Artificial Intelligence Performance in Cardiac Magnetic Resonance Strain Analysis for Aortic Stenosis: Validation with Echocardiography and Healthy Controls
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Imaging Protocols
2.3. Data Analysis
3. Results
3.1. Performance of AI-Based CMR Strain Analysis
3.2. Comparison of the Performance of AI-Based CMR Feature Tracking Technique Between Aortic Stenosis and Healthy Control Groups
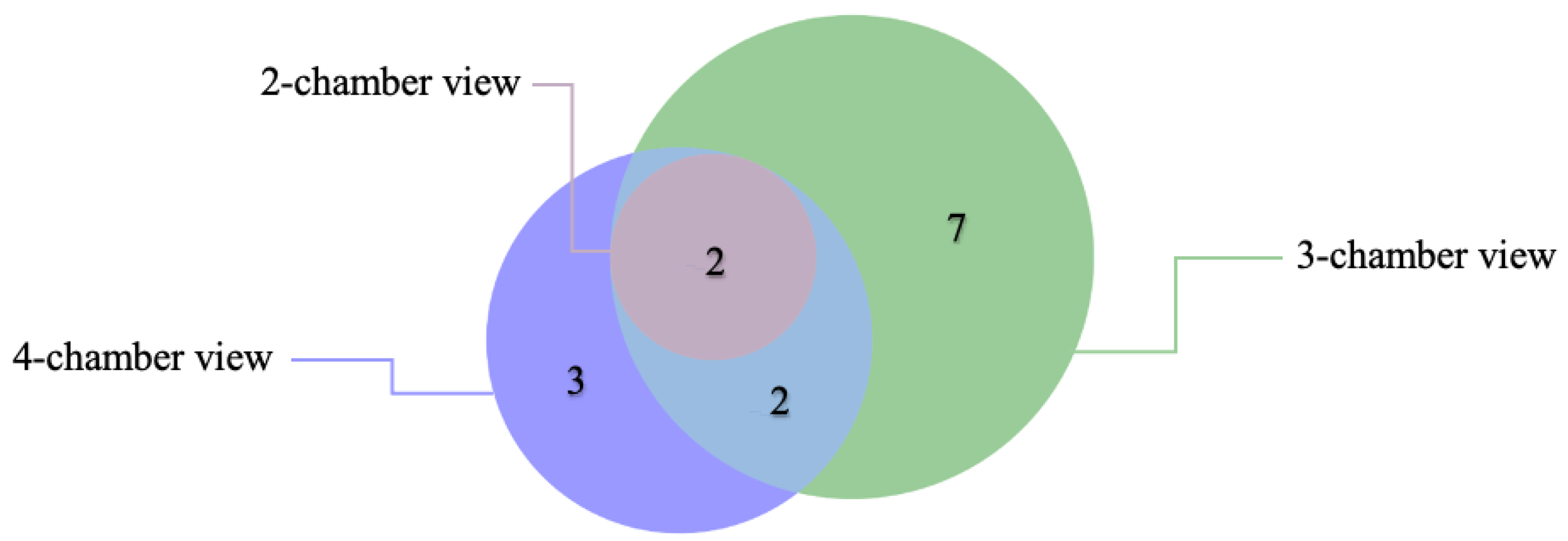
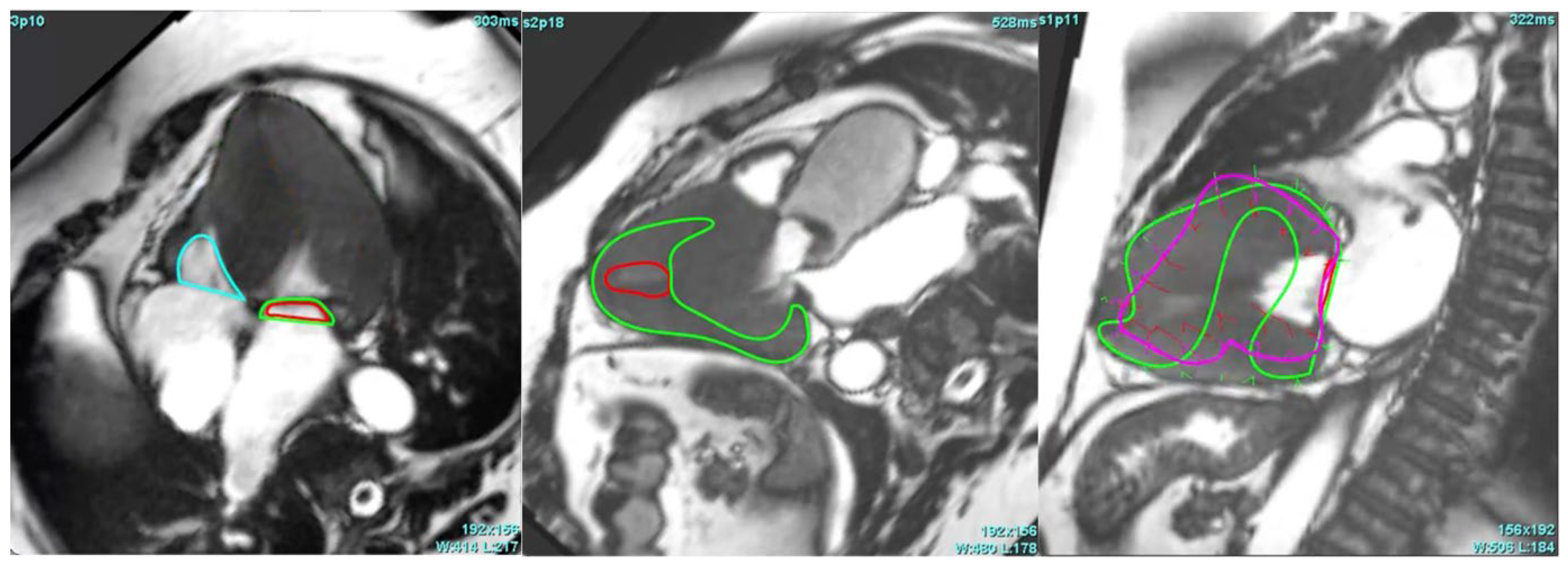
3.3. Critical Limitation in AI Performance
3.4. Segmental and Global AI-Based CMR Myocardial Strain Analysis in AS Patients vs. Controls
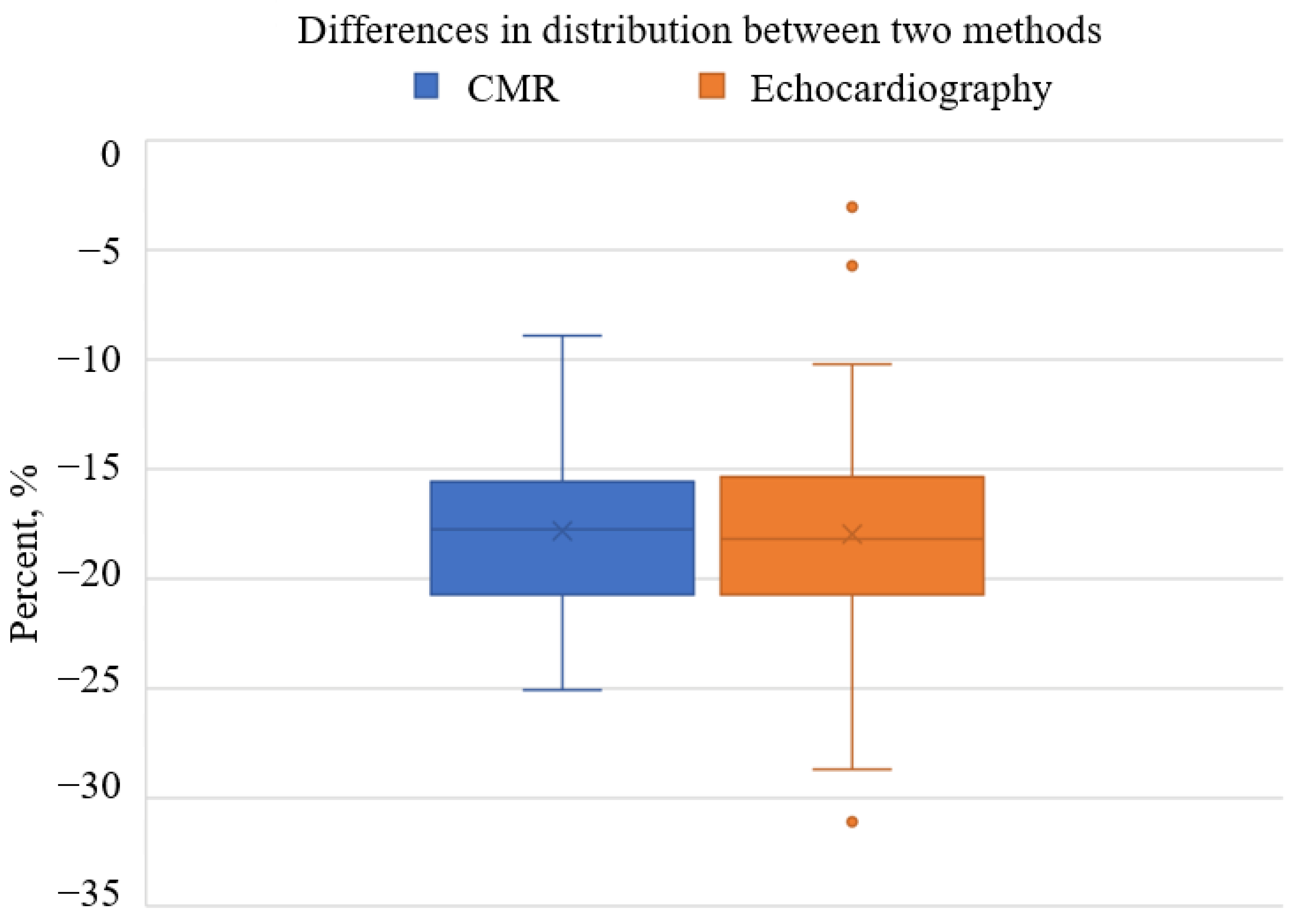
3.5. Comparison of AI-Based CMR GLS with Echocardiography-Based GLS in AS Patient Cohort
4. Discussion
Limitations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Eveborn, G.W.; Schirmer, H.; Heggelund, G.; Lunde, P.; Rasmussen, K. The Evolving Epidemiology of Valvular Aortic Stenosis. The Tromsø Study. Heart 2013, 99, 396–400. [Google Scholar] [CrossRef] [PubMed]
- Iung, B.; Vahanian, A. Epidemiology of Acquired Valvular Heart Disease. Can. J. Cardiol. 2014, 30, 962–970. [Google Scholar] [CrossRef] [PubMed]
- Joseph, J.; Naqvi, S.Y.; Giri, J.; Goldberg, S. Aortic Stenosis: Pathophysiology, Diagnosis, and Therapy. Am. J. Med. 2017, 130, 253–263. [Google Scholar] [CrossRef]
- Zakkar, M.; Bryan, A.J.; Angelini, G.D. Aortic Stenosis: Diagnosis and Management. BMJ 2016, 355, i5425. [Google Scholar] [CrossRef]
- Kumar, A.; Majmundar, M.; Doshi, R.; Kansara, T.; Shariff, M.; Shah, P.; Adalja, D.; Gullapalli, N.; Vallabhajosyula, S.; Panaich, S.S.; et al. Meta-Analysis of Early Intervention Versus Conservative Management for Asymptomatic Severe Aortic Stenosis. Am. J. Cardiol. 2021, 138, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Vahanian, A.; Beyersdorf, F.; Praz, F.; Milojevic, M.; Baldus, S.; Bauersachs, J.; Capodanno, D.; Conradi, L.; De Bonis, M.; De Paulis, R.; et al. 2021 ESC/EACTS Guidelines for the Management of Valvular Heart Disease: Developed by the Task Force for the Management of Valvular Heart Disease of the European Society of Cardiology (ESC) and the European Association for Cardio-Thoracic Surgery (EACTS). Rev. Esp. Cardiol. 2022, 75, 524. [Google Scholar] [CrossRef]
- Repanas, T.I.; Papanastasiou, C.A.; Efthimiadis, G.K.; Fragkakis, N.; Sachpekidis, V.; Klein, R.M.; Karvounis, H.; Karamitsos, T.D. Cardiovascular Magnetic Resonance as a Complementary Method to Transthoracic Echocardiography for Aortic Valve Area Estimation in Patients with Aortic Stenosis: A Systematic Review and Meta-Analysis. Hell. J. Cardiol. 2021, 62, 107–111. [Google Scholar] [CrossRef]
- Shah, S.M.; Shah, J.; Lakey, S.M.; Garg, P.; Ripley, D.P. Pathophysiology, Emerging Techniques for the Assessment and Novel Treatment of Aortic Stenosis. Open Heart 2023, 10, e002244. [Google Scholar] [CrossRef]
- Villegas-Martinez, M.; De Villedon De Naide, V.; Muthurangu, V.; Bustin, A. The Beating Heart: Artificial Intelligence for Cardiovascular Application in the Clinic. Magn. Reson. Mater. Phys. Biol. Med. 2024, 37, 369–382. [Google Scholar] [CrossRef]
- Ramanauskaitė, D.; Balčiūnaitė, G.; Palionis, D.; Besusparis, J.; Žurauskas, E.; Janušauskas, V.; Zorinas, A.; Valevičienė, N.; Sogaard, P.; Glaveckaitė, S. The Relative Apical Sparing Strain Pattern in Severe Aortic Valve Stenosis: A Marker of Adverse Cardiac Remodeling. JPM 2024, 14, 707. [Google Scholar] [CrossRef]
- Cameli, M. Echocardiography Strain: Why Is It Used More and More? Eur. Heart J. Suppl. 2022, 24, I38–I42. [Google Scholar] [CrossRef] [PubMed]
- Mor-Avi, V.; Lang, R.M.; Badano, L.P.; Belohlavek, M.; Cardim, N.M.; Derumeaux, G.; Galderisi, M.; Marwick, T.; Nagueh, S.F.; Sengupta, P.P.; et al. Current and Evolving Echocardiographic Techniques for the Quantitative Evaluation of Cardiac Mechanics: ASE/EAE Consensus Statement on Methodology and Indications. J. Am. Soc. Echocardiogr. 2011, 24, 277–313. [Google Scholar] [CrossRef] [PubMed]
- Claus, P.; Omar, A.M.S.; Pedrizzetti, G.; Sengupta, P.P.; Nagel, E. Tissue Tracking Technology for Assessing Cardiac Mechanics. JACC Cardiovasc. Imaging 2015, 8, 1444–1460. [Google Scholar] [CrossRef]
- Smiseth, O.A.; Rider, O.; Cvijic, M.; Valkovič, L.; Remme, E.W.; Voigt, J.-U. Myocardial Strain Imaging. JACC Cardiovasc. Imaging 2025, 18, 340–381. [Google Scholar] [CrossRef] [PubMed]
- Obokata, M.; Nagata, Y.; Wu, V.C.-C.; Kado, Y.; Kurabayashi, M.; Otsuji, Y.; Takeuchi, M. Direct Comparison of Cardiac Magnetic Resonance Feature Tracking and 2D/3D Echocardiography Speckle Tracking for Evaluation of Global Left Ventricular Strain. Eur. Heart J. Cardiovasc. Imaging 2016, 17, 525–532. [Google Scholar] [CrossRef]
- Onishi, T.; Saha, S.K.; Ludwig, D.R.; Onishi, T.; Marek, J.J.; Cavalcante, J.L.; Schelbert, E.B.; Schwartzman, D.; Gorcsan, J. Feature Tracking Measurement of Dyssynchrony from Cardiovascular Magnetic Resonance Cine Acquisitions: Comparison with Echocardiographic Speckle Tracking. J. Cardiovasc. Magn. Reson. 2013, 15, 95. [Google Scholar] [CrossRef]
- Yang, C.-H.; Takeuchi, M.; Nabeshima, Y.; Yamashita, E.; Izumo, M.; Ishizu, T.; Seo, Y. Prognostic Value of Apical Sparing of Longitudinal Strain in Patients with Symptomatic Aortic Stenosis. Acta Cardiol. Sin. 2022, 38, 341–351. [Google Scholar] [CrossRef]
- Abecasis, J.; Lopes, P.; Santos, R.R.; Maltês, S.; Guerreiro, S.; Ferreira, A.; Freitas, P.; Ribeiras, R.; Andrade, M.J.; Manso, R.T.; et al. Prevalence and Significance of Relative Apical Sparing in Aortic Stenosis: Insights from an Echo and Cardiovascular Magnetic Resonance Study of Patients Referred for Surgical Aortic Valve Replacement. Eur. Heart J. Cardiovasc. Imaging 2023, 24, 1033–1042. [Google Scholar] [CrossRef]
- Büchi, M.; Hess, O.H.; Murakami, T.; Krayenbuehl, H.P. Left Ventricular Wall Stress Distribution in Chronic Pressure and Volume Overload: Effect of Normal and Depressed Contractility on Regional Stress-Velocity Relations. Basic. Res. Cardiol. 1990, 85, 367–383. [Google Scholar] [CrossRef]
- Pryds, K.; Larsen, A.H.; Hansen, M.S.; Grøndal, A.Y.K.; Tougaard, R.S.; Hansson, N.H.; Clemmensen, T.S.; Løgstrup, B.B.; Wiggers, H.; Kim, W.Y.; et al. Myocardial Strain Assessed by Feature Tracking Cardiac Magnetic Resonance in Patients with a Variety of Cardiovascular Diseases—A Comparison with Echocardiography. Sci. Rep. 2019, 9, 11296. [Google Scholar] [CrossRef]
- Pedrizzetti, G.; Claus, P.; Kilner, P.J.; Nagel, E. Principles of Cardiovascular Magnetic Resonance Feature Tracking and Echocardiographic Speckle Tracking for Informed Clinical Use. J. Cardiovasc. Magn. Reson. 2016, 18, 51. [Google Scholar] [CrossRef] [PubMed]
- Onishi, T.; Saha, S.K.; Delgado-Montero, A.; Ludwig, D.R.; Onishi, T.; Schelbert, E.B.; Schwartzman, D.; Gorcsan, J. Global Longitudinal Strain and Global Circumferential Strain by Speckle-Tracking Echocardiography and Feature-Tracking Cardiac Magnetic Resonance Imaging: Comparison with Left Ventricular Ejection Fraction. J. Am. Soc. Echocardiogr. 2015, 28, 587–596. [Google Scholar] [CrossRef]
- Reichek, N.; Devereux, R.B. Left Ventricular Hypertrophy: Relationship of Anatomic, Echocardiographic and Electrocardiographic Findings. Circulation 1981, 63, 1391–1398. [Google Scholar] [CrossRef] [PubMed]
- Gröschel, J.; Kuhnt, J.; Viezzer, D.; Hadler, T.; Hormes, S.; Barckow, P.; Schulz-Menger, J.; Blaszczyk, E. Comparison of Manual and Artificial Intelligence Based Quantification of Myocardial Strain by Feature Tracking—A Cardiovascular MR Study in Health and Disease. Eur. Radiol. 2023, 34, 1003–1015. [Google Scholar] [CrossRef]
- Dobrovie, M.; Bėzy, S.; Ünlü, S.; Chakraborty, B.; Petrescu, A.; Duchenne, J.; Beela, A.S.; Voigt, J.-U. How Does Regional Hypertrophy Affect Strain Measurements with Different Speckle-Tracking Methods? J. Am. Soc. Echocardiogr. 2019, 32, 1444–1450. [Google Scholar] [CrossRef] [PubMed]
- Muscogiuri, G.; Volpato, V.; Cau, R.; Chiesa, M.; Saba, L.; Guglielmo, M.; Senatieri, A.; Chierchia, G.; Pontone, G.; Dell’Aversana, S.; et al. Application of AI in Cardiovascular Multimodality Imaging. Heliyon 2022, 8, e10872. [Google Scholar] [CrossRef] [PubMed]
- Van Assen, M.; Muscogiuri, G.; Caruso, D.; Lee, S.J.; Laghi, A.; De Cecco, C.N. Artificial Intelligence in Cardiac Radiology. Radiol. Med. 2020, 125, 1186–1199. [Google Scholar] [CrossRef]
- Wang, S.; Patel, H.; Miller, T.; Ameyaw, K.; Narang, A.; Chauhan, D.; Anand, S.; Anyanwu, E.; Besser, S.A.; Kawaji, K.; et al. AI Based CMR Assessment of Biventricular Function. JACC Cardiovasc. Imaging 2022, 15, 413–427. [Google Scholar] [CrossRef]
- Elvas, L.B.; Águas, P.; Ferreira, J.C.; Oliveira, J.P.; Dias, M.S.; Rosário, L.B. AI-Based Aortic Stenosis Classification in MRI Scans. Electronics 2023, 12, 4835. [Google Scholar] [CrossRef]
- Evertz, R.; Lange, T.; Backhaus, S.J.; Schulz, A.; Beuthner, B.E.; Topci, R.; Toischer, K.; Puls, M.; Kowallick, J.T.; Hasenfuß, G.; et al. Artificial Intelligence Enabled Fully Automated CMR Function Quantification for Optimized Risk Stratification in Patients Undergoing Transcatheter Aortic Valve Replacement. J. Interv. Cardiol. 2022, 2022, 1–9. [Google Scholar] [CrossRef]
- Augusto, J.B.; Davies, R.H.; Bhuva, A.N.; Knott, K.D.; Seraphim, A.; Alfarih, M.; Lau, C.; Hughes, R.K.; Lopes, L.R.; Shiwani, H.; et al. Diagnosis and Risk Stratification in Hypertrophic Cardiomyopathy Using Machine Learning Wall Thickness Measurement: A Comparison with Human Test-Retest Performance. Lancet Digit. Health 2021, 3, e20–e28. [Google Scholar] [CrossRef] [PubMed]
- Le, Y.; Zhao, C.; An, J.; Zhou, J.; Deng, D.; He, Y. Progress in the Clinical Application of Artificial Intelligence for Left Ventricle Analysis in Cardiac Magnetic Resonance. Rev. Cardiovasc. Med. 2024, 25, 447. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Guo, Z.; Yuan, J.; Xia, K.; Yu, H. Fine-Grained Calibrated Double-Attention Convolutional Network for Left Ventricular Segmentation. Phys. Med. Biol. 2022, 67, 055013. [Google Scholar] [CrossRef]
- Mariscal-Harana, J.; Kifle, N.; Razavi, R.; King, A.P.; Ruijsink, B.; Puyol-Antón, E. Improved AI-Based Segmentation of Apical and Basal Slices from Clinical Cine CMR; Springer: Berlin/Heidelberg, Germany, 2021. [Google Scholar]
- Assadi, H.; Alabed, S.; Li, R.; Matthews, G.; Karunasaagarar, K.; Kasmai, B.; Nair, S.; Mehmood, Z.; Grafton-Clarke, C.; Swoboda, P.P.; et al. Development and Validation of AI-Derived Segmentation of Four-Chamber Cine Cardiac Magnetic Resonance. Eur. Radiol. Exp. 2024, 8, 77. [Google Scholar] [CrossRef] [PubMed]
- Kuruvilla, S.; Adenaw, N.; Katwal, A.B.; Lipinski, M.J.; Kramer, C.M.; Salerno, M. Late Gadolinium Enhancement on Cardiac Magnetic Resonance Predicts Adverse Cardiovascular Outcomes in Nonischemic Cardiomyopathy: A Systematic Review and Meta-Analysis. Circ. Cardiovasc. Imaging 2014, 7, 250–258. [Google Scholar] [CrossRef]
- Cau, R.; Pisu, F.; Suri, J.S.; Mannelli, L.; Scaglione, M.; Masala, S.; Saba, L. Artificial Intelligence Applications in Cardiovascular Magnetic Resonance Imaging: Are We on the Path to Avoiding the Administration of Contrast Media? Diagnostics 2023, 13, 2061. [Google Scholar] [CrossRef]
- Xu, C.; Howey, J.; Ohorodnyk, P.; Roth, M.; Zhang, H.; Li, S. Segmentation and Quantification of Infarction without Contrast Agents via Spatiotemporal Generative Adversarial Learning. Med. Image Anal. 2020, 59, 101568. [Google Scholar] [CrossRef]
- Zhang, Q.; Burrage, M.K.; Shanmuganathan, M.; Gonzales, R.A.; Lukaschuk, E.; Thomas, K.E.; Mills, R.; Leal Pelado, J.; Nikolaidou, C.; Popescu, I.A.; et al. Artificial Intelligence for Contrast-Free MRI: Scar Assessment in Myocardial Infarction Using Deep Learning–Based Virtual Native Enhancement. Circulation 2022, 146, 1492–1503. [Google Scholar] [CrossRef]
- Mayerhoefer, M.E.; Materka, A.; Langs, G.; Häggström, I.; Szczypiński, P.; Gibbs, P.; Cook, G. Introduction to Radiomics. J. Nucl. Med. 2020, 61, 488–495. [Google Scholar] [CrossRef]
- Raisi-Estabragh, Z.; Izquierdo, C.; Campello, V.M.; Martin-Isla, C.; Jaggi, A.; Harvey, N.C.; Lekadir, K.; Petersen, S.E. Cardiac Magnetic Resonance Radiomics: Basic Principles and Clinical Perspectives. Eur. Heart J. Cardiovasc. Imaging 2020, 21, 349–356. [Google Scholar] [CrossRef]
- Neisius, U.; El-Rewaidy, H.; Nakamori, S.; Rodriguez, J.; Manning, W.J.; Nezafat, R. Radiomic Analysis of Myocardial Native T1 Imaging Discriminates Between Hypertensive Heart Disease and Hypertrophic Cardiomyopathy. JACC Cardiovasc. Imaging 2019, 12, 1946–1954. [Google Scholar] [CrossRef] [PubMed]
- Raisi-Estabragh, Z.; Martin-Isla, C.; Nissen, L.; Szabo, L.; Campello, V.M.; Escalera, S.; Winther, S.; Bøttcher, M.; Lekadir, K.; Petersen, S.E. Radiomics Analysis Enhances the Diagnostic Performance of CMR Stress Perfusion: A Proof-of-Concept Study Using the Dan-NICAD Dataset. Front. Cardiovasc. Med. 2023, 10, 1141026. [Google Scholar] [CrossRef] [PubMed]
- Shinuo, L.; Lu, T. The Clinical Application of Radiomics Models Based on Cardiac Magnetic Resonance (CMR) Non-Contrast-Enhanced T1 Mapping for Discriminating Acute and Chronic Myocardial Infarction. J. Cardiovasc. Magn. Reson. 2025, 27, 101218. [Google Scholar] [CrossRef]
- Avard, E.; Shiri, I.; Hajianfar, G.; Abdollahi, H.; Kalantari, K.R.; Houshmand, G.; Kasani, K.; Bitarafan-rajabi, A.; Deevband, M.R.; Oveisi, M.; et al. Non-Contrast Cine Cardiac Magnetic Resonance Image Radiomics Features and Machine Learning Algorithms for Myocardial Infarction Detection. Comput. Biol. Med. 2022, 141, 105145. [Google Scholar] [CrossRef]
- Nakamori, S.; Amyar, A.; Fahmy, A.S.; Ngo, L.H.; Ishida, M.; Nakamura, S.; Omori, T.; Moriwaki, K.; Fujimoto, N.; Imanaka-Yoshida, K.; et al. Cardiovascular Magnetic Resonance Radiomics to Identify Components of the Extracellular Matrix in Dilated Cardiomyopathy. Circulation 2024, 150, 7–18. [Google Scholar] [CrossRef]
- Mushari, N.A.; Soultanidis, G.; Duff, L.; Trivieri, M.G.; Fayad, Z.A.; Robson, P.; Tsoumpas, C. An Assessment of PET and CMR Radiomic Features for the Detection of Cardiac Sarcoidosis. Front. Nucl. Med. 2024, 4, 1324698. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Mui, D.; Chirinos, J.A.; Zamani, P.; Ferrari, V.A.; Chen, Y.; Han, Y. Comparing Cardiovascular Magnetic Resonance Strain Software Packages by Their Abilities to Discriminate Outcomes in Patients with Heart Failure with Preserved Ejection Fraction. J. Cardiovasc. Magn. Reson. 2021, 23, 55. [Google Scholar] [CrossRef]
- Chudgar, P.D.; Burkule, N.J.; Kamat, N.V.; Rege, G.M.; Jantre, M.N. Myocardial Strain Imaging Using Feature Tracking Method of Cardiac MRI: Our Initial Experience of This Novel Parameter as an Additional Diagnostic Tool. Indian. J. Radiol. Imaging 2022, 32, 479–487. [Google Scholar] [CrossRef]
- Van Der Ven, J.P.G.; Van Genuchten, W.; Sadighy, Z.; Valsangiacomo Buechel, E.R.; Sarikouch, S.; Boersma, E.; Helbing, W.A. Multivendor Evaluation of Automated MRI Postprocessing of Biventricular Size and Function for Children with and Without Congenital Heart Defects. Magn. Reson. Imaging 2023, 58, 794–804. [Google Scholar] [CrossRef]
- Gao, X.; Abdi, M.; Auger, D.A.; Sun, C.; Hanson, C.A.; Robinson, A.A.; Schumann, C.; Oomen, P.J.; Ratcliffe, S.; Malhotra, R.; et al. Cardiac Magnetic Resonance Assessment of Response to Cardiac Resynchronization Therapy and Programming Strategies. JACC Cardiovasc. Imaging 2021, 14, 2369–2383. [Google Scholar] [CrossRef]
- Gil, K.E.; Truong, V.; Liu, C.; Ibrahim, D.Y.; Mikrut, K.; Satoskar, A.; Varghese, J.; Kahwash, R.; Han, Y. Distinguishing Hypertensive Cardiomyopathy from Cardiac Amyloidosis in Hypertensive Patients with Heart Failure: A CMR Study with Histological Confirmation. Int. J. Cardiovasc. Imaging 2024, 40, 2559–2570. [Google Scholar] [CrossRef] [PubMed]
- Backhaus, S.J.; Schuster, A.; Lange, T.; Stehning, C.; Billing, M.; Lotz, J.; Pieske, B.; Hasenfuß, G.; Kelle, S.; Kowallick, J.T. Impact of Fully Automated Assessment on Interstudy Reproducibility of Biventricular Volumes and Function in Cardiac Magnetic Resonance Imaging. Sci. Rep. 2021, 11, 11648. [Google Scholar] [CrossRef] [PubMed]
| Group Statistics and Independent Samples T-Test for Equality of Means: Mean GLS in Cardiac Segments Between Aortic Stenosis Patients and Healthy Controls | ||||||
|---|---|---|---|---|---|---|
| Aortic Stenosis | N | Mean | Mean Difference | Std, Deviation | Two-Sided p | |
| Basal Anterior | No | 40 | −24.89 | 3.23 | 4.85 | 0.004 |
| Yes | 68 | −21.66 | 6.48 | |||
| Basal Anteroseptal | No | 40 | −21.03 | 6.11 | 5.40 | <0.001 |
| Yes | 68 | −14.92 | 5.83 | |||
| Basal Inferoseptal | No | 40 | −28.09 | 7.86 | 3.58 | <0.001 |
| Yes | 68 | −20.23 | 5.77 | |||
| Basal Inferior | No | 40 | −36.12 | 6.41 | 3.73 | <0.001 |
| Yes | 68 | −29.72 | 5.05 | |||
| Basal Inferolateral | No | 40 | −36.10 | 6.20 | 3.51 | <0.001 |
| Yes | 68 | −29.90 | 5.69 | |||
| Basal Anterolateral | No | 40 | −31.41 | 3.05 | 4.86 | 0.009 |
| Yes | 68 | −28.36 | 6.16 | |||
| Mid Anterior | No | 40 | −18.65 | 4.60 | 5.10 | <0.001 |
| Yes | 68 | −14.05 | 5.11 | |||
| Mid Anteroseptal | No | 40 | −16.23 | −0.23 | 4.63 | 0.837 |
| Yes | 68 | −16.46 | 6.09 | |||
| Mid Inferoseptal | No | 40 | −13.64 | −0.87 | 5.52 | 0.473 |
| Yes | 68 | −14.50 | 6.31 | |||
| Mid Inferior | No | 40 | −16.84 | 3.22 | 5.16 | 0.007 |
| Yes | 68 | −13.62 | 6.19 | |||
| Mid Inferolateral | No | 40 | −15.35 | 1.09 | 5.36 | 0.385 |
| Yes | 68 | −14.26 | 6.75 | |||
| Mid Anterolateral | No | 40 | −19.29 | 2.29 | 5.38 | 0.079 |
| Yes | 68 | −17.00 | 7.04 | |||
| Apical Anterior | No | 40 | −15.08 | −0.64 | 5.04 | 0.605 |
| Yes | 68 | −15.72 | 6.81 | |||
| Apical Septal | No | 40 | −16.79 | 0.57 | 4.34 | 0.592 |
| Yes | 70 | −16.23 | 6.69 | |||
| Apical Inferior | No | 40 | −10.59 | −0.96 | 6.27 | 0.423 |
| Yes | 68 | −11.55 | 5.80 | |||
| Apical Lateral | No | 40 | −14.92 | 3.48 | 5.68 | 0.002 |
| Yes | 70 | −11.45 | 5.38 | |||
| Apex | No | 40 | −16.90 | 2.35 | 4.13 | 0.013 |
| Yes | 70 | −14.55 | 5.54 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Published by MDPI on behalf of the Lithuanian University of Health Sciences. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abramikas, Ž.; Jasiukevičiūtė, I.; Balčiūnaitė, G.; Glaveckaitė, S.; Palionis, D.; Valevičienė, N. Artificial Intelligence Performance in Cardiac Magnetic Resonance Strain Analysis for Aortic Stenosis: Validation with Echocardiography and Healthy Controls. Medicina 2025, 61, 950. https://doi.org/10.3390/medicina61060950
Abramikas Ž, Jasiukevičiūtė I, Balčiūnaitė G, Glaveckaitė S, Palionis D, Valevičienė N. Artificial Intelligence Performance in Cardiac Magnetic Resonance Strain Analysis for Aortic Stenosis: Validation with Echocardiography and Healthy Controls. Medicina. 2025; 61(6):950. https://doi.org/10.3390/medicina61060950
Chicago/Turabian StyleAbramikas, Žygimantas, Ieva Jasiukevičiūtė, Giedrė Balčiūnaitė, Sigita Glaveckaitė, Darius Palionis, and Nomeda Valevičienė. 2025. "Artificial Intelligence Performance in Cardiac Magnetic Resonance Strain Analysis for Aortic Stenosis: Validation with Echocardiography and Healthy Controls" Medicina 61, no. 6: 950. https://doi.org/10.3390/medicina61060950
APA StyleAbramikas, Ž., Jasiukevičiūtė, I., Balčiūnaitė, G., Glaveckaitė, S., Palionis, D., & Valevičienė, N. (2025). Artificial Intelligence Performance in Cardiac Magnetic Resonance Strain Analysis for Aortic Stenosis: Validation with Echocardiography and Healthy Controls. Medicina, 61(6), 950. https://doi.org/10.3390/medicina61060950





