Antimicrobial and Defense Proteins in Chronic Rhinosinusitis with Nasal Polyps
Abstract
:1. Introduction
2. Materials and Methods
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fokkens, W.J.; Lund, V.J.; Hopkins, C.; Hellings, P.W.; Kern, R.; Reitsma, S.; Toppila-Salmi, S.; Bernal-Sprekelsen, M.; Mullol, J.; Alobid, I.; et al. European Position Paper on Rhinosinusitis and Nasal Polyps 2020. Rhinology 2020, 58 (Suppl. S29), 1–464. [Google Scholar] [CrossRef]
- Chaaban, M.R.; Walsh, E.M.; Woodworth, B.A. Epidemiology and Differential Diagnosis of Nasal Polyps. Am. J. Rhinol. Allergy 2013, 27, 473–478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sedaghat, A.R.; Kuan, E.C.; Scadding, G.K. Epidemiology of Chronic Rhinosinusitis: Prevalence and Risk Factors. J. Allergy Clin. Immunol. Pract. 2022, 10, 1395–1403. [Google Scholar] [CrossRef] [PubMed]
- Cho, D.-Y.; Hunter, R.C.; Ramakrishnan, V.R. The Microbiome and Chronic Rhinosinusitis. Immunol. Allergy Clin. N. Am. 2020, 40, 251–263. [Google Scholar] [CrossRef] [PubMed]
- Stevens, W.W.; Lee, R.J.; Schleimer, R.P.; Cohen, N.A. Chronic rhinosinusitis pathogenesis. J. Allergy Clin. Immunol. 2015, 136, 1442–1453. [Google Scholar] [CrossRef] [Green Version]
- Carothers, D.G.; Graham, S.M.; Jia, H.P.; Ackermann, M.R.; Tack, B.F.; McCray, P.B. Production of β-Defensin Antimicrobial Peptides by Maxillary Sinus Mucosa. Am. J. Rhinol. 2001, 15, 175–180. [Google Scholar] [CrossRef]
- Luo, Y.; Song, Y. Mechanism of Antimicrobial Peptides: Antimicrobial, Anti-Inflammatory and Antibiofilm Activities. Int. J. Mol. Sci. 2021, 22, 11401. [Google Scholar] [CrossRef]
- Kumar, P.; Kizhakkedathu, J.; Straus, S. Antimicrobial Peptides: Diversity, Mechanism of Action and Strategies to Improve the Activity and Biocompatibility In Vivo. Biomolecules 2018, 8, 4. [Google Scholar] [CrossRef] [Green Version]
- Bin Hafeez, A.; Jiang, X.; Bergen, P.J.; Zhu, Y. Antimicrobial Peptides: An Update on Classifications and Databases. Int. J. Mol. Sci. 2021, 22, 11691. [Google Scholar] [CrossRef]
- Dhople, V.; Krukemeyer, A.; Ramamoorthy, A. The human beta-defensin-3, an antibacterial peptide with multiple biological functions. Biochim. Biophys. Acta BBA—Biomembr. 2006, 1758, 1499–1512. [Google Scholar] [CrossRef] [Green Version]
- Cieślik, M.; Bagińska, N.; Górski, A.; Jończyk-Matysiak, E. Human β-Defensin 2 and Its Postulated Role in Modulation of the Immune Response. Cells 2021, 10, 2991. [Google Scholar] [CrossRef]
- Chen, P.-H.; Fang, S.-Y. Expression of human β-defensin 2 in human nasal mucosa. Eur. Arch. Otorhinolaryngol. 2004, 261, 238–241. [Google Scholar] [CrossRef]
- Xiao, C.; He, G.; Deng, W.; Zhang, H.; Sun, W. Expression of human beta-defensin after endoscopic sinus surgery for chronic sinusitis. Nan Fang Yi Ke Da Xue Xue Bao 2010, 30, 1580–1583. [Google Scholar]
- Chen, X.; Niyonsaba, F.; Ushio, H.; Hara, M.; Yokoi, H.; Matsumoto, K.; Saito, H.; Nagaoka, I.; Ikeda, S.; Okumura, K.; et al. Antimicrobial peptides human β-defensin (hBD)-3 and hBD-4 activate mast cells and increase skin vascular permeability. Eur. J. Immunol. 2007, 37, 434–444. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Tan, H.; Cheng, T.; Shen, H.; Shao, J.; Guo, Y.; Shi, S.; Zhang, X. Human β-defensin 3 inhibits antibiotic-resistant Staphylococcus biofilm formation. J. Surg. Res. 2013, 183, 204–213. [Google Scholar] [CrossRef]
- Sutton, J.M.; Pritts, T.A. Human beta-defensin 3: A novel inhibitor of Staphylococcus-Produced biofilm production. Commentary on “Human β-defensin 3 inhibits antibiotic-resistant Staphylococcus biofilm formation”. J. Surg. Res. 2014, 186, 99–100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biswas, L.; Götz, F. Molecular Mechanisms of Staphylococcus and Pseudomonas Interactions in Cystic Fibrosis. Front. Cell. Infect. Microbiol. 2022, 11, 1383. [Google Scholar] [CrossRef]
- Batoni, G.; Maisetta, G.; Esin, S.; Campa, M. Human Beta-Defensin-3: A Promising Antimicrobial Peptide. Mini-Rev. Med. Chem. 2006, 6, 1063–1073. [Google Scholar] [CrossRef] [PubMed]
- Schneider, J.J.; Unholzer, A.; Schaller, M.; Schäfer-Korting, M.; Korting, H.C. Human defensins. J. Mol. Med. 2005, 83, 587–595. [Google Scholar] [CrossRef]
- Lohova, E.; Vitenberga-Verza, Z.; Kazoka, D.; Pilmane, M. Local Defence System in Healthy Lungs. Clin. Pract. 2021, 11, 728–746. [Google Scholar] [CrossRef]
- Bandurska, K.; Berdowska, A.; Barczyńska-Felusiak, R.; Krupa, P. Unique features of human cathelicidin LL-37: Unique Features of Human Cathelicidin LL-37. BioFactors 2015, 41, 289–300. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.T.; Cha, H.E.; Kim, D.Y.; Han, G.C.; Chung, Y.; Young, J.L.; Hwang, Y.J.; Lee, H. Antimicrobial Peptide LL-37 is Upregulated in Chronic Nasal Inflammatory Disease. Acta Oto-Laryngol. 2003, 123, 81–85. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Chen, F.; Sun, Y.; Hong, H.; Wen, Y.; Lai, Y.; Xu, Z.; Luo, X.; Chen, Y.; Shi, J.; et al. LL-37 promotes neutrophil extracellular trap formation in chronic rhinosinusitis with nasal polyps. Clin. Exp. Allergy 2019, 49, 990–999. [Google Scholar] [CrossRef]
- Ooi, E.H.; Wormald, P.-J.; Carney, A.S.; James, C.L.; Tan, L.W. Fungal Allergens Induce Cathelicidin LL-37 Expression in Chronic Rhinosinusitis Patients in a Nasal Explant Model. Am. J. Rhinol. 2007, 21, 367–372. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-W.; Roh, J.; Park, C.-S. Immunohistochemistry for Pathologists: Protocols, Pitfalls, and Tips. J. Pathol. Transl. Med. 2016, 50, 411–418. [Google Scholar] [CrossRef] [Green Version]
- Pilmane, M.; Luts, A.; Sundler, F. Changes in neuroendocrine elements in bronchial mucosa in chronic lung disease in adults. Thorax 1995, 50, 551–554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tobin, G.; Luts, A.; Sundler, F.; Ekström, J. Peptidergic innervation of the major salivary glands of the ferret. Peptides 1990, 11, 863–867. [Google Scholar] [CrossRef]
- Dambergs, K.; Sumeraga, G.; Pilmane, M. Morphopathogenesis of Adult Acquired Cholesteatoma. Medicina 2023, 59, 306. [Google Scholar] [CrossRef] [PubMed]
- Konopecka, V.; Pilmane, M.; Sumerags, D.; Sumeraga, G. Distribution and Appearance of Ki-67, IL-1α, IL-10, and PGP 9.5 in Reinke’s Oedema-Affected Larynx Tissue Compared with Control Tissue. Life 2021, 11, 1379. [Google Scholar] [CrossRef]
- Viksne, R.J.; Sumeraga, G.; Pilmane, M. Characterization of Cytokines and Proliferation Marker Ki67 in Chronic Rhinosinusitis with Nasal Polyps: A Pilot Study. Medicina 2021, 57, 607. [Google Scholar] [CrossRef]
- Hulse, K.E.; Stevens, W.W.; Tan, B.K.; Schleimer, R.P. Pathogenesis of nasal polyposis. Clin. Exp. Allergy 2015, 45, 328–346. [Google Scholar] [CrossRef] [Green Version]
- Martens, K.; Seys, S.F.; Alpizar, Y.A.; Schrijvers, R.; Bullens, D.M.A.; Breynaert, C.; Lebeer, S.; Steelant, B. Staphylococcus aureus enterotoxin B disrupts nasal epithelial barrier integrity. Clin. Exp. Allergy 2021, 51, 87–98. [Google Scholar] [CrossRef]
- Zhang, R.; Zhang, L.; Li, P.; Pang, K.; Liu, H.; Tian, L. Epithelial Barrier in the Nasal Mucosa, Related Risk Factors and Diseases. Int. Arch. Allergy Immunol. 2023, 184, 481–501. [Google Scholar] [CrossRef]
- Prince, A.A.; Steiger, J.D.; Khalid, A.N.; Dogrhamji, L.; Reger, C.; Claire, S.E.; Chiu, A.G.; Kennedy, D.W.; Palmer, J.N.; Cohen, N.A. Prevalence of Biofilm-forming Bacteria in Chronic Rhinosinusitis. Am. J. Rhinol. 2008, 22, 239–245. [Google Scholar] [CrossRef]
- Thienhaus, M.L.; Wohlers, J.; Podschun, R.; Hedderich, J.; Ambrosch, P.; Laudien, M. Antimicrobial peptides in nasal secretion and mucosa with respect to Staphylococcus aureus colonization in chronic rhinosinusitis with nasal polyps. Rhinol. J. 2011, 49, 554–561. [Google Scholar] [CrossRef] [PubMed]
- Pácová, H.; Astl, J.; Martínek, J. The incidence of β-defensin-1, 2, 3 in human healthy and chronically inflamed nasal and tonsillar mucosa. J. Appl. Biomed. 2010, 8, 81–86. [Google Scholar] [CrossRef] [Green Version]
- Ou, J.; Bassiouni, A.; Drilling, A.; Psaltis, A.J.; Vreugde, S.; Wormald, P.J. The persistence of intracellular Staphylococcus aureus in the sinuses: A longitudinal study. Rhinol. J. 2017, 55, 305–311. [Google Scholar] [CrossRef] [PubMed]
- Altunbulakli, C.; Costa, R.; Lan, F.; Zhang, N.; Akdis, M.; Bachert, C.; Akdis, C.A. Staphylococcus aureus enhances the tight junction barrier integrity in healthy nasal tissue, but not in nasal polyps. J. Allergy Clin. Immunol. 2018, 142, 665–668.e8. [Google Scholar] [CrossRef] [Green Version]
- Tomassen, P.; Vandeplas, G.; Van Zele, T.; Cardell, L.-O.; Arebro, J.; Olze, H.; Förster-Ruhrmann, U.; Kowalski, M.L.; Olszewska-Ziąber, A.; Holtappels, G.; et al. Inflammatory endotypes of chronic rhinosinusitis based on cluster analysis of biomarkers. J. Allergy Clin. Immunol. 2016, 137, 1449–1456.e4. [Google Scholar] [CrossRef] [Green Version]
- Coromina, J.; Sauret, J. Nasal Mucociliary Clearance in Patients with Nasal Polyposis. ORL 1990, 52, 311–315. [Google Scholar] [CrossRef]
- Chiu, A.G.; Palmer, J.N.; Woodworth, B.A.; Doghramji, L.; Cohen, M.B.; Prince, A.; Cohen, N.A. Baby Shampoo Nasal Irrigations for the Symptomatic Post-functional Endoscopic Sinus Surgery Patient. Am. J. Rhinol. 2008, 22, 34–37. [Google Scholar] [CrossRef] [PubMed]
- Meyer, J.E.; Harder, J.; Görögh, T.; Schröder, J.M.; Maune, S. hBD-2 gene expression in nasal mucosa. Laryngo-Rhino-Otologie 2000, 79, 400–403. [Google Scholar] [CrossRef] [PubMed]
- Xie, D.; Guo, Y.; Wu, D.; Xie, D. Expressions of LL-37 and IL-8 in chronic sinusitis with nasal polyps. Lin Chuang Er Bi Yan Hou Tou Jing Wai Ke Za Zhi J. Clin. Otorhinolaryngol. Head Neck Surg. 2010, 24, 337–340. [Google Scholar]
- Perestam, A.T.; Fujisaki, K.K.; Nava, O.; Hellberg, R.S. Comparison of real-time PCR and ELISA-based methods for the detection of beef and pork in processed meat products. Food Control 2017, 71, 346–352. [Google Scholar] [CrossRef] [Green Version]
- Georgakopoulos, B.; Hohman, M.H.; Le, P.H. Anatomy, Head and Neck, Nasal Concha. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2023. Available online: http://www.ncbi.nlm.nih.gov/books/NBK546636/ (accessed on 26 June 2023).
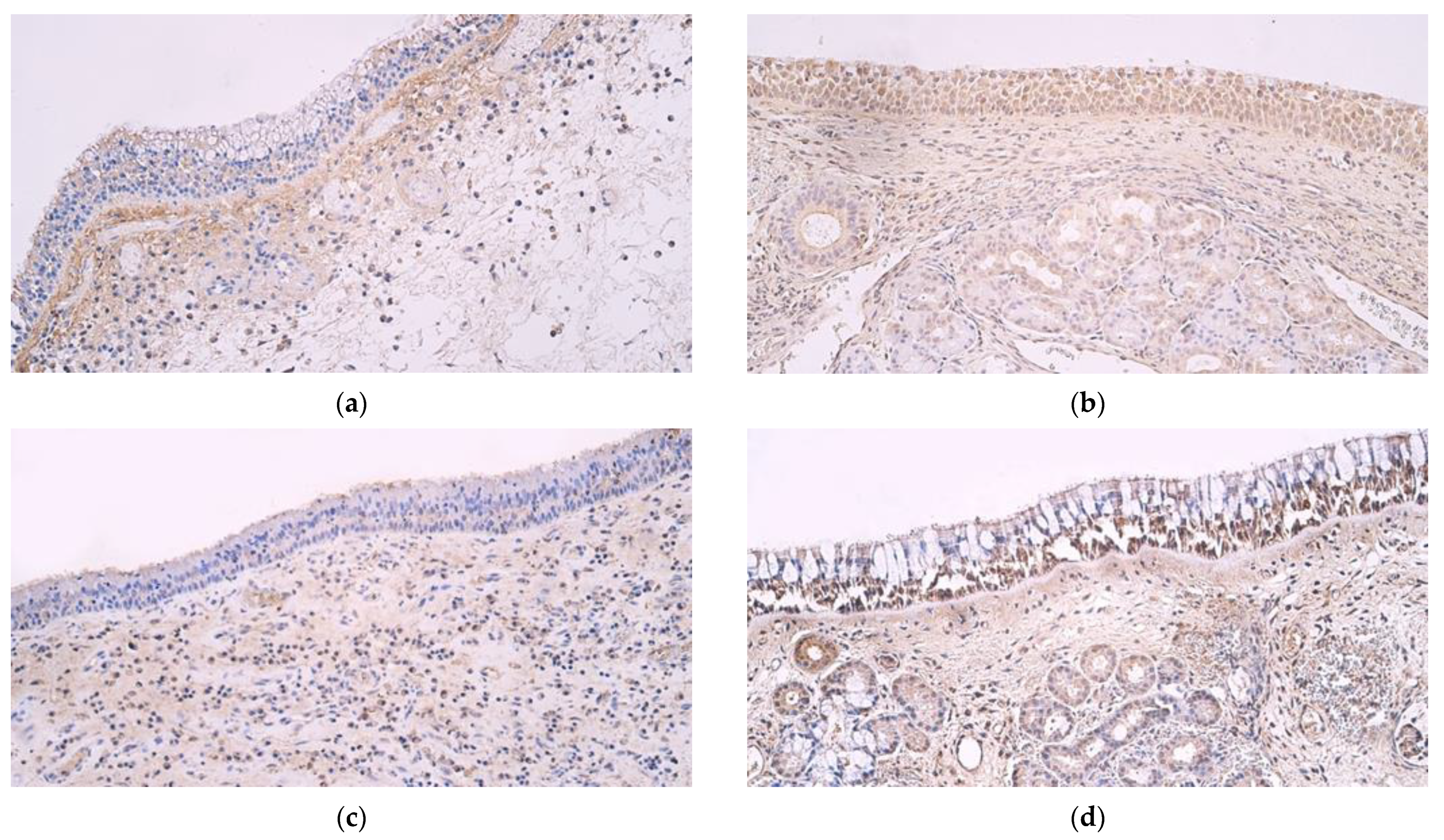
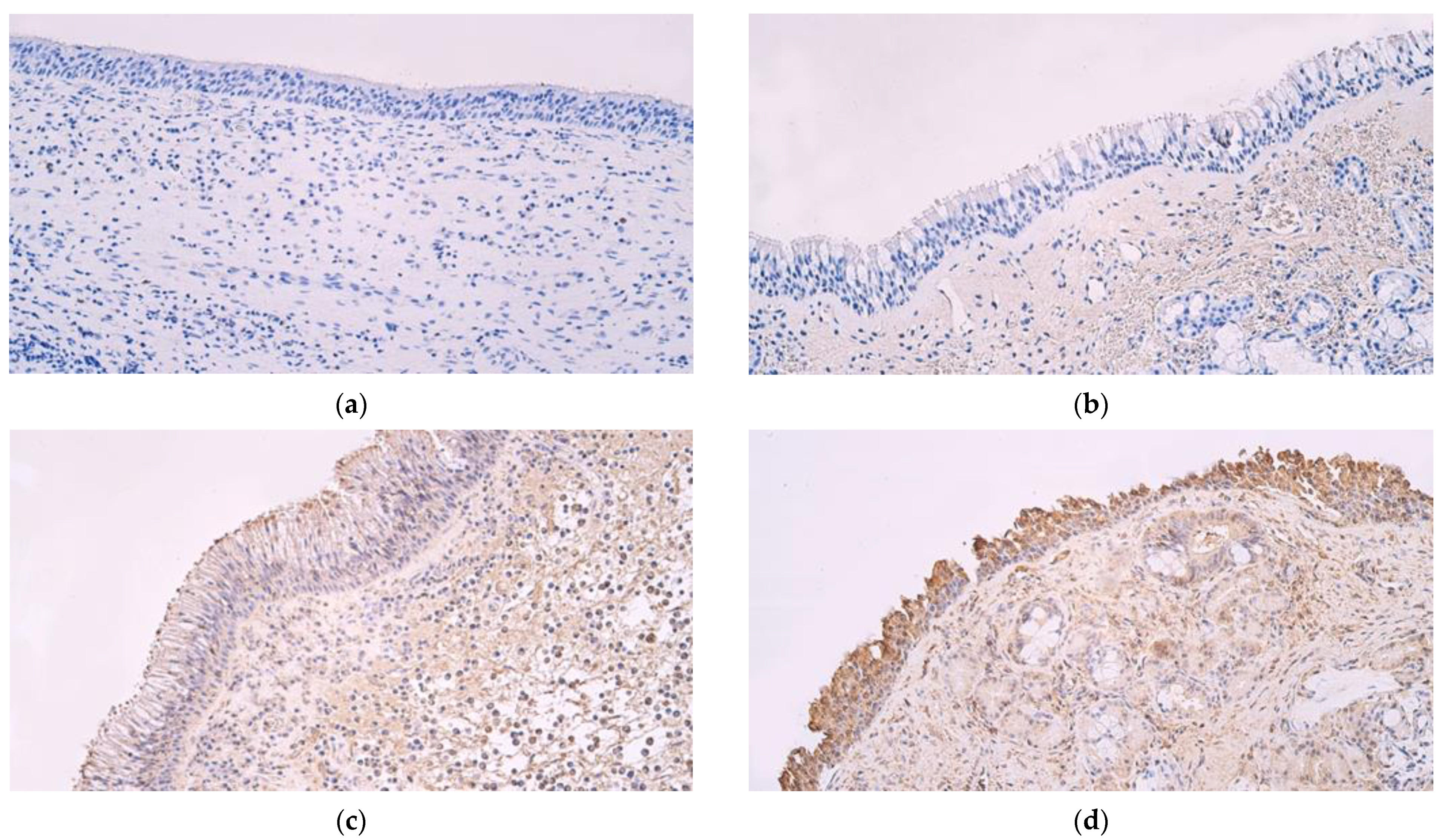
| Epithelial β Defensin 2 | Connective Tissue β Defensin 2 | Epithelial β Defensin 3 | Connective Tissue β Defensin 3 | Epithelial β Defensin 4 | Connective Tissue β Defensin 4 | Epithelial Cathelicidin LL 37 | Connective Tissue Cathelicidin LL 37 | |
|---|---|---|---|---|---|---|---|---|
| Primary polyps | 0.328 (SD ± 0.603) | 1.052 (SD ± 0.939) | 0.603 (SD ± 0.588) | 1.552 (SD ± 1.080) | 0 | 0 | 0.534 (SD ± 0.597) | 1.534 (SD ± 1.008) |
| Recurrent polyps | 0.526 (SD ± 0.716) | 1.632 (SD ± 0.970) | 0.342 (SD ± 0.473) | 1.263 (SD ± 1.147) | 0 | 0 | 0.816 (SD ± 0.691) | 1.632 (SD ± 0.831) |
| Control group | 3.059 (SD ± 0.659) | 1.647 (SD ± 0.386) | 1.853 (SD ± 0.843) | 0.912 (SD ± 0.618) | 0 | 0 | 2.176 (SD ± 0.809) | 1.294 (SD ± 0.811) |
| Primary vs. recurrent group | p = 0.283 | p = 0.050 | p = 0.096 | p = 0.387 | p > 0.999 | p > 0.999 | p = 0.145 | p = 0.568 |
| Primary vs. control group | p < 0.001 | p = 0.026 | p < 0.001 | p = 0.049 | p > 0.999 | p > 0.999 | p < 0.001 | p = 0.610 |
| Recurrent vs. control group | p < 0.001 | p = 0.950 | p < 0.001 | p = 0.531 | p > 0.999 | p > 0.999 | p < 0.001 | p = 0.315 |
| Factor 1 | Factor 2 | R | p Value |
|---|---|---|---|
| Connective tissue Cathelicidin LL 37 | Epithelial Cathelicidin LL 37 | 0.758 ** | <0.001 |
| Connective tissue Cathelicidin LL 37 | Connective tissue β defensin 3 | 0.584 ** | 0.001 |
| Connective tissue β defensin 2 | Epithelial β defensin 2 | 0.561 ** | 0.002 |
| Epithelial Cathelicidin LL 37 | Connective tissue β defensin 3 | 0.556 ** | 0.002 |
| Factor 1 | Factor 2 | R | p Value |
|---|---|---|---|
| Epithelial β defensin 2 | Connective tissue β defensin 2 | 0.635 ** | 0.004 |
| Epithelial β defensin 2 | Epithelial β defensin 3 | 0.505 * | 0.027 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Viksne, R.J.; Sumeraga, G.; Pilmane, M. Antimicrobial and Defense Proteins in Chronic Rhinosinusitis with Nasal Polyps. Medicina 2023, 59, 1259. https://doi.org/10.3390/medicina59071259
Viksne RJ, Sumeraga G, Pilmane M. Antimicrobial and Defense Proteins in Chronic Rhinosinusitis with Nasal Polyps. Medicina. 2023; 59(7):1259. https://doi.org/10.3390/medicina59071259
Chicago/Turabian StyleViksne, Rudolfs Janis, Gunta Sumeraga, and Mara Pilmane. 2023. "Antimicrobial and Defense Proteins in Chronic Rhinosinusitis with Nasal Polyps" Medicina 59, no. 7: 1259. https://doi.org/10.3390/medicina59071259
APA StyleViksne, R. J., Sumeraga, G., & Pilmane, M. (2023). Antimicrobial and Defense Proteins in Chronic Rhinosinusitis with Nasal Polyps. Medicina, 59(7), 1259. https://doi.org/10.3390/medicina59071259






