Machine-Learning-Driven Phenotyping in Heart Failure with Preserved Ejection Fraction: Current Approaches and Future Directions
Abstract
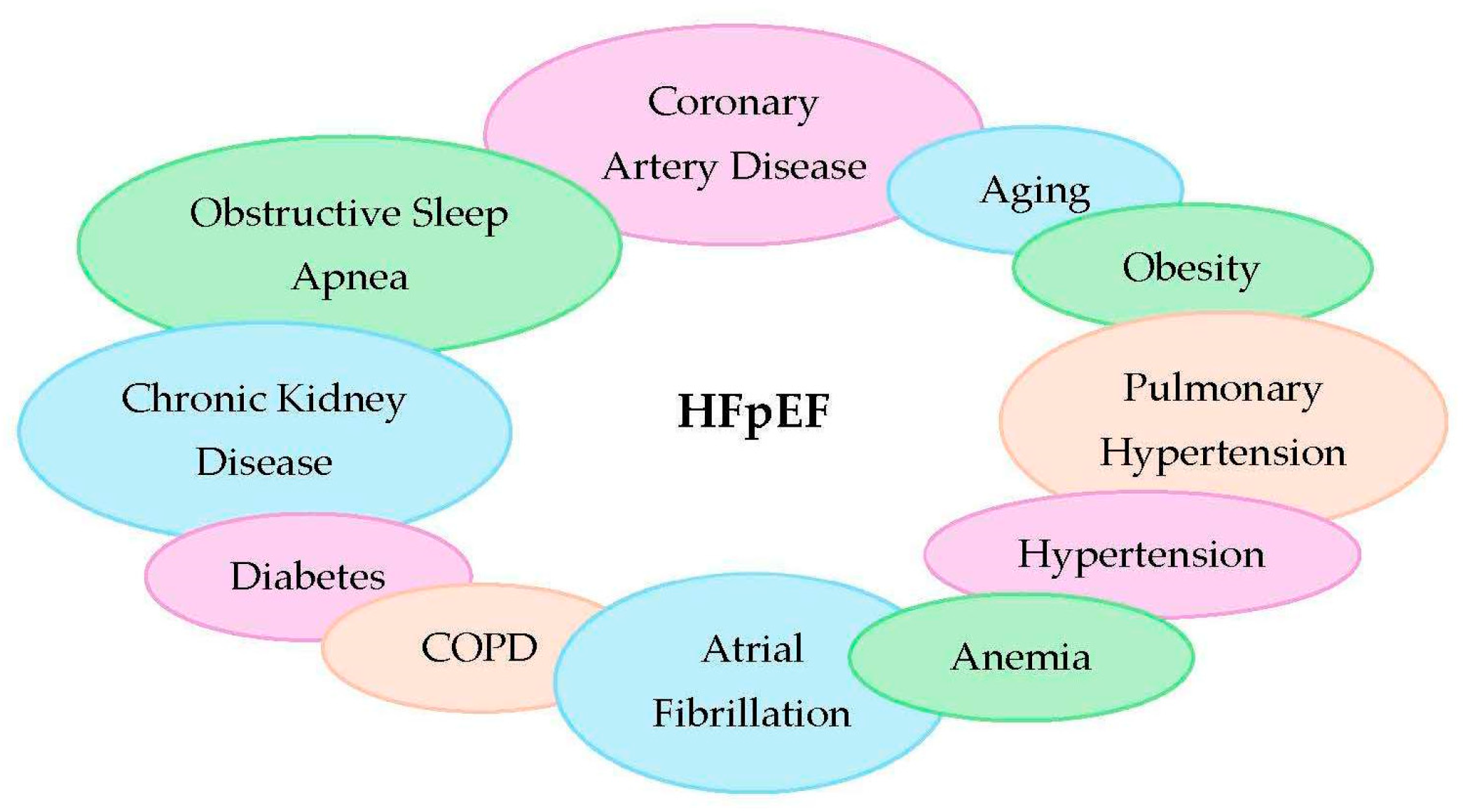
1. Introduction
2. Challenges of Traditional Methods in HFpEF Phenotyping
3. Machine Learning Techniques in HFpEF Phenotyping
4. Major Machine Learning-Based Phenotypes
5. Tailored Therapy Based on ML Phenotyping
6. Advancements in ML Techniques
7. Future Directions and Areas of Improvement
8. Conclusions and Limitations
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| AI | Artificial intelligence |
| ARB | Angiotensin receptor blocker |
| ARNI | Angiotensin receptor/neprilysin inhibitor |
| BNP | B-type natriuretic peptide |
| CDSS | Clinical decision support system |
| CMR | Cardiac magnetic resonance |
| CNN | Convolutional neural network |
| COPD | Chronic obstructive pulmonary disease |
| CRP | C-reactive protein |
| EHR | Electronic health record |
| GDF-15 | Growth differentiation factor-15 |
| GMM | Gaussian mixture model |
| HF | Heart failure |
| HFpEF | Heart failure with preserved ejection fraction |
| HFrEF | Heart failure with reduced ejection fraction |
| IL-6 | Interleukin-6 |
| LSTM | Long Short-Term Memory |
| LV | Left Ventricle |
| LVEF | Left Ventricular Ejection Fraction |
| LVH | Left Ventricular Hypertrophy |
| ML | Machine Learning |
| MRA | Mineralocorticoid Receptor Antagonist |
| nt-proBNP | N-terminal pro-BNP |
| RAAS | Renin-Angiotensin-Aldosterone System |
| RNN | Recurrent Neural Network |
| RV | Right Ventricle |
| SGLT2 | Sodium-Glucose Cotransporter-2 |
| SVM | Support Vector Machine |
| T2DM | Type 2 Diabetes Mellitus |
| VBGMM | Variational Bayesian–Gaussian Mixture Model |
References
- Virani, S.S.; Alonso, A.; Benjamin, E.J.; Bittencourt, M.S.; Callaway, C.W.; Carson, A.P.; Chamberlain, A.M.; Chang, A.R.; Cheng, S.; Delling, F.N.; et al. Heart Disease and Stroke Statistics—2020 Update: A Report From the American Heart Association. Circulation 2020, 141, e139–e596. [Google Scholar] [CrossRef]
- Bashir, Z.; Chen, E.W.; Tori, K.; Ghosalkar, D.; Aurigemma, G.P.; Dickey, J.B.; Haines, P. Insight into different phenotypic presentations of heart failure with preserved ejection fraction. Prog. Cardiovasc. Dis. 2023, 79, 80–88. [Google Scholar] [CrossRef]
- Lin, C.-Y.; Sung, H.-Y.; Chen, Y.-J.; Yeh, H.-I.; Hou, C.J.-Y.; Tsai, C.-T.; Hung, C.-L. Personalized Management for Heart Failure with Preserved Ejection Fraction. J. Pers. Med. 2023, 13, 746. [Google Scholar] [CrossRef]
- Zawadzka, M.; Grabowski, M.; Kapłon-Cieślicka, A. Phenotyping in heart failure with preserved ejection fraction: A key to find effective treatment. Adv. Clin. Exp. Med. 2022, 31, 1163–1172. [Google Scholar] [CrossRef]
- Gevaert, A.B.; Tibebu, S.; Mamas, M.A.; Ravindra, N.G.; Lee, S.F.; Ahmad, T.; Ko, D.T.; Jr, J.L.J.; Van Spall, H.G. Clinical phenogroups are more effective than left ventricular ejection fraction categories in stratifying heart failure outcomes. ESC Heart Fail. 2021, 8, 2741–2754. [Google Scholar] [CrossRef]
- Arnold, J.R.; McCann, G.P. Cardiovascular magnetic resonance: Applications and practical considerations for the general cardiologist. Heart 2020, 106, 174–181. [Google Scholar] [CrossRef] [PubMed]
- Suthahar, N.; Lau, E.S.; Blaha, M.J.; Paniagua, S.M.; Larson, M.G.; Psaty, B.M.; Benjamin, E.J.; Allison, M.A.; Bartz, T.M.; Januzzi, J.L.; et al. Sex-Specific Associations of Cardiovascular Risk Factors and Biomarkers with Incident Heart Failure. J. Am. Coll. Cardiol. 2020, 76, 1455–1465. [Google Scholar] [CrossRef]
- Reddy, Y.N.; Carter, R.E.; Obokata, M.; Redfield, M.M.; Borlaug, B.A. A Simple, Evidence-Based Approach to Help Guide Diagnosis of Heart Failure With Preserved Ejection Fraction. Circulation 2018, 138, 861–870. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.J.; Katz, D.H.; Selvaraj, S.; Burke, M.A.; Yancy, C.W.; Gheorghiade, M.; Bonow, R.O.; Huang, C.-C.; Deo, R.C. Phenomapping for novel classification of heart failure with preserved ejection fraction. Circulation 2015, 131, 269–279. [Google Scholar] [CrossRef]
- Lundberg, S.M.; Lee, S.-I. A unified approach to interpreting model predictions. In Proceedings of the 31st International Conference on Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017; Curran Associates Inc.: Long Beach, CA, USA, 2017; pp. 4768–4777. [Google Scholar]
- Kyodo, A.; Kanaoka, K.; Keshi, A.; Nogi, M.; Nogi, K.; Ishihara, S.; Kamon, D.; Hashimoto, Y.; Nakada, Y.; Ueda, T.; et al. Heart failure with preserved ejection fraction phenogroup classification using machine learning. ESC Heart Fail. 2023, 10, 2019–2030. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, F.S.; Luo, Y.; Wehbe, R.M.; Thomas, J.D.; Shah, S.J. Advances in Machine Learning Approaches to Heart Failure with Preserved Ejection Fraction. Heart Fail. Clin. 2022, 18, 287–300. [Google Scholar] [CrossRef]
- Schober, P.; Vetter, T.R. Logistic Regression in Medical Research. Anesth. Analg. 2021, 132, 365–366. [Google Scholar] [CrossRef]
- Blockeel, H.; Devos, L.; Frénay, B.; Nanfack, G.; Nijssen, S. Decision trees: From efficient prediction to responsible AI. Front. Artif. Intell. 2023, 6, 1124553. [Google Scholar] [CrossRef] [PubMed]
- Angraal, S.; Mortazavi, B.J.; Gupta, A.; Khera, R.; Ahmad, T.; Desai, N.R.; Jacoby, D.L.; Masoudi, F.A.; Spertus, J.A.; Krumholz, H.M. Machine Learning Prediction of Mortality and Hospitalization in Heart Failure With Preserved Ejection Fraction. JACC Heart Fail. 2020, 8, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhu, J.; Li, H.; Wu, S.; Li, S.; Yao, Z.; Zhu, T.; Tang, B.; Tang, S.; Liu, J. Multimodal Visualization and Explainable Machine Learning-Driven Markers Enable Early Identification and Prognosis Prediction for Symptomatic Aortic Stenosis and Heart Failure With Preserved Ejection Fraction After Transcatheter Aortic Valve Replacement: Multicenter Cohort Study. J. Med. Internet Res. 2025, 27, e70587. [Google Scholar]
- Khyathi, G.; Indumathi, K.P.; Jumana Hasin, A.; Lisa Flavin Jency, M.; Krishnaprakash, G.; Lisa, F.J.M. Support Vector Machines: A Literature Review on Their Application in Analyzing Mass Data for Public Health. Cureus 2025, 17, e77169. [Google Scholar] [CrossRef] [PubMed]
- Segar, M.W.; Patel, K.V.; Ayers, C.; Basit, M.; Tang, W.W.; Willett, D.; Berry, J.; Grodin, J.L.; Pandey, A. Phenomapping of patients with heart failure with preserved ejection fraction using machine learning-based unsupervised cluster analysis. Eur. J. Heart Fail. 2020, 22, 148–158. [Google Scholar] [CrossRef]
- Shah, A.M.; Cikes, M.; Prasad, N.; Li, G.; Getchevski, S.; Claggett, B.; Rizkala, A.; Lukashevich, I.; O’meara, E.; Ryan, J.J.; et al. Echocardiographic Features of Patients With Heart Failure and Preserved Left Ventricular Ejection Fraction. J. Am. Coll. Cardiol. 2019, 74, 2858–2873. [Google Scholar] [CrossRef]
- McDonagh, T.A.; Metra, M.; Adamo, M.; Gardner, R.S.; Baumbach, A.; Böhm, M.; Burri, H.; Butler, J.; Čelutkienė, J.; Chioncel, O.; et al. 2023 Focused Update of the 2021 ESC Guidelines for the diagnosis and treatment of acute and chronic heart failure: Developed by the task force for the diagnosis and treatment of acute and chronic heart failure of the European Society of Cardiology (ESC) With the special contribution of the Heart Failure Association (HFA) of the ESC. Eur. Heart J. 2023, 44, 3627–3639. [Google Scholar]
- Awan, S.E.; Sohel, F.; Sanfilippo, F.M.; Bennamoun, M.; Dwivedi, G. Machine learning in heart failure: Ready for prime time. Curr. Opin. Cardiol. 2018, 33, 190–195. [Google Scholar] [CrossRef]
- Woolley, R.J.; Ceelen, D.; Ouwerkerk, W.; Tromp, J.; Figarska, S.M.; Anker, S.D.; Dickstein, K.; Filippatos, G.; Zannad, F.; Metra, M.; et al. Machine learning based on biomarker profiles identifies distinct subgroups of heart failure with preserved ejection fraction. Eur. J. Heart Fail. 2021, 23, 983–991. [Google Scholar] [CrossRef] [PubMed]
- Contreras, J.; Winterfeld, A.; Popp, J.; Bocklitz, T. Spectral Zones-Based SHAP/LIME: Enhancing Interpretability in Spectral Deep Learning Models Through Grouped Feature Analysis. Anal. Chem. 2024, 96, 15588–15597. [Google Scholar] [CrossRef]
- Farajidavar, N.; O’gAllagher, K.; Bean, D.; Nabeebaccus, A.; Zakeri, R.; Bromage, D.; Kraljevic, Z.; Teo, J.T.H.; Dobson, R.J.; Shah, A.M. Diagnostic signature for heart failure with preserved ejection fraction (HFpEF): A machine learning approach using multi-modality electronic health record data. BMC Cardiovasc. Disord. 2022, 22, 567. [Google Scholar] [CrossRef]
- Galli, E.; Bourg, C.; Kosmala, W.; Oger, E.; Donal, E. Phenomapping Heart Failure with Preserved Ejection Fraction Using Machine Learning Cluster Analysis: Prognostic and Therapeutic Implications. Heart Fail. Clin. 2021, 17, 499–518. [Google Scholar] [CrossRef]
- Pierre-Jean, M.; Marut, B.; Curtis, E.; Galli, E.; Cuggia, M.; Bouzillé, G.; Donal, E. Phenotyping of heart failure with preserved ejection faction using electronic health records and echocardiography. Eur. Hear. J. Open 2024, 4, oead133. [Google Scholar] [CrossRef]
- Murray, E.M.; Greene, S.J.; Rao, V.N.; Sun, J.-L.; Alhanti, B.A.; Blumer, V.; Butler, J.; Ahmad, T.; Mentz, R.J. Machine learning to define phenotypes and outcomes of patients hospitalized for heart failure with preserved ejection fraction: Findings from ASCEND-HF. Am. Heart J. 2022, 254, 112–121. [Google Scholar] [CrossRef]
- Jones, E.; Randall, E.B.; Hummel, S.L.; Cameron, D.M.; Beard, D.A.; Carlson, B.E. Phenotyping heart failure using model-based analysis and physiology-informed machine learning. J. Physiol. 2021, 599, 4991–5013. [Google Scholar] [CrossRef]
- Rabkin, S.W. Evaluating the adverse outcome of subtypes of heart failure with preserved ejection fraction defined by machine learning: A systematic review focused on defining high risk phenogroups. Excli. J. 2022, 21, 487–518. [Google Scholar]
- Lewis, G.A.; Dodd, S.; Clayton, D.; Bedson, E.; Eccleson, H.; Schelbert, E.B.; Naish, J.H.; Jimenez, B.D.; Williams, S.G.; Cunnington, C.; et al. Pirfenidone in heart failure with preserved ejection fraction: A randomized phase 2 trial. Nat. Med. 2021, 27, 1477–1482. [Google Scholar] [CrossRef] [PubMed]
- Lewis, G.A.; Rosala-Hallas, A.; Dodd, S.; Schelbert, E.B.; Williams, S.G.; Cunnington, C.; McDonagh, T.; Miller, C.A. Predictors of myocardial fibrosis and response to anti-fibrotic therapy in heart failure with preserved ejection fraction. Int. J. Cardiovasc. Imaging 2022, 38, 1569–1578. [Google Scholar] [CrossRef] [PubMed]
- Albulushi, A.; Askari, K.M.; Al-Abedi, A.M.; Al-Kulaibi, M.A.; Hasan, M.S.; Hosseini, Z.; Al-Rahman, M.T.; Tanoh, D.B.; Hasan, A.S.; Al-Helli, Y.; et al. Impact of SGLT2 inhibitors on myocardial fibrosis in diabetic HFpEF: A longitudinal study. Eur. J. Med. Res. 2025, 30, 592. [Google Scholar] [CrossRef]
- Aimo, A.; Spitaleri, G.; Panichella, G.; Lupón, J.; Emdin, M.; Bayes-Genis, A. Pirfenidone as a novel cardiac protective treatment. Heart Fail. Rev. 2022, 27, 525–532. [Google Scholar] [CrossRef]
- Ridker, P.M.; Everett, B.M.; Thuren, T.; MacFadyen, J.G.; Chang, W.H.; Ballantyne, C.; Fonseca, F.; Nicolau, J.; Koenig, W.; Anker, S.D.; et al. Antiinflammatory Therapy with Canakinumab for Atherosclerotic Disease. N. Engl. J. Med. 2017, 377, 1119–1131. [Google Scholar] [CrossRef]
- Chia, Y.C.; Kieneker, L.M.; van Hassel, G.; Binnenmars, S.H.; Nolte, I.M.; van Zanden, J.J.; van der Meer, P.; Navis, G.; Voors, A.A.; Bakker, S.J.L.; et al. Interleukin 6 and Development of Heart Failure With Preserved Ejection Fraction in the General Population. J. Am. Heart Assoc. 2021, 10, e018549. [Google Scholar] [CrossRef] [PubMed]
- Segar, M.W.; Coulter, S.A. Inflammatory Pathways and Their Implications in Heart Failure With Preserved Ejection Fraction. Tex. Heart Inst. J. 2023, 50. [Google Scholar] [CrossRef] [PubMed]
- Bonnet, F.; Scheen, A.J. Effects of SGLT2 inhibitors on systemic and tissue low-grade inflammation: The potential contribution to diabetes complications and cardiovascular disease. Diabetes Metab. 2018, 44, 457–464. [Google Scholar] [CrossRef] [PubMed]
- Solomon, S.D.; McMurray, J.J.; Claggett, B.; de Boer, R.A.; DeMets, D.; Hernandez, A.F.; Inzucchi, S.E.; Kosiborod, M.N.; Lam, C.S.; Martinez, F.; et al. Dapagliflozin in Heart Failure with Mildly Reduced or Preserved Ejection Fraction. N. Engl. J. Med. 2022, 387, 1089–1098. [Google Scholar] [CrossRef]
- Anker, S.D.; Butler, J.; Filippatos, G.; Ferreira, J.P.; Bocchi, E.; Böhm, M.; Packer, M. Empagliflozin in Heart Failure with a Preserved Ejection Fraction. N. Engl. J. Med. 2021, 385, 1451–1461. [Google Scholar] [CrossRef]
- La Fazia, V.M.; Pierucci, N.; Mohanty, S.; Chiricolo, G.; Natale, A. Atrial Fibrillation Ablation in Heart Failure with Preserved Ejection Fraction. Card. Electrophysiol. Clin. 2025, 17, 53–62. [Google Scholar] [CrossRef]
- Kairouz, P.; McMahan, H.B.; Avent, B.; Bellet, A.; Bennis, M.; Bhagoji, A.N.; Bonawitz, K.; Charles, Z.; Cormode, G.; Cummings, R.; et al. Advances and Open Problems in Federated Learning. Found. Trends Mach. Learn. 2021, 14, 1–210. [Google Scholar] [CrossRef]
- Hasin, Y.; Seldin, M.; Lusis, A. Multi-omics approaches to disease. Genome Biol. 2017, 18, 83. [Google Scholar] [CrossRef]
- Esteva, A.; Robicquet, A.; Ramsundar, B.; Kuleshov, V.; DePristo, M.; Chou, K.; Dean, J. A guide to deep learning in healthcare. Nat. Med. 2019, 25, 24–29. [Google Scholar] [CrossRef]
- Cersosimo, A.; Zito, E.; Pierucci, N.; Matteucci, A.; La Fazia, V.M. A Talk with ChatGPT: The Role of Artificial Intelligence in Shaping the Future of Cardiology and Electrophysiology. J. Pers. Med. 2025, 15, 205. [Google Scholar] [CrossRef]
- Alimbayeva, Z.; Alimbayev, C.; Ozhikenov, K.; Bayanbay, N.; Ozhikenova, A. Wearable ECG Device and Machine Learning for Heart Monitoring. Sensors 2024, 24, 4201. [Google Scholar] [CrossRef]
- Cheema, B.; Tibrewala, A. SEISMIC-HF 1: Key findings from AHA24 and implications for remote cardiac monitoring. Heart Fail. Rev. 2025, 30, 1099–1101. [Google Scholar] [CrossRef] [PubMed]
- Shinzato, M.H.; Santos, N.; Nishida, G.; Moriya, H.; Assef, J.; Feres, F.; Hortegal, R.A. Left ventricular and atrial myocardial strain in heart failure with preserved ejection fraction: The evidence so far and prospects for phenotyping strategy. Cardiovasc. Ultrasound 2024, 22, 4. [Google Scholar] [CrossRef]
- Kraus, M.J.; Veshapeli, A.; Reich, C.; Hund, H.; Hamed, S.; Raake, P.W.; Kreusser, M.M.; Frey, N.; Lehmann, L. Risk prediction in heart failure using invasive hemodynamics. Clin. Res. Cardiol. 2025, 114, 1388–1399. [Google Scholar] [CrossRef]
- Choi, K.H.; Yang, J.H.; Seo, J.H.; Hong, D.; Youn, T.; Joh, H.S.; Lee, S.H.; Kim, D.; Park, T.K.; Lee, J.M.; et al. Discriminative Role of Invasive Left Heart Catheterization in Patients Suspected of Heart Failure With Preserved Ejection Fraction. J. Am. Heart Assoc. 2023, 12, e027581. [Google Scholar] [CrossRef]
- Rajkomar, A.; Dean, J.; Kohane, I. Machine Learning in Medicine. N. Engl. J. Med. 2019, 380, 1347–1358. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Casalino, L.P.; Khullar, D. Deep Learning in Medicine-Promise, Progress, and Challenges. JAMA Intern. Med. 2019, 179, 293–294. [Google Scholar] [CrossRef] [PubMed]
- Topol, E.J. High-performance medicine: The convergence of human and artificial intelligence. Nat. Med. 2019, 25, 44–56. [Google Scholar] [CrossRef] [PubMed]
| Technique | Interpretability | Handles Complexity | Application in HFpEF | Advantages | Limitations |
|---|---|---|---|---|---|
| Logistic regression | High | Low–Moderate | Classifying HFpEF vs. other HF types using clinical predictors | Simple, interpretable, well-understood | May underperform with complex/non-linear relationships |
| Decision trees | High | Moderate | Clear rule-based HFpEF classification | Easy to interpret, generates clear rules | Prone to overfitting |
| Random forests | Moderate | High | Identifying HFpEF subgroups via ensemble decision trees | Robust, handles non-linearities, reduces overfitting | Less interpretable than single trees |
| Support vector machines (SVMs) | Low | High | Classifying HFpEF based on complex feature interactions | Effective in high-dimensional spaces, handles non-linearity well | Poor interpretability, sensitive to parameter tuning |
| k-means clustering | Moderate | Moderate | Grouping patients with similar phenotypes | Simple, fast, intuitive clustering | Assumes spherical clusters, sensitive to initialization |
| Hierarchical clustering | High | Moderate | Creating tree-structured patient subgroup hierarchies | Reveals subgroup relationships, no need to predefine number of clusters | Computationally expensive for large datasets |
| Gaussian mixture models (GMMs) | Moderate | High | Probabilistic clustering of overlapping HFpEF phenotypes | Captures uncertainty and soft clustering | Assumes Gaussian distribution, may converge to local minima |
| CNNs (deep learning) | Low | Very High | Extracting features from echocardiographic images for phenotype classification | Automatic feature extraction, high accuracy in image tasks | Requires large datasets, low interpretability |
| RNNs (deep learning) | Low | Very High | Analyzing time-series data from EHRs and wearables | Captures temporal dynamics, good for sequential data | Training complexity, vanishing gradient issues (mitigated by LSTM/GRU) |
| Autoencoders | Low | High | Dimensionality reduction and latent phenotype discovery | Identifies hidden patterns, reduces noise | Requires tuning, difficult to interpret latent features |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Published by MDPI on behalf of the Lithuanian University of Health Sciences. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Potoupni, V.; Samaras, A.; Papadopoulos, C.; Boulmpou, A.; Moysiadis, T.; Zormpas, G.; Tzikas, A.; Fragakis, N.; Giannakoulas, G.; Vassilikos, V. Machine-Learning-Driven Phenotyping in Heart Failure with Preserved Ejection Fraction: Current Approaches and Future Directions. Medicina 2025, 61, 1937. https://doi.org/10.3390/medicina61111937
Potoupni V, Samaras A, Papadopoulos C, Boulmpou A, Moysiadis T, Zormpas G, Tzikas A, Fragakis N, Giannakoulas G, Vassilikos V. Machine-Learning-Driven Phenotyping in Heart Failure with Preserved Ejection Fraction: Current Approaches and Future Directions. Medicina. 2025; 61(11):1937. https://doi.org/10.3390/medicina61111937
Chicago/Turabian StylePotoupni, Victoria, Athanasios Samaras, Christodoulos Papadopoulos, Aristi Boulmpou, Theodoros Moysiadis, Georgios Zormpas, Apostolos Tzikas, Nikolaos Fragakis, George Giannakoulas, and Vassilios Vassilikos. 2025. "Machine-Learning-Driven Phenotyping in Heart Failure with Preserved Ejection Fraction: Current Approaches and Future Directions" Medicina 61, no. 11: 1937. https://doi.org/10.3390/medicina61111937
APA StylePotoupni, V., Samaras, A., Papadopoulos, C., Boulmpou, A., Moysiadis, T., Zormpas, G., Tzikas, A., Fragakis, N., Giannakoulas, G., & Vassilikos, V. (2025). Machine-Learning-Driven Phenotyping in Heart Failure with Preserved Ejection Fraction: Current Approaches and Future Directions. Medicina, 61(11), 1937. https://doi.org/10.3390/medicina61111937









