Terpinen-4-ol Targets HIF-1α/TGF-β1/TNF-α Axis to Attenuate Ethanol-Induced Hepatotoxicity: Network Pharmacology and In Vitro Validation
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. In Silico Study—Network Pharmacology Analysis
2.2.1. T4OL and ALD-Associated Targets Prediction
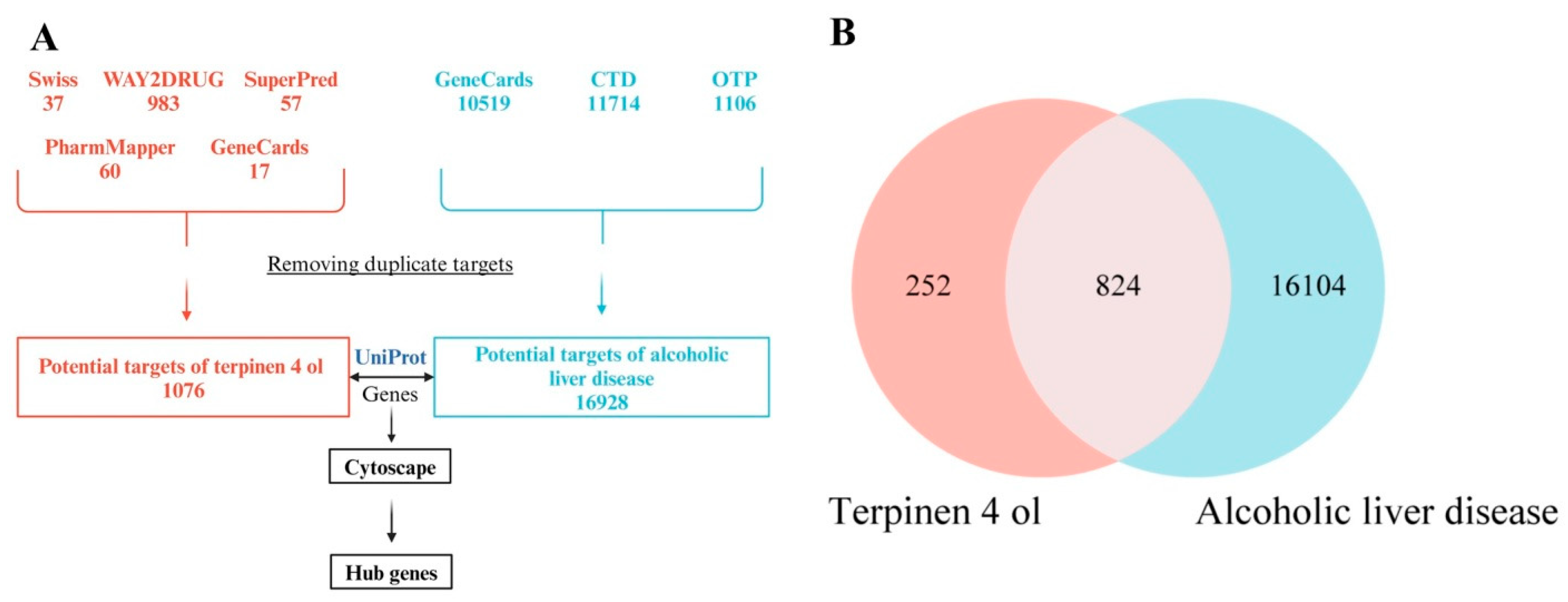
2.2.2. Gene Matching and Venn Diagram Construction
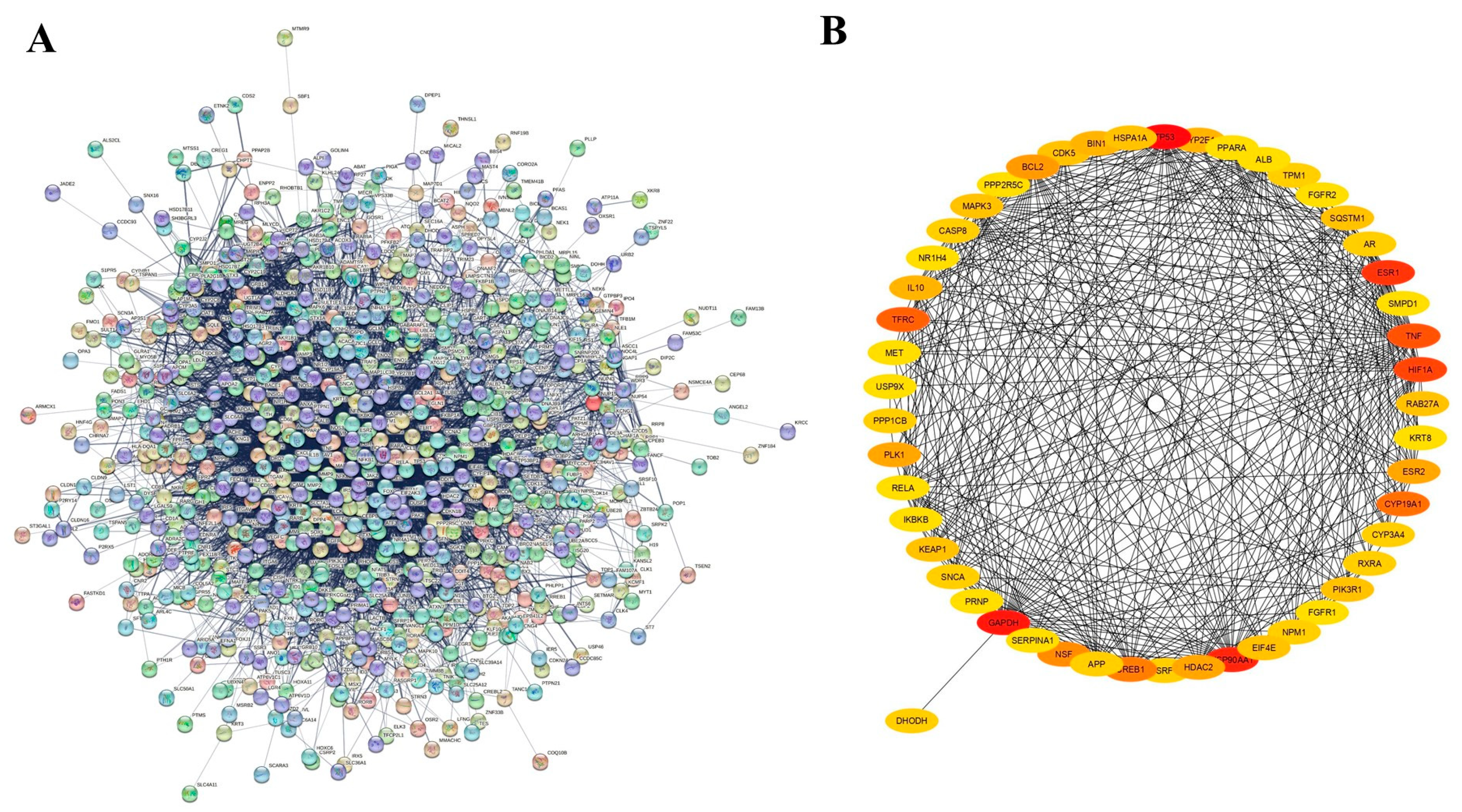
2.2.3. Protein–Protein Interaction (PPI) Network
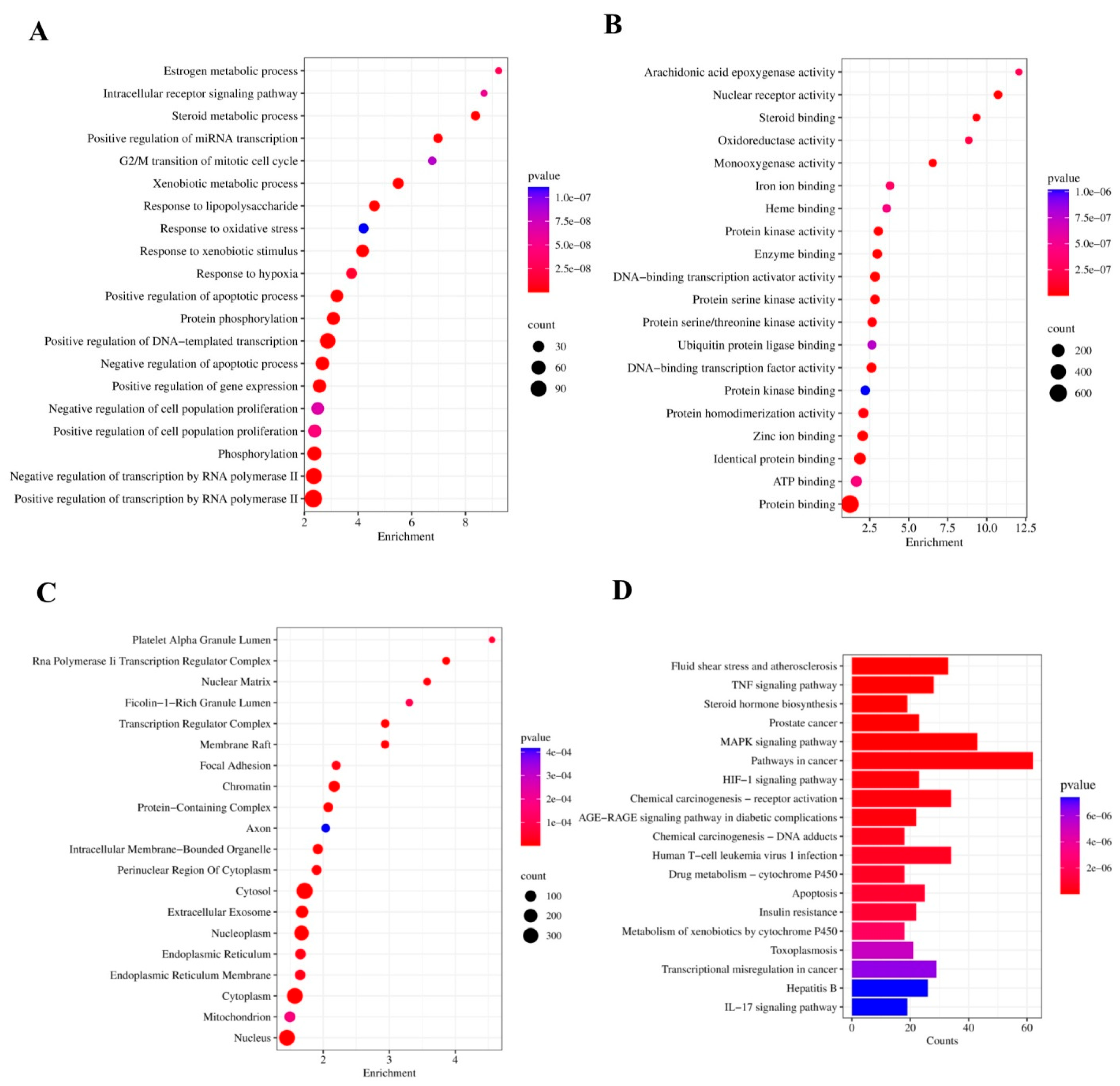
2.2.4. Gene Ontology and Functional Enrichment Analysis
2.3. Molecular Docking
2.3.1. Preparation of Ligand and Target Proteins
2.3.2. Active Binding Site Estimation
2.3.3. Docking of Ligand with Target Proteins
2.4. HepG2 Cell Culture
2.4.1. Cytotoxic Evaluation of T4OL and Ethanol
2.4.2. Determination of Hepatoprotective Effect of T4OL
- Control—Complete growth medium;
- Disease group—8% ethanol in complete growth medium;
- SIL 200 µg/mL—200 µg/mL of SIL in complete growth medium;
- T4OL (13 µM)—13 µM of T4OL in complete growth medium;
- T4OL (130 µM)—130 µM of T4OL in complete growth medium;
- T4OL (650 µM)—650 µM of T4OL in complete growth medium;
- T4OL (1.3 mM)—1.3 mM of T4OL in complete growth medium.
2.5. Cell Viability Assays
2.5.1. MTT Assay
2.5.2. Crystal Violet Test
2.6. Antioxidant Assays
2.6.1. Glutathione Reductase (GSH) Assay
2.6.2. Superoxide Dismutase (SOD) Assay
2.6.3. Real-Time PCR Analysis
2.7. Statistical Analysis
3. Results
3.1. Prediction of the Biological Spectrum
3.2. Strong Binding Interactions of T4OL with Target Proteins
3.3. Target Proteins of T4OL Linked with ALD
3.4. GO Analysis
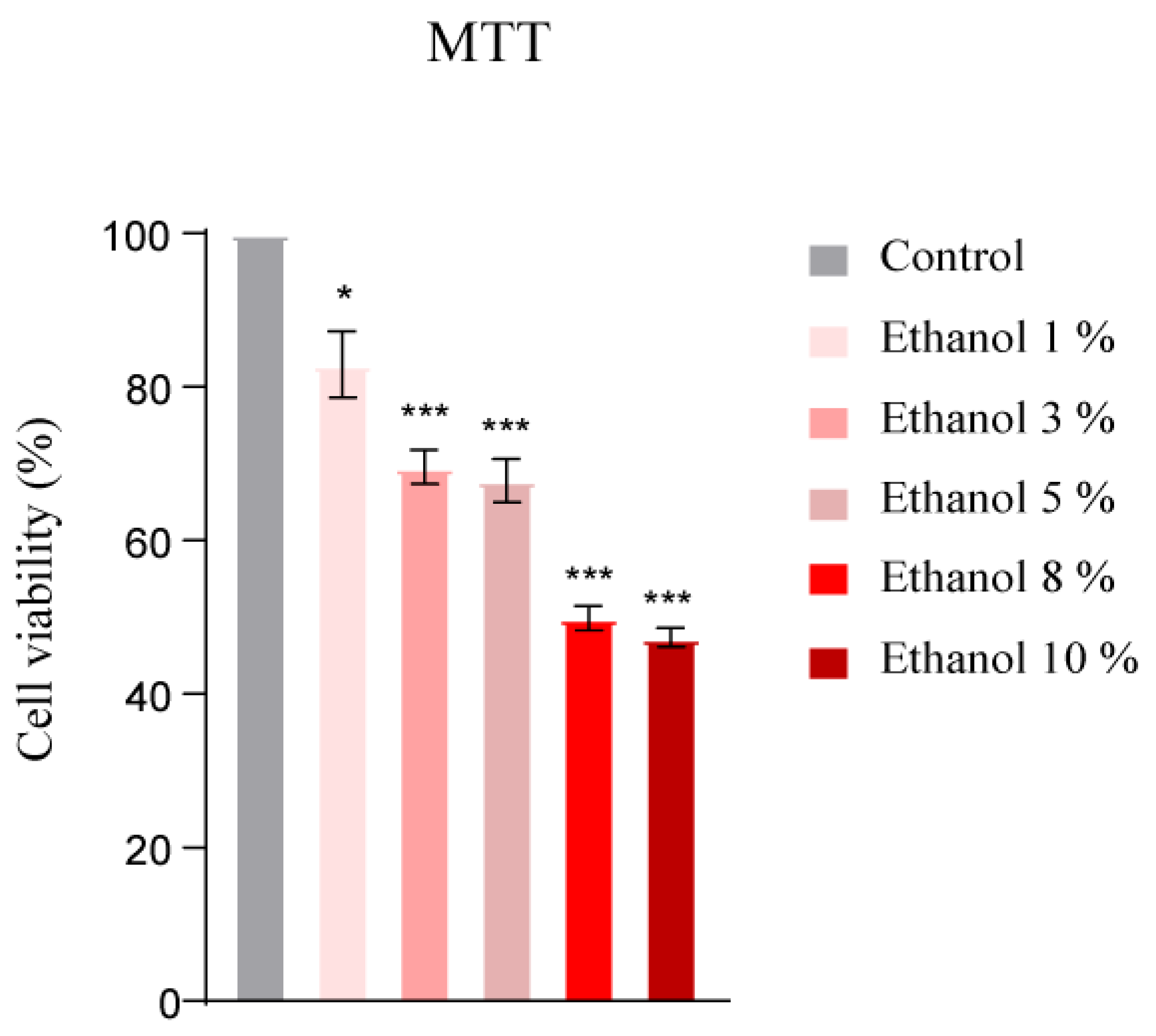
3.5. Cytotoxic Dose Determination of Ethanol and T4OL by MTT Assay
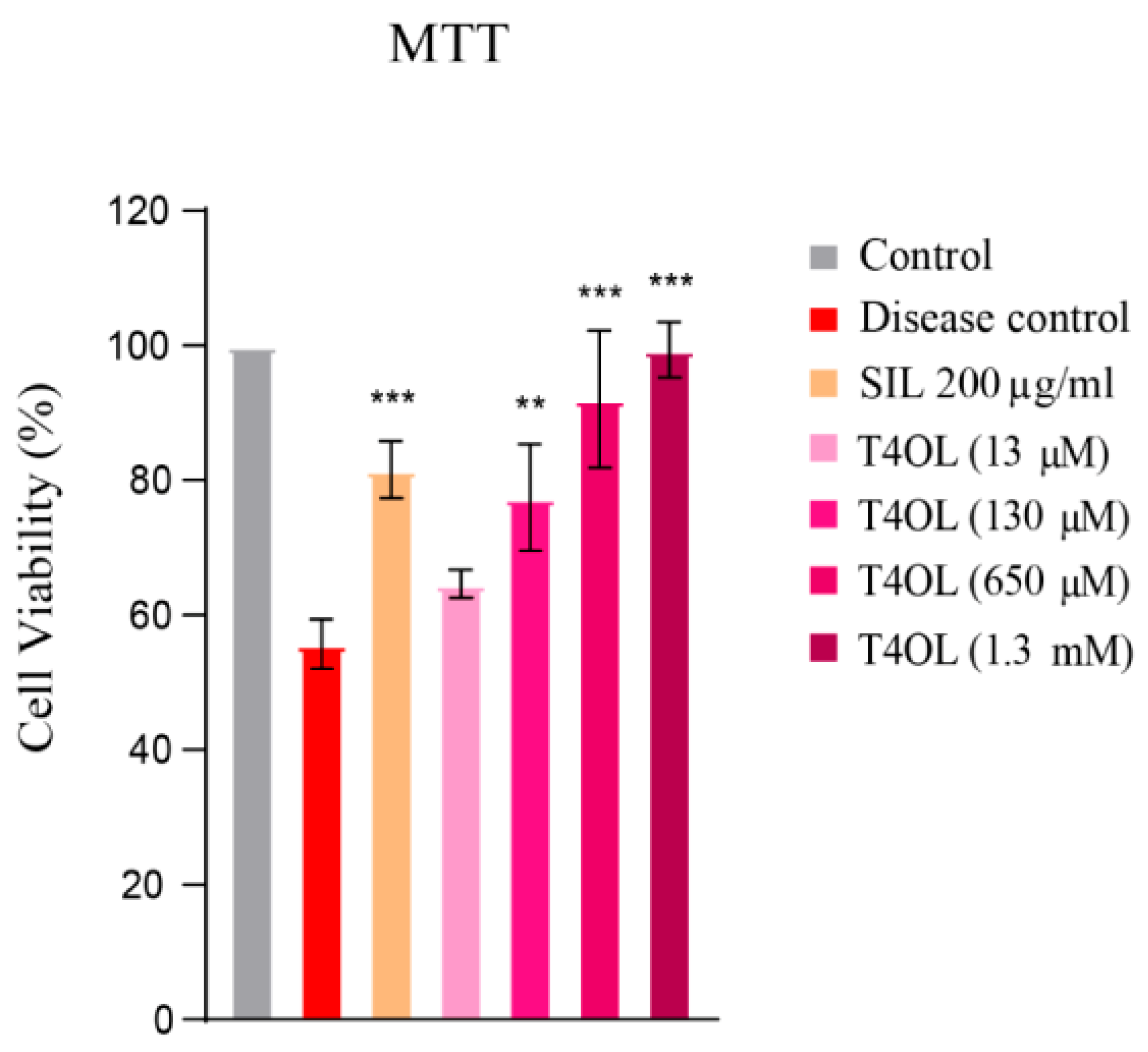
3.6. T4OL’s Hepatoprotective Response to Ethanol-Induced Cellular Damage
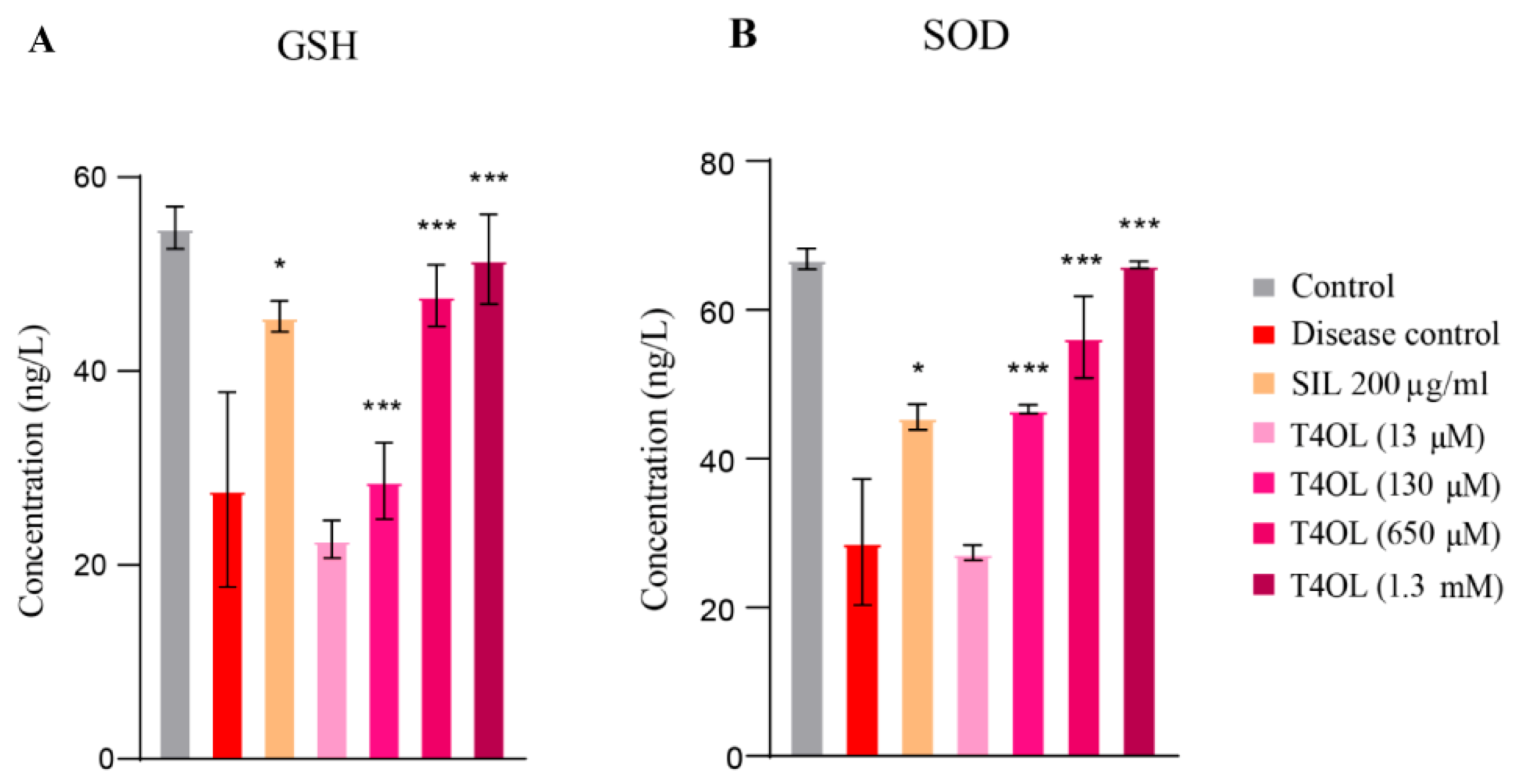
3.7. T4OL Restored GSH and SOD Levels
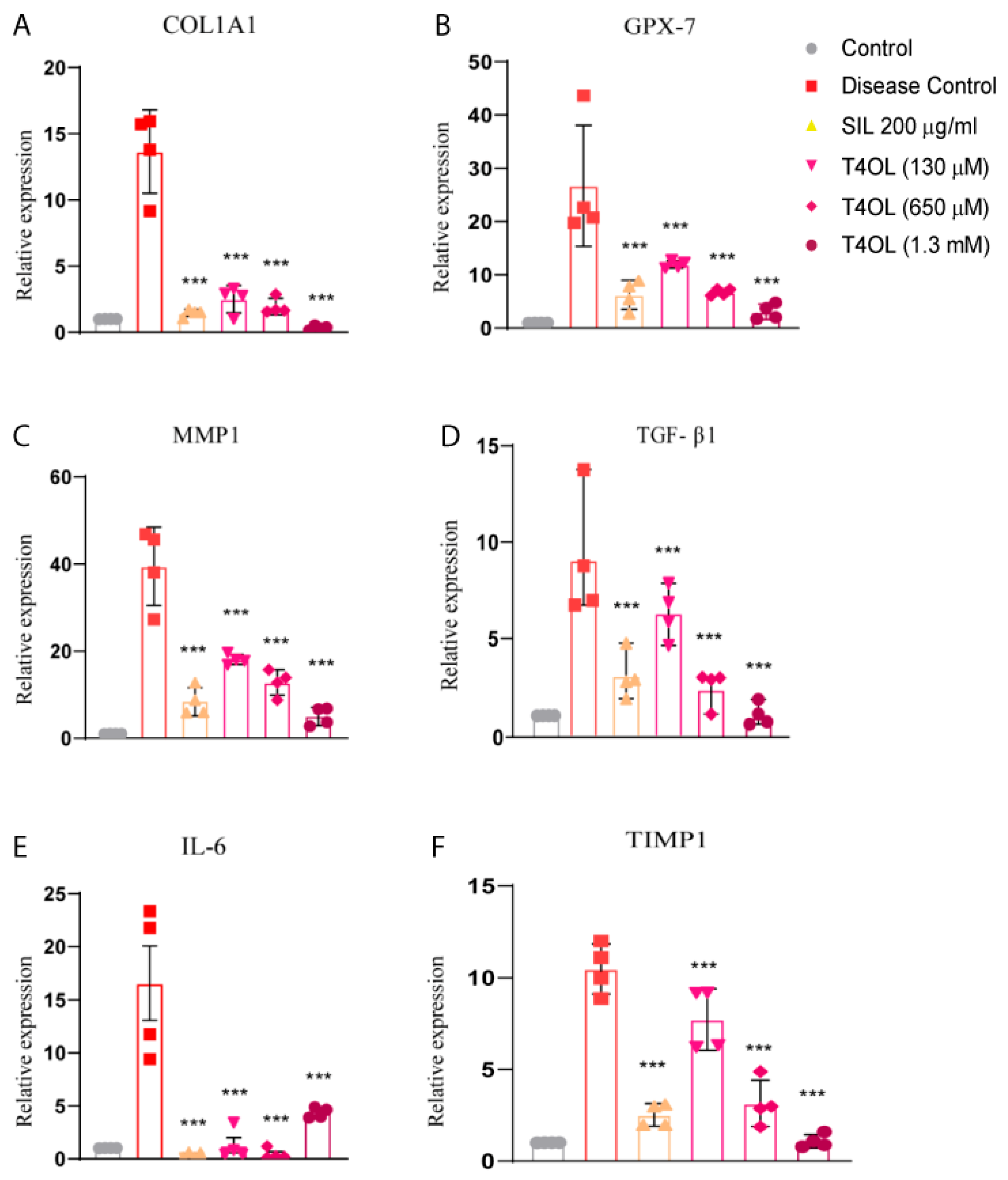
3.8. T4OL Reduced mRNA Levels of Inflammatory and Fibrotic Markers
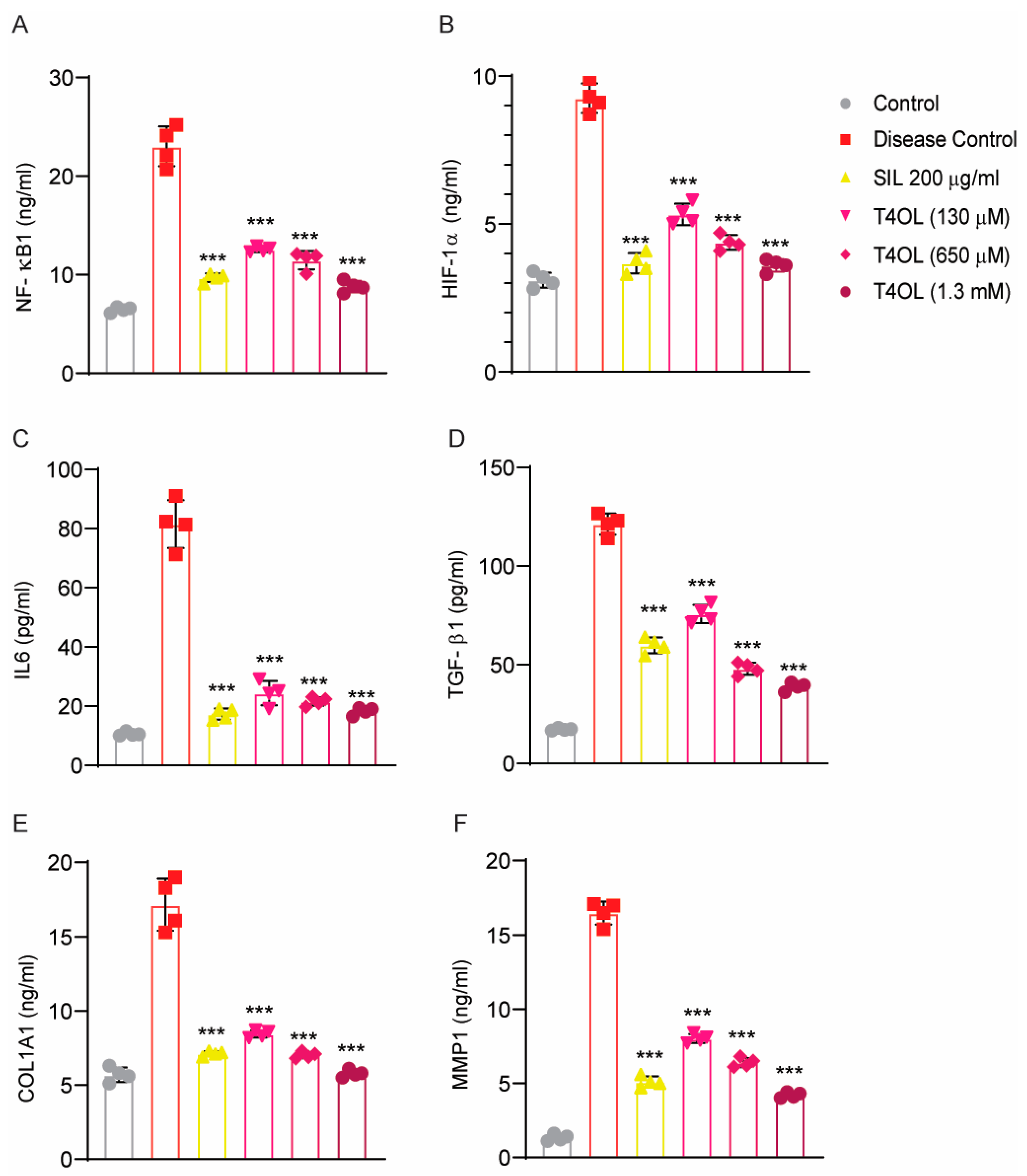
3.9. Dose-Dependent Amelioration of Alcohol-Induced Liver Injury by T40L via Inhibition of Inflammatory, Hypoxic, and Fibrotic Signaling
4. Discussion
- (1)
- Anti-fibrotic—by suppressing COL1A1, TGF-β1, and TIMP-1;
- (2)
- Anti-inflammatory—by attenuating IL-6 expression;
- (3)
- Antioxidant—by normalizing GPX7 and HIF-1α levels and increasing GSH and SOD.
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Navarro, V.J.; Senior, J.R. Drug-related hepatotoxicity. N. Engl. J. Med. 2006, 354, 731–739. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.K.; Balamurugan, A.; Saha, P.K.; Pandey, R.M.; Mehra, N.K. Evaluation of clinical and immunogenetic risk factors for the development of hepatotoxicity during antituberculosis treatment. Am. J. Respir. Crit. Care Med. 2002, 166, 916–919. [Google Scholar] [CrossRef]
- Seitz, H.K.; Bataller, R.; Cortez-Pinto, H.; Gao, B.; Gual, A.; Lackner, C.; Mathurin, P.; Mueller, S.; Szabo, G.; Tsukamoto, H. Alcoholic liver disease. Nat. Rev. Dis. Primer 2018, 4, 16. [Google Scholar] [CrossRef] [PubMed]
- Seitz, H.K.; Neuman, M.G. The history of alcoholic liver disease: From an unrecognized disease to one of the most frequent diseases in hepatology. J. Clin. Med. 2021, 10, 858. [Google Scholar] [CrossRef]
- Kong, L.Z.; Chandimali, N.; Han, Y.H.; Lee, D.H.; Kim, J.S.; Kim, S.U.; Kim, T.D.; Jeong, D.K.; Sun, H.N.; Lee, D.S.; et al. Pathogenesis, Early Diagnosis, and Therapeutic Management of Alcoholic Liver Disease. Int. J. Mol. Sci. 2019, 20, 2712. [Google Scholar] [CrossRef] [PubMed]
- Singal, A.K.; Bataller, R.; Ahn, J.; Kamath, P.S.; Shah, V.H. ACG Clinical Guideline: Alcoholic Liver Disease. Am. J. Gastroenterol. 2018, 113, 175–194. [Google Scholar] [CrossRef]
- Gao, B.; Bataller, R. Alcoholic liver disease: Pathogenesis and new therapeutic targets. Gastroenterology 2011, 141, 1572–1585. [Google Scholar] [CrossRef]
- Morley, K.C.; Leung, S.; Baillie, A.; Haber, P.S. The efficacy and biobehavioural basis of baclofen in the treatment of alcoholic liver disease (BacALD): Study protocol for a randomised controlled trial. Contemp. Clin. Trials 2013, 36, 348–355. [Google Scholar] [CrossRef]
- Chen, Y.; Singh, S.; Matsumoto, A.; Manna, S.K.; Abdelmegeed, M.A.; Golla, S.; Murphy, R.C.; Dong, H.; Song, B.J.; Gonzalez, F.J.; et al. Chronic Glutathione Depletion Confers Protection against Alcohol-induced Steatosis: Implication for Redox Activation of AMP-activated Protein Kinase Pathway. Sci. Rep. 2016, 6, 29743. [Google Scholar] [CrossRef]
- Zhao, H.; Zhao, C.; Dong, Y.; Zhang, M.; Wang, Y.; Li, F.; Li, X.; McClain, C.; Yang, S.; Feng, W. Inhibition of miR122a by Lactobacillus rhamnosus GG culture supernatant increases intestinal occludin expression and protects mice from alcoholic liver disease. Toxicol. Lett. 2015, 234, 194–200. [Google Scholar] [CrossRef]
- Muriel, P.; Rivera-Espinoza, Y. Beneficial drugs for liver diseases. J. Appl. Toxicol. 2008, 28, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Osna, N.A.; Donohue, T.M., Jr.; Kharbanda, K.K. Alcoholic Liver Disease: Pathogenesis and Current Management. Alcohol. Res. 2017, 38, 147–161. [Google Scholar]
- Nault, J.C.; Bioulac–Sage, P.; Zucman–Rossi, J. Reviews in basic and clinical gastroenterology and hepatology. Gastroenterology 2013, 144, 888–902. [Google Scholar] [CrossRef]
- Abe, R.; Ohtani, K. An ethnobotanical study of medicinal plants and traditional therapies on Batan Island, the Philippines. J. Ethnopharmacol. 2013, 145, 554–565. [Google Scholar] [CrossRef]
- Atanasov, A.G.; Waltenberger, B.; Pferschy-Wenzig, E.M.; Linder, T.; Wawrosch, C.; Uhrin, P.; Temml, V.; Wang, L.; Schwaiger, S.; Heiss, E.H.; et al. Discovery and resupply of pharmacologically active plant-derived natural products: A review. Biotechnol. Adv. 2015, 33, 1582–1614. [Google Scholar] [CrossRef]
- Yadav, E.; Rao, R. A promising bioactive component terpinen-4-ol: A review. Int. J. Pharmacog. 2016, 1, 336–345. [Google Scholar]
- Bakkali, F.; Averbeck, S.; Averbeck, D.; Idaomar, M. Biological effects of essential oils—A review. Food Chem. Toxicol. 2008, 46, 446–475. [Google Scholar] [CrossRef]
- Brand, C.; Ferrante, A.; Prager, R.H.; Riley, T.V.; Carson, C.F.; Finlay-Jones, J.J.; Hart, P.H. The water-soluble components of the essential oil of Melaleuca alternifolia (tea tree oil) suppress the production of superoxide by human monocytes, but not neutrophils, activated in vitro. Inflamm. Res. 2001, 50, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Calcabrini, A.; Stringaro, A.; Toccacieli, L.; Meschini, S.; Marra, M.; Colone, M.; Salvatore, G.; Mondello, F.; Arancia, G.; Molinari, A. Terpinen-4-ol, the main component of Melaleuca alternifolia (tea tree) oil inhibits the in vitro growth of human melanoma cells. J. Invest. Dermatol. 2004, 122, 349–360. [Google Scholar] [CrossRef]
- Gfeller, D.; Grosdidier, A.; Wirth, M.; Daina, A.; Michielin, O.; Zoete, V. SwissTargetPrediction: A web server for target prediction of bioactive small molecules. Nucleic Acids Res. 2014, 42, W32–W38. [Google Scholar] [CrossRef]
- Lagunin, A.; Ivanov, S.; Rudik, A.; Filimonov, D.; Poroikov, V. DIGEP-Pred: Web service for in silico prediction of drug-induced gene expression profiles based on structural formula. Bioinformatics 2013, 29, 2062–2063. [Google Scholar] [CrossRef] [PubMed]
- Pogodin, P.V.; Lagunin, A.A.; Filimonov, D.A.; Poroikov, V.V. PASS Targets: Ligand-based multi-target computational system based on a public data and naive Bayes approach. SAR QSAR Env. Res. 2015, 26, 783–793. [Google Scholar] [CrossRef] [PubMed]
- Gallo, K.; Goede, A.; Preissner, R.; Gohlke, B.O. SuperPred 3.0: Drug classification and target prediction-a machine learning approach. Nucleic Acids Res. 2022, 50, W726–W731. [Google Scholar] [CrossRef]
- Wang, X.; Shen, Y.; Wang, S.; Li, S.; Zhang, W.; Liu, X.; Lai, L.; Pei, J.; Li, H. PharmMapper 2017 update: A web server for potential drug target identification with a comprehensive target pharmacophore database. Nucleic Acids Res. 2017, 45, W356–W360. [Google Scholar] [CrossRef] [PubMed]
- Ochoa, D.; Hercules, A.; Carmona, M.; Suveges, D.; Gonzalez-Uriarte, A.; Malangone, C.; Miranda, A.; Fumis, L.; Carvalho-Silva, D.; Spitzer, M.; et al. Open Targets Platform: Supporting systematic drug-target identification and prioritisation. Nucleic Acids Res. 2021, 49, D1302–D1310. [Google Scholar] [CrossRef]
- Jia, A.; Xu, L.; Wang, Y. Venn diagrams in bioinformatics. Brief. Bioinform. 2021, 22, bbab108. [Google Scholar] [CrossRef]
- Tang, D.; Chen, M.; Huang, X.; Zhang, G.; Zeng, L.; Zhang, G.; Wu, S.; Wang, Y. SRplot: A free online platform for data visualization and graphing. PLoS ONE 2023, 18, e0294236. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Mering, C.v.; Huynen, M.; Jaeggi, D.; Schmidt, S.; Bork, P.; Snel, B. STRING: A database of predicted functional associations between proteins. Nucleic Acids Res. 2003, 31, 258–261. [Google Scholar] [CrossRef]
- Consortium, G.O. The gene ontology (GO) project in 2006. Nucleic Acids Res. 2006, 34, D322–D326. [Google Scholar] [CrossRef]
- Gao, Y.-D.; Huang, J.-F. An extension strategy of Discovery Studio 2.0 for non-bonded interaction energy automatic calculation at the residue level. Dongwuxue Yanjiu 2011, 32, 262–266. [Google Scholar] [PubMed]
- Huxford, T.; Huang, D.B.; Malek, S.; Ghosh, G. The crystal structure of the IkappaBalpha/NF-kappaB complex reveals mechanisms of NF-kappaB inactivation. Cell 1998, 95, 759–770. [Google Scholar] [CrossRef] [PubMed]
- Sharma, U.; Carrique, L.; Vadon-Le Goff, S.; Mariano, N.; Georges, R.N.; Delolme, F.; Koivunen, P.; Myllyharju, J.; Moali, C.; Aghajari, N.; et al. Structural basis of homo- and heterotrimerization of collagen I. Nat. Commun. 2017, 8, 14671. [Google Scholar] [CrossRef]
- Tian, W.; Chen, C.; Lei, X.; Zhao, J.; Liang, J. CASTp 3.0: Computed atlas of surface topography of proteins. Nucleic Acids Res. 2018, 46, W363–W367. [Google Scholar] [CrossRef]
- Maqbool, T.; Awan, S.J.; Malik, S.; Hadi, F.; Shehzadi, S.; Tariq, K. In-Vitro Anti-Proliferative, Apoptotic and Antioxidative Activities of Medicinal Herb Kalonji (Nigella sativa). Curr. Pharm. Biotechnol. 2019, 20, 1288–1308. [Google Scholar] [CrossRef]
- Lee, J.Y.; Kim, H.; Jeong, Y.; Kang, C.H. Lactic Acid Bacteria Exert a Hepatoprotective Effect against Ethanol-Induced Liver Injury in HepG2 Cells. Microorganisms 2021, 9, 1844. [Google Scholar] [CrossRef]
- Pouresmaeil, V.; Al Abudi, A.H.; Mahimid, A.H.; Sarafraz Yazdi, M.; Es-Haghi, A. Evaluation of Serum Selenium and Copper Levels with Inflammatory Cytokines and Indices of Oxidative Stress in Type 2 Diabetes. Biol. Trace Elem. Res. 2023, 201, 617–626. [Google Scholar] [CrossRef] [PubMed]
- Valko, M.; Leibfritz, D.; Moncol, J.; Cronin, M.T.; Mazur, M.; Telser, J. Free radicals and antioxidants in normal physiological functions and human disease. Int. J. Biochem. Cell Biol. 2007, 39, 44–84. [Google Scholar] [CrossRef]
- Kumar, C.H.; Ramesh, A.; Kumar, J.S.; Ishaq, B.M. A review on hepatoprotective activity of medicinal plants. Int. J. Pharm. Sci. Res. 2011, 2, 501. [Google Scholar]
- Gan, Y.; Tong, J.; Zhou, X.; Long, X.; Pan, Y.; Liu, W.; Zhao, X. Hepatoprotective Effect of Lactobacillus plantarum HFY09 on Ethanol-Induced Liver Injury in Mice. Front. Nutr. 2021, 8, 684588. [Google Scholar] [CrossRef]
- Zhang, S.; Gong, Y.; Xiao, J.; Chai, Y.; Lei, J.; Huang, H.; Xiang, T.; Shen, W. A COL1A1 promoter-controlled expression of TGF-β soluble receptor inhibits hepatic fibrosis without triggering autoimmune responses. Dig. Dis. Sci. 2018, 63, 2662–2672. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, M.; Nomoto, S.; Hishida, M.; Inokawa, Y.; Kanda, M.; Okamura, Y.; Nishikawa, Y.; Tanaka, C.; Kobayashi, D.; Yamada, S. Identification of the collagen type 1 alpha 1 gene (COL1A1) as a candidate survival-related factor associated with hepatocellular carcinoma. BMC Cancer 2014, 14, 108. [Google Scholar] [CrossRef] [PubMed]
- Gressner, A.; Weiskirchen, R. Modern pathogenetic concepts of liver fibrosis suggest stellate cells and TGF-β as major players and therapeutic targets. J. Cell Mol. Med. 2006, 10, 76–99. [Google Scholar] [CrossRef] [PubMed]
- Dooley, S.; ten Dijke, P. TGF-beta in progression of liver disease. Cell Tissue Res. 2012, 347, 245–256. [Google Scholar] [CrossRef]
- Hemmann, S.; Graf, J.; Roderfeld, M.; Roeb, E. Expression of MMPs and TIMPs in liver fibrosis—A systematic review with special emphasis on anti-fibrotic strategies. J. Hepatol. 2007, 46, 955–975. [Google Scholar] [CrossRef]
- Lin, J.; Deng, C.; Peng, Y.; Zheng, J.; Wei, L.; Shi, Y.; Gong, Z.; Hu, G. Dynamic Changes in MMP1 and TIMP1 in the Antifibrotic Process of Dahuang Zhechong Pill in Rats with Liver Fibrosis. Open Chem. 2019, 17, 346–356. [Google Scholar] [CrossRef]
- Hou, X.; Yin, S.; Ren, R.; Liu, S.; Yong, L.; Liu, Y.; Li, Y.; Zheng, M.H.; Kunos, G.; Gao, B.; et al. Myeloid-Cell-Specific IL-6 Signaling Promotes MicroRNA-223-Enriched Exosome Production to Attenuate NAFLD-Associated Fibrosis. Hepatology 2021, 74, 116–132. [Google Scholar] [CrossRef]
- Scarlata, G.G.M.; Colaci, C.; Scarcella, M.; Dallio, M.; Federico, A.; Boccuto, L.; Abenavoli, L. The Role of Cytokines in the Pathogenesis and Treatment of Alcoholic Liver Disease. Diseases 2024, 12, 69. [Google Scholar] [CrossRef]
- Schmidt-Arras, D.; Rose-John, S. IL-6 pathway in the liver: From physiopathology to therapy. J. Hepatol. 2016, 64, 1403–1415. [Google Scholar] [CrossRef]
- Yao, J.; Chen, X.; Liu, Z.; Zhang, R.; Zhang, C.; Yang, Q.; Yao, P.; Jiang, Q.; Wu, J.; Zhao, S. The increasing expression of GPX7 related to the malignant clinical features leading to poor prognosis of glioma patients. Chin. Neurosurg. J. 2021, 7, 21. [Google Scholar] [CrossRef]
- Xiao, Z.; Yu, X.; Zhang, S.; Liang, A. The Expression Levels and Significance of GSH, MDA, SOD, and 8-OHdG in Osteochondral Defects of Rabbit Knee Joints. Biomed. Res. Int. 2022, 2022, 6916179. [Google Scholar] [CrossRef] [PubMed]
- Bostrom, P.; Magnusson, B.; Svensson, P.A.; Wiklund, O.; Boren, J.; Carlsson, L.M.; Stahlman, M.; Olofsson, S.O.; Hulten, L.M. Hypoxia converts human macrophages into triglyceride-loaded foam cells. Arter. Thromb. Vasc. Biol. 2006, 26, 1871–1876. [Google Scholar] [CrossRef] [PubMed]
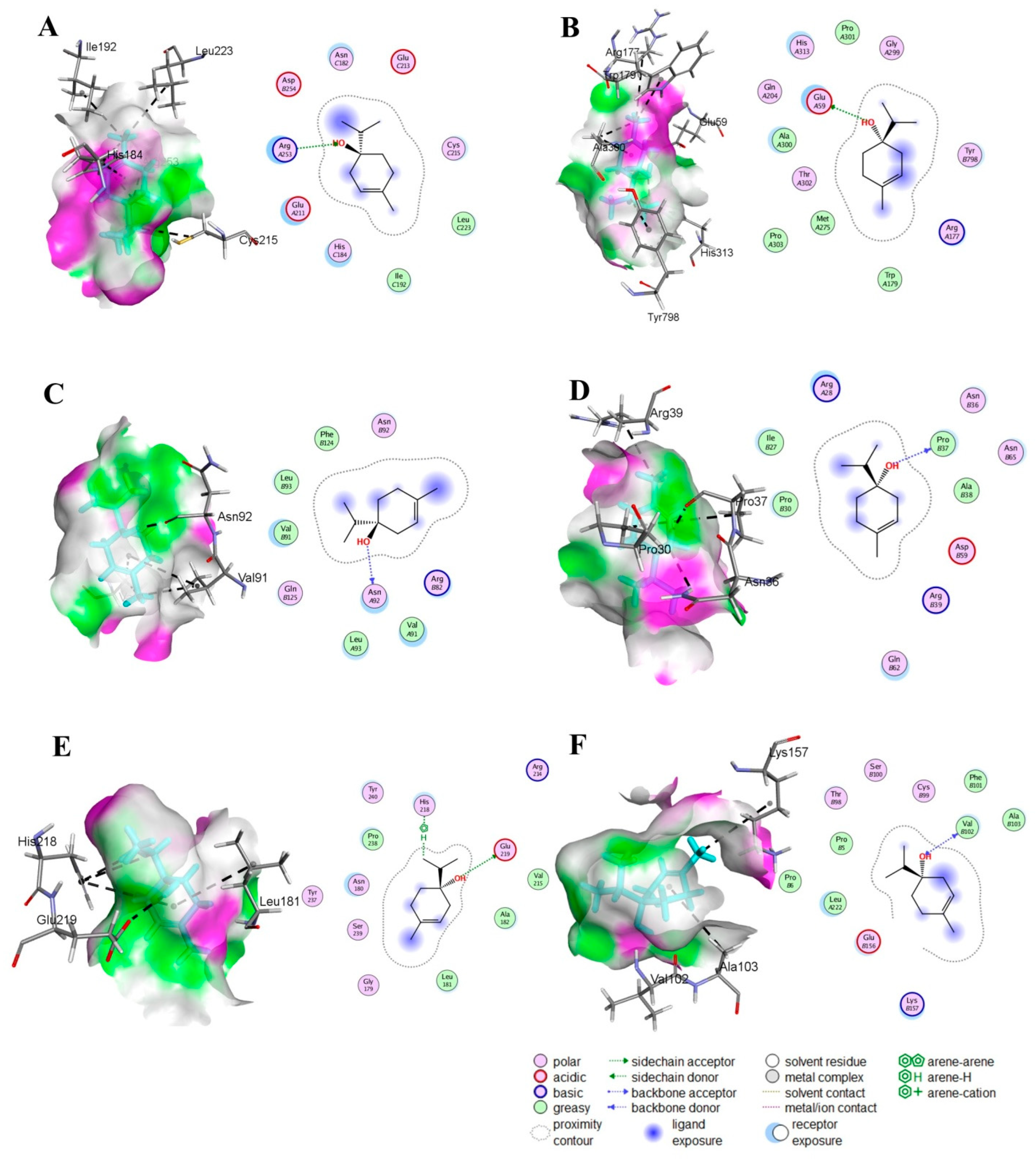
| Compound | S Score (kcal/mol) | RMSD (Å) | Atom of Compounds | Atom of Receptors | Residue of Receptor | Type of Interaction Bond | Distance (Å) | E (kcal/mol) |
|---|---|---|---|---|---|---|---|---|
| NF-KB (PDB ID: 1IKN) | ||||||||
| T4OL | −5.05 | 1.19 | O-28 | NH2 | ARG 253 (A) | H-acceptor | 3.26 | −1.3 |
| SIL | −7.98 | 1.14 | O-54 6-ring | O CA | PRO 275 (A) THR 52 (A) | H-donor pi-H | 3.08 3.93 | −0.9 −0.5 |
| HIF-1α (PDB ID: 1H2M) | ||||||||
| T4OL | −4.84 | 0.61 | O-28 | OE1 | GLU 59 (A) | H-donor | 2.89 | −2.5 |
| SIL | −6.52 | 1.86 | O-47 O-49 O-54 O-56 6-ring | OD1 OE1 OE1 OE1 NE2 | ASP 104 (A) GLU 105 (A) GLN 148 (A) GLN 814 (S) GLN 148 (A) | H-donor H-donor H-donor H-donor pi-H | 2.82 3.13 3.09 2.84 4.27 | −3.1 −1.8 −0.9 −2.9 −0.6 |
| TNF-α (PDB ID: 2AZ5) | ||||||||
| Terpinen-4-ol | −4.63 | 1.35 | O-28 | O | ASN 92 (A) | H-donor | 2.70 | −2.1 |
| Silymarin | −7.13 | 1.46 | O-49 6-ring 6-ring | O CB CA | ARG 32 (C) ALA 35 (C) GLY 148 (C) | H-donor pi-H pi-H | 2.85 3.69 3.84 | −2.4 −0.5 −0.5 |
| COL1A1 (PDB ID: 5K31) | ||||||||
| T4OL | −4.90 | 1.35 | O-28 | O | PRO 37 (F) | H-donor | 2.82 | −1.3 |
| SIL | −8.42 | 2.02 | O-49 O-54 | O O | ILE 27 (F) ASP 59 (F) | H-donor H-donor | 2.88 2.79 | −0.5 −1.3 |
| MMP-1 (PDB ID: 3SHI) | ||||||||
| T4OL | −5.40 | 1.85 | O-28 C-24 | OE2 5-ring | GLU 219 (A) HIS 218 (A) | H-donor H-pi | 3.43 3.68 | −0.8 −0.8 |
| SIL | −7.86 | 1.72 | O-47 O-49 O-49 | OE1 OE1 N | GLU 209 (A) GLU 201 (A) ARG 202 (A) | H-donor H-donor H-acceptor | 2.89 2.87 3.35 | −4.5 −4.1 −0.9 |
| TIMP-1 (PDB ID: 1UEA) | ||||||||
| T4OL | −4.86 | 1.73 | O-28 O-28 | O N | VAL 102 (D) VAL 102 (D) | H-donor H-acceptor | 2.84 3.05 | −1.6 −2.2 |
| SIL | −8.33 | 1.33 | O-54 O-56 6-ring 6-ring 6-ring 6-ring | O O N CB CD2 N | GLU 216 (A) TYR 220 (A) HIS 224 (A) LEU 226 (A) LEU 226 (A) THR 227 (A) | H-donor H-donor pi-H pi-H pi-H pi-H | 2.73 3.02 4.30 4.36 4.15 4.43 | −2.3 −3.6 −0.8 −0.7 −0.5 −0.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Published by MDPI on behalf of the Lithuanian University of Health Sciences. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alsahli, T.G.; Khalid, M.; Malik, M.N.H.; Alshammari, S.O. Terpinen-4-ol Targets HIF-1α/TGF-β1/TNF-α Axis to Attenuate Ethanol-Induced Hepatotoxicity: Network Pharmacology and In Vitro Validation. Medicina 2025, 61, 1048. https://doi.org/10.3390/medicina61061048
Alsahli TG, Khalid M, Malik MNH, Alshammari SO. Terpinen-4-ol Targets HIF-1α/TGF-β1/TNF-α Axis to Attenuate Ethanol-Induced Hepatotoxicity: Network Pharmacology and In Vitro Validation. Medicina. 2025; 61(6):1048. https://doi.org/10.3390/medicina61061048
Chicago/Turabian StyleAlsahli, Tariq G., Maryam Khalid, Muhammad Nasir Hayat Malik, and Saud O. Alshammari. 2025. "Terpinen-4-ol Targets HIF-1α/TGF-β1/TNF-α Axis to Attenuate Ethanol-Induced Hepatotoxicity: Network Pharmacology and In Vitro Validation" Medicina 61, no. 6: 1048. https://doi.org/10.3390/medicina61061048
APA StyleAlsahli, T. G., Khalid, M., Malik, M. N. H., & Alshammari, S. O. (2025). Terpinen-4-ol Targets HIF-1α/TGF-β1/TNF-α Axis to Attenuate Ethanol-Induced Hepatotoxicity: Network Pharmacology and In Vitro Validation. Medicina, 61(6), 1048. https://doi.org/10.3390/medicina61061048







