0 (0%) | 10 (6.8%) | 0.014 * | ||
| rs374987145 | A20 | GG | 129 (89.58%) | 136 (92.52%) | |
| GT | 15 (10.42%) | 11 (7.48%) | 0.613 | ||
| rs214496684 | A20 | CC | 133 (92.36%) | 143 (97.28%) | |
| CT | 11 (7.64%) | 4 (2.72%) | 0.121 | ||
| rs751096907 | A20 | GG | 144 (100%) | 144 (97.96%) | |
| GT | 0 (0%) | 3 (2.04%) | 0.497 | ||
| c.1239-437 | Cezanne | TT | 144 (100%) | 142 (96.6%) | |
| TA | 0 (0%) | 5 (3.4%) | 0.246 | ||
| rs587631702 | Cezanne | TT | 141 (97.92%) | 144 (97.96%) | |
| TA | 3 (2.08%) | 3 (2.04%) | 1 | ||
| rs1230581026 | Cezanne | GG | 142 (98.61%) | 123 (83.67%) | |
| GA | 2 (1.39%) | 24 (16.33%) | <0.001 *** | ||
| p.E747K | CYLD | GG | 91 (63.2%) | 125 (85.03%) | |
| GA | 53 (36.8%) | 22 (14.97%) | 0.001 ** | ||
| c.2242+53 | CYLD | GG | 55 (38.19%) | 39 (26.53%) | |
| GA | 89 (61.81%) | 108 (73.47% | 0.131 | ||
| c.2242+121 | CYLD | GG | 105 (72.92%) | 113 (76.87%) | |
| GA | 39 (27.08%) | 34 (23.13%) | 0.514 | ||
| c.2242+169 | CYLD | GG | 85 (59.03%) | 115 (78.23%) | |
| GA | 59 (40.94%) | 32 (21.77%) | 0.006 ** | ||
| c.2242+188 | CYLD | GG | 99 (68.75%) | 117 (79.59%) | |
| GA | 45 (31.25%) | 30 (20.41%) | 0.104 |
| Gene/SNP | Type of Variant | Allele | MAF | HWE (p-Value) | |||
|---|---|---|---|---|---|---|---|
| ALL Patients | Controls | Controls | ALL Patients | All Population | |||
| A20/rs2114496205 | Missense | T/C | 0.010 | 0.000 | N/A | 0.9006 | 0.9296 |
| A20/p.K337Q | Missense | A/C | 0.024 | 0.000 | N/A | 0.7675 | 0.8355 |
| A20/p.P348L | Missense | T/C | 0.034 | 0.000 | N/A | 0.6694 | 0.7656 |
| A20/rs374987145 | Missense | G/T | 0.037 | 0.052 | 0.5097 | 0.6374 | 0.4251 |
| A20/rs2114496684 | Stop-gained | C/T | 0.014 | 0.038 | 0.6337 | 0.8671 | 0.5721 |
| A20/rs751096907 | Stop-gained | G/T | 0.010 | 0.000 | N/A | 0.9006 | 0.9296 |
| Cezanne/c.1239-437 | Intron | T/A | 0.017 | 0.000 | N/A | 0.8339 | 0.8824 |
| Cezanne/rs587631702 | Intron | T/A | 0.010 | 0.010 | 0.8993 | 0.9006 | 0.8589 |
| Cezanne/rs1230581026 | Intron | GA | 0.082 | 0.007 | 0.9333 | 0.2812 | 0.4251 |
| CYLD/p.E747K | Missense | GA | 0.075 | 0.184 | 0.006802 | 0.3268 | 0.01162 |
| CYLD/c.2242+53 | Intron | GA | 0.367 | 0.309 | 0.000 | 0.000 | 0.000 |
| CYLD/c.2242+121 | Intron | GA | 0.116 | 0.135 | 0.06017 | 0.1129 | 0.01442 |
| CYLD/c.2242+169 | Intron | GA | 0.109 | 0.205 | 0.00199 | 0.1386 | 0.001569 |
| CYLD/c.2242+188 | Intron | GA | 0.102 | 0.156 | 0.02627 | 0.1683 | 0.01162 |
| A20 Expression | Cezanne Expression | |||||
|---|---|---|---|---|---|---|
| Characteristics | Low (n = 119) | High (n = 28) | p Value | Low (n = 132) | High (n = 15) | p Value |
| Age (years) | 31.69 ± 20.7 | 28.34 ± 23.65 | 0.476 | 31.35 ± 21.12 | 29.92 ± 25.01 | 0.825 |
| Sex. male (n. %) | 75 (63) | 15 (53.6) | 0.384 | 80 (60.6) | 8 (53.33) | 0.484 |
| Urea (mmol/L) | 6.1 ± 3.52 | 6.85 ± 4.4 | 0.405 | 6.21 ± 3.7 | 7.55 ± 2.47 | 0.266 |
| Glucose (mmol/L) | 7.92 ± 9.12 | 6.21 ± 2.6 | 0.407 | 7.09 ± 7.67 | 6.96 ± 3.39 | 0.958 |
| Creatinine (µmol/L) | 91.09 ± 62.5 | 90.15 ± 64.2 | 0.951 | 87.04 ± 48.34 | 83.6 ± 38.44 | 0.827 |
| Uric acid (µmol/L) | 470.45 ± 203.1 | 451.8 ± 256.2 | 0.725 | 463.1 ± 193.2 | 562.9 ± 335.5 | 0.146 |
| Total bilirubin (µmol/L) | 14.83 ± 15.94 | 28.72 ± 58.07 | 0.044 * | 17.8 ± 28.9 | 15.76 ± 10.45 | 0.825 |
| Direct bilirunbin (µmol/L) | 4.43 ± 9.97 | 14.6 ± 44.4 | 0.042 * | 6.38 ± 21.1 | 3.9 ± 4.21 | 0.712 |
| Indirect bilirubin (µmol/L) | 11.14 ± 8.5 | 14.1 ± 14.4 | 0.218 | 11.44 ± 8.68 | 11.86 ± 7.08 | 0.882 |
| Total protein (g/L) | 72.07 ± 7.43 | 112.5 ± 175.45 | 0.025 * | 79.14 ± 74.25 | 71.8 ± 6.68 | 0.756 |
| Albumin (g/L) | 38.58 ± 4.9 | 37.46 ± 6.12 | 0.383 | 38.43 ± 5.17 | 39.85 ± 5.09 | 0.407 |
| Globulin (g/L) | 33.4 ± 6 | 35.42 ± 9.98 | 0.242 | 33.67 ± 6.91 | 31.95 ± 6.03 | 0.447 |
| Ferritin (µg/L) | 1008.3 ± 728.1 | 1127.8 ± 987.5 | 0.532 | 1085.1 ± 765.6 | 1026 ± 1038.9 | 0.812 |
| AST (GOT) (U/L) | 60.46 ± 68.04 | 71.35 ± 62.6 | 0.51 | 61.16 ± 65.7 | 112.3 ± 81 | 0.022 * |
| ALT (GPT) (U/L) | 43.19 ± 43.3 | 46.65 ± 40.8 | 0.743 | 46.14 ± 46.5 | 60.7 ± 39.66 | 0.339 |
| GGT (UI/L) | 134.04 ± 143 | 150.94 ± 147.5 | 0.727 | 135.8 ± 142.7 | 196.5 ± 59.3 | 0.469 |
| LDH (IU/L) | 2895.8 ± 4426.7 | 2813.2 ± 3693.9 | 0.938 | 2733.7 ± 4174.8 | 5706.3 ± 5921.4 | 0.039 * |
| Erythrocyte count (T/L) | 4.2 ± 9.08 | 3.34 ± 0.95 | 0.675 | 4.05 ± 8.52 | 3.47 ± 0.75 | 0.828 |
| Hemoglobin (g/L) | 92.54 ± 25.3 | 92.8 ± 20.7 | 0.966 | 92.08 ± 23.43 | 97 ± 18.06 | 0.519 |
| Hematocrit (%) | 28.5 ± 7.54 | 28.7 ± 6.85 | 0.928 | 28.28 ± 7.2 | 29.47 ± 5.86 | 0.613 |
| Nucleated erythrocyte count (G/L) | 0.23 ± 0.54 | 0.13 ± 0.18 | 0.416 | 0.21 ± 0.49 | 0.1 ± 0.12 | 0.508 |
| Reticulocytes (%) | 1.44 ± 1.05 | 1.46 ± 1.42 | 0.859 | 1.52 ± 1.27 | 1.63 ± 1.68 | 0.824 |
| Platelet count (G/L) | 102.2 ± 110.35 | 47.92 ± 52.6 | 0.034 * | 97.98 ± 92.5 | 40.1 ± 20.6 | 0.045 * |
| WBC count (G/L) | 70.63 ± 104.7 | 94.87 ± 138.4 | 0.373 | 67.6 ± 100.7 | 74.1 ± 121.5 | 0.849 |
| BM blasts (%) | 72.57 ± 26.53 | 64.72 ± 36.01 | 0.279 | 70.31 ± 28.89 | 65.69 ± 37.74 | 0.653 |
| Neutrophil count (G/L) | 6.27 ± 20.07 | 6.73 ± 16.01 | 0.923 | 5.78 ± 18.8 | 3.76 ± 4.59 | 0.737 |
| Eosinophil count (G/L) | 0.29 ± 0.97 | 0.11 ± 0.25 | 0.406 | 0.27 ± 0.91 | 0.05 ± 0.11 | 0.456 |
| Basophil count (G/L) | 0.15 ± 0.48 | 0.07 ± 0.16 | 0.427 | 0.13 ± 0.44 | 0.005 ± 0.01 | 0.368 |
| Monocyte count (G/L) | 1.51 ± 4.28 | 0.67 ± 1.04 | 0.385 | 1.42 ± 4.05 | 0.87 ± 1.18 | 0.668 |
| Lymphocyte count (G/L) | 12.55 ± 16.2 | 21.88 ± 15.6 | 0.055 | 13.31 ± 39.9 | 21.36 ± 54.7 | 0.554 |
| PLR | 50.78 ± 84.75 | 15.93 ± 22.47 | 0.046 * | 45.04 ± 78.14 | 20.62 ± 29.88 | 0.329 |
| Relative expression of A20/GAPDH | 2.22 ± 2.53 | 2850.8 ± 13945.2 | 0.035 * | 18.97 ± 59.5 | 7222.9 ± 22486.6 | <0.001 *** |
| Relative expression of CYLD/GAPDH | 1.92 ± 7.02 | 126.7 ± 494.5 | 0.009 ** | 2.87 ± 10.1 | 322.6 ± 780.9 | <0.001 *** |
| Relative expression of Cezanne/GAPDH | 0.09 ± 0.2 | 14.82 ± 54.5 | 0.007 ** | 0.22 ± 1.61 | 57.2 ± 109.01 | <0.001 *** |
| SNP | Gene | Test Model | A20 Expression | Cezanne Expression | ||||
|---|---|---|---|---|---|---|---|---|
| Low (n = 119) | High (n = 28) | p Value | Low (n = 132) | High (n = 15) | p Value | |||
| rs2114496205 | A20 | TT | 116 (97.48%) | 28 (100%) | 129 (97.73%) | 15 (100%) | ||
| TC | 3 (2.52%) | 0 (0%) | 0.246 | 3 (2.27%) | 0 (0%) | 0.497 | ||
| p.K337Q | A20 | AA | 112 (94.42%) | 28 (100%) | 125 (94.7%) | 15 (100%) | ||
| AC | 7 (5.88%) | 0 (0%) | 0.029 * | 7 (5.3%) | 0 (0%) | 0.059 | ||
| p.P348L | A20 | TT | 109 (91.6%) | 28 (100%) | 122 (92.42%) | 15 (100%) | ||
| TC | 10 (8.4%) | 0 (0%) | 0.007 ** | 10 (7.58%) | 0 (0%) | 0.007 ** | ||
| rs374987145 | A20 | GG | 108 (90.76%) | 28 (100%) | 121 (91.67%) | 15 (100%) | ||
| GT | 11 (9.24%) | 0 (0%) | 0.003 ** | 11 (8.33%) | 0 (0%) | 0.007 ** | ||
| rs214496684 | A20 | CC | 115 (96.64%) | 28 (100%) | 128 (96.97%) | 15 (100%) | ||
| CT | 4 (3.36%) | 0 (0%) | 0.246 | 4 (3.03%) | 0 (0%) | 0.246 | ||
| rs751096907 | A20 | GG | 116 (97.48%) | 28 (100%) | 129 (97.73%) | 15 (100%) | ||
| GT | 3 (2.52%) | 0 (0%) | 0.246 | 3 (2.27%) | 0 (0%) | 0.497 | ||
| c.1239-437 | Cezanne | TT | 114 (95.8%) | 28 (100%) | 127 (96.21%) | 15 (100%) | ||
| TA | 5 (4.2%) | 0 (0%) | 0.121 | 5 (3.79%) | 0 (0%) | 0.121 | ||
| rs587631702 | Cezanne | TT | 117 (98.32%) | 27 (96.43%) | 129 (97.73%) | 15 (100%) | ||
| TA | 2 (1.68%) | 1 (3.57%) | 0.683 | 3 (2.27%) | 0 (0%) | 0.497 | ||
| rs1230581026 | Cezanne | GG | 97 (81.5%) | 28 (92.86%) | 109 (82.58%) | 14 (93.33%) | ||
| GA | 22 (18.5%) | 2 (7.14%) | 0.019 * | 23 (17.42%) | 1 (6.67%) | 0.048 * | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Published by MDPI on behalf of the Lithuanian University of Health Sciences. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ha, L.T.; Giang, N.H.; Toan, N.L.; Giang, N.V.; Mao, C.V.; Nhat, N.Q.; Quan, T.D.; Hoang, N.H.; Hang, N.T.; Xuan, N.T. Cross-Sectional Study: Associations of A20 and Cezanne with Leukocyte Accumulation in B-Cell Acute Lymphoblastic Leukemia. Medicina 2025, 61, 1166. https://doi.org/10.3390/medicina61071166
Ha LT, Giang NH, Toan NL, Giang NV, Mao CV, Nhat NQ, Quan TD, Hoang NH, Hang NT, Xuan NT. Cross-Sectional Study: Associations of A20 and Cezanne with Leukocyte Accumulation in B-Cell Acute Lymphoblastic Leukemia. Medicina. 2025; 61(7):1166. https://doi.org/10.3390/medicina61071166
Chicago/Turabian StyleHa, Le Thuy, Nguyen Hoang Giang, Nguyen Linh Toan, Nguyen Van Giang, Can Van Mao, Nguyen Quoc Nhat, Tran Dang Quan, Nguyen Huy Hoang, Ngo Thu Hang, and Nguyen Thi Xuan. 2025. "Cross-Sectional Study: Associations of A20 and Cezanne with Leukocyte Accumulation in B-Cell Acute Lymphoblastic Leukemia" Medicina 61, no. 7: 1166. https://doi.org/10.3390/medicina61071166
APA StyleHa, L. T., Giang, N. H., Toan, N. L., Giang, N. V., Mao, C. V., Nhat, N. Q., Quan, T. D., Hoang, N. H., Hang, N. T., & Xuan, N. T. (2025). Cross-Sectional Study: Associations of A20 and Cezanne with Leukocyte Accumulation in B-Cell Acute Lymphoblastic Leukemia. Medicina, 61(7), 1166. https://doi.org/10.3390/medicina61071166






