Viruses Infecting Trees and Herbs That Produce Edible Fleshy Fruits with a Prominent Value in the Global Market: An Evolutionary Perspective
Abstract
1. Introduction
2. Edible Fruits of the World: Which of Them Feed the World?
3. A Co-Evolutionary Arms′ Race between Plants and Viruses: Major Families of Plant Viruses Affecting EFF Crops
4. Beyond the Visible Symptoms Caused by Plant Viruses: Biochemical, Cellular, and Physiological Changes
5. EFF Yield Losses Caused by Viral Diseases
5.1. Tomato
5.2. Bananas and Plantains
5.3. Cucurbitaceae
5.4. Apples
5.5. Grapes
5.6. Citrus
6. Diagnosis of Plant Virus Diseases
6.1. Serological Assays
6.1.1. ELISA
6.1.2. Lateral Flow Assay (LFA)
6.1.3. Dot Immunobinding Assays
6.1.4. Computer-Assisted Epitope Identification to Improve Antibody Production
6.2. Nucleic Acid-Based Assays
6.2.1. PCR
6.2.2. Multiplex PCR
6.2.3. Real-Time PCR
6.2.4. Immunocapture-PCR (IC-PCR)
6.2.5. Loop-Mediated Isothermal Amplification (LAMP)
6.2.6. Recombinase Polymerase Amplification (RPA)
6.2.7. Rolling Circle Amplification (RCA)
6.2.8. Microarray
6.2.9. Next-Generation Sequencing (NGS)
6.3. Biosensors
7. Disease Management
7.1. Horticultural Practices
7.1.1. Use of Disease-Free Propagating Materials and Seeds
7.1.2. Nutrition
7.1.3. Intercropping
7.1.4. Nucellar Embryony
7.1.5. Orchard Roguing
7.1.6. Destruction and Avoidance of Reservoir Plants
7.2. Vector Control
7.3. Thermotherapy
7.4. Biological Control
7.5. Chemical Control
7.6. Use of Disease Resistant Varieties
7.7. Quarantine and Legislations
8. Concluding Remarks and Perspectives
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gepts, P. Origins of plant agriculture and major crop plants. In Our Fragile World: Challenges and Opportunities for Sustainable Development; Tolba, M., Ed.; EOLSS Publishers: Oxford, UK, 2001; pp. 629–637. [Google Scholar]
- Barker, G. The Agricultural Revolution in Prehistory. Why Did Foragers Become Farmers? Oxford Univ Press: Oxford, UK, 2006; p. 616. [Google Scholar]
- Larson, G.; Piperno, D.R.; Allaby, R.G.; Purugganan, M.D.; Andersson, L.; Arrollo-Kalin, M.; Barton, L.; Vigueira, C.C.; Denham, T.; Dobney, K.; et al. Current perspectives and the future of domestication studies. Proc. Natl. Acad. Sci. USA 2014, 111, 6139–6146. [Google Scholar] [CrossRef]
- Harris, D.R. An evolutionary continuum of people-plant interaction. In Foraging and Farming, the Evolution of Plant Exploitation; Harris, D.R., Hillman, G.C., Eds.; Unwin Hyman: London, UK, 1989; pp. 11–26. [Google Scholar]
- Gepts, P. Crop domestication as a long-term selection experiment. Plant. Breed. Rev. 2004, 24, 1–44. [Google Scholar]
- Khoshbakht, K.; Hammer, K. Species richness in relation to the presence of crop plants in families of higher plants. J. Agric. Rural Dev. Trop. Subtrop. 2008, 109, 181–190. [Google Scholar]
- Meyer, R.S.; DuVal, A.E.; Jensen, H.R. Patterns and processes in crop domestication: An historical review and quantitative analysis of 203 global food crops. New Phytol. 2012, 196, 29–48. [Google Scholar] [CrossRef]
- Milla, R.; Bastida, J.M.; Turcotte, M.M.; Jones, G.; Violle, C.; Osborne, C.P.; Chacon-Labella, J.; Sosinski, E.E.; Kattge, J.; Laughlin, D.C.; et al. Phylogenetic patterns and phenotypic profiles of the species of plants and mammals farmed for food. Nature Ecol. Evol. 2018, 2, 1808–1817. [Google Scholar] [CrossRef]
- RBG Kew. The State of the World’s Plants Report, 1st ed.; Willis, K.J., Bachman, S., Eds.; Royal Botanic Gardens Kew: Richmond, UK, 2016; p. 86. [Google Scholar]
- Wiersum, K.F. From natural forests to tree crops, co-domestication of forests and tree species, an overview. Neth. J. Agri. Sci. 1997, 45, 425–438. [Google Scholar] [CrossRef]
- Janick, J. The origin of fruits, fruit growing and fruit breeding. Plant. Breed. Rev. 2005, 25, 255–320. [Google Scholar]
- Blancke, R. Tropical Fruits and other Edible Plants of the World: An Illustrated Guide; Cornell University Press: New York, NY, USA, 2016; p. 350. [Google Scholar]
- Zohary, D.; Hopf, M. Domestication of Plants in the Old World, 3rd ed.; Oxford University Press: New York, NY, USA, 2001; p. 316. [Google Scholar]
- Spengler, R.N. Anthropogenic seed dispersal: Rethinking the origins of plant domestication. Trends Plant. Sci. 2020, 25, 340–348. [Google Scholar] [CrossRef]
- Spengler, R.N.; Petraglia, M.; Roberts, P.; Ashastina, K.; Kistler, L.; Mueller, N.G.; Boivin, N. Exaptation traits for megafaunal mutualisms as a factor in plant domestication. Front. Plant. Sci. 2021, 24, 649394. [Google Scholar] [CrossRef]
- Statista.com. Available online: https://www.statista.com/statistics/264001/worldwide-production-of-fruit-by-variety/ (accessed on 29 July 2021).
- ITC News. Available online: http://www.intracen.org/news/What-are-the-worlds-favourite-fruits/ (accessed on 29 July 2021).
- Halliwell, B.; Rafter, J.; Jenner, A. Health promotion by flavonoids, tocopherols, tocotrienols, and other phenols: Direct or indirect effects? Antioxidant or not? Am. J. Clin. Nutr. 2005, 81, 268S–276S. [Google Scholar] [CrossRef]
- Boeing, H.; Bechthold, A.; Bub, A.; Ellinger, S.; Haller, D.; Kroke, A.; Leschik-Bonnet, E.; Müller, M.J.; Oberritter, H.; Schulze, M. Critical review: Vegetables and fruit in the prevention of chronic diseases. Eur. J. Nutr. 2012, 51, 637–663. [Google Scholar] [CrossRef]
- Aune, D.; Giovannucci, E.; Boffetta, P.; Fadnes, L.T.; Keum, N.; Norat, T.; Greenwood, D.C.; Riboli, E.; Vatten, L.J.; Tonstad, S. Fruit and vegetable intake and the risk of cardiovascular disease, total cancer and all-cause mortality-a systematic review and dose-response meta-analysis of prospective studies. Int. J. Epidemiol. 2017, 46, 1029–1056. [Google Scholar] [CrossRef] [PubMed]
- Rekhy, R.; McConchie, R. Promoting consumption of fruit and vegetables for better health. Have campaigns delivered on the goals? Appetite 2014, 79, 113–123. [Google Scholar] [CrossRef]
- Strange, R.N.; Scott, P.R. Plant disease: A threat to global food security. Annu. Rev. Phytopathol. 2005, 43, 83–116. [Google Scholar] [CrossRef]
- Oerke, E.C. Crop losses to pests. J. Agric. Sci. 2006, 144, 31–43. [Google Scholar] [CrossRef]
- Agrios, G.N. Plant pathogens and disease: General introduction. In Encyclopedia of Microbiology, 3rd ed.; Schaechter, M., Ed.; Academic Press: Burlington, MA, USA, 2009; pp. 613–646. [Google Scholar]
- Jones, R.A.C.; Naidu, R.A. Global dimensions of plant virus diseases: Current status and future perspectives. Annu. Rev. Virol. 2019, 6, 387–409. [Google Scholar] [CrossRef]
- Jones, R.A.C. Global plant virus disease pandemics and epidemics. Plants 2021, 10, 233. [Google Scholar] [CrossRef]
- Jones, R.A.C. Plant virus emergence and evolution: Origins, new encounter scenarios, factors driving emergence, effects of changing world conditions, and prospects for control. Virus. Res. 2009, 141, 113–130. [Google Scholar] [CrossRef]
- DeFries, R.S.; Rudel, T.; Uriarte, M.; Hansen, M. Deforestation driven by urban population growth and agricultural trade in the twenty-first century. Nat. Geosci. 2010, 3, 178–181. [Google Scholar] [CrossRef]
- Godfray, H.C.J.; Beddington, J.R.; Crute, I.R.; Haddad, L.; Lawrence, D.; Muir, J.F.; Pretty, J.; Robinson, S.; Thomas, S.M.; Toulmin, C. Food security: The challenge of feeding nine billion people. Science 2010, 327, 812–818. [Google Scholar] [CrossRef]
- Chakraborty, S.; Newton, A.C. Climate change, plant diseases and food security: An overview. Plant. Pathol. 2011, 60, 2–14. [Google Scholar] [CrossRef]
- Chichester, C.O.; Mrak, E.M.; Stewart, G.F. Advances in Food Research., 1st ed.; Academic Press: Burlington, MA, USA, 1988; p. 299. [Google Scholar]
- Martin, F.W.; Campbell, C.W.; Ruberté, R.M. Perennial Edible Fruits of the Tropics: An Inventory; USDA Agricultural Research Service Agriculture Handbook No. 642; US Government Printing Office: Washington, DC, USA, 1987; p. 251. [Google Scholar]
- Srivastava, A.K.; Hu, C. Fruit Crops: Diagnosis and Management of Nutrient Constraints, 1st ed.; Elsevier Science: Cambridge, MA, USA, 2019; p. 776. [Google Scholar]
- Ploetz, R.C. Diseases of tropical perennial crops: Challenging problems in diverse environments. Plant. Dis. 2007, 91, 644–663. [Google Scholar] [CrossRef]
- Miller, A.J.; Gross, B.L. From forest to field: Perennial fruit crop domestication. Am. J. Bot. 2011, 98, 1389–1414. [Google Scholar] [CrossRef]
- Warren, J. The Nature of Crops: How We Came to Eat the Plants We Do; CABI: Wallingford, UK, 2015; p. 192. [Google Scholar]
- Fleming, T.H.; Kress, W.J. A brief history of fruits and frugivores. Acta Oecol. 2011, 37, 521–530. [Google Scholar] [CrossRef]
- Xiang, Y.; Huang, C.H.; Hu, Y.; Wen, J.; Li, S.; Yi, T.; Chen, H.; Xiang, J.; Ma, H. Evolution of Rosaceae fruit types based on nuclear phylogeny in the context of geological times and genome duplication. Mol. Biol. Evol. 2017, 34, 262–281. [Google Scholar] [CrossRef]
- Sousa-Paz, F.; Pinto, C.E.; Melo-de Brito, R.; Imperatriz-Fonseca, V.L.; Giannini, T.C. Edible fruit plant species in the Amazon forest rely mostly on bees and beetles as pollinators. J. Econ. Entomol. 2021, 114, 710–722. [Google Scholar]
- WFO. Available online: http://www.worldfloraonline.org (accessed on 29 July 2021).
- Chen, J.; Hao, Z.; Guang, X.; Zhao, C.; Wang, P.; Xue, L.; Zhu, Q.; Yang, L.; Sheng, Y.; Zhou, Y.; et al. Liriodendron genome sheds light on angiosperm phylogeny and species-pair differentiation. Nat. Plants 2019, 5, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Judd, W.S.; Campbell, C.S.; Kellogg, E.A.; Stevens, P.F.; Donoghue, M.J. Plant. Systematics: A Phylogenetic Approach, 4th ed.; Sinauer Associates Inc.: Sunderland, MA, USA, 2015; p. 696. [Google Scholar]
- APG III. An update of the Angiosperm phylogeny group classification for the orders and families of flowering plants: APG III. Bot. J. Linn. Soc. 2009, 161, 105–121. [Google Scholar] [CrossRef]
- Tang, H.; Lyons, E.; Schnable, J.C. Early history of the angiosperms. Adv. Bot. Res. 2014, 69, 195–222. [Google Scholar]
- FAOSTAT. Available online: http://www.fao.org/faostat/en/#data/QC (accessed on 29 July 2021).
- OEC. Available online: https://oec.world/en (accessed on 29 July 2021).
- Expert Market Research. Available online: https://www.researchandmarkets.com/ (accessed on 29 July 2021).
- FAO. Available online: www.fao.org/fileadmin/templates/est/COMM_MARKETS_MONITORING/Bananas/Documents/web_Banana_Review_2018_Final_DV.pdf (accessed on 29 July 2021).
- Statista.com. Available online: https://www.statista.com/statistics/716037/global-banana-market-volume/ (accessed on 29 July 2021).
- Fresh Plaza. Available online: https://www.freshplaza.com/article/9299699/overview-global-melon-and-watermelon-market/ (accessed on 29 July 2021).
- Vasylieva, N.; James, H. Production and trade patterns in the world apple market. Innov. Mark. 2021, 17, 16–25. [Google Scholar] [CrossRef]
- FAO. FAO forecasts strong growth prospects for global production and trade of tropical fruits. Available online: http://www.fao.org/americas/informations/ver/fr/c/1193642/ (accessed on 29 July 2021).
- Boyer, J.S. Plant productivity and environment. Science 1982, 218, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Lobell, D.B.; Cassman, K.G.; Field, C.B. Crop yield gaps: Their importance, magnitudes, and causes. Annu. Rev. Environ. Resour. 2009, 34, 179–204. [Google Scholar] [CrossRef]
- Oerke, E.C.; Dehne, H.W.; Schonbeck, F.; Weber, A. Crop. Production and Crop. Protection—Estimated Losses in Major Food and Cash Crops; Elsevier Science: Amsterdam, The Netherlands, 1994; p. 808. [Google Scholar]
- Lovisolo, O.; Hull, R.; Rosler, O. Coevolution of viruses with hosts and vectors and possible paleontology. Adv. Virus Res. 2003, 62, 326–379. [Google Scholar]
- Andret-Link, P.; Fuchs, M. Transmission specificity of plant viruses by vectors. J. Plant. Pathol. 2005, 87, 153–165. [Google Scholar]
- Sastry, K.S.; Mandal, B.; Hammond, J.; Scott, S.W.; Briddon, R.W. Encyclopedia of Plant. Viruses and Viroids; Springer: New Delhi, India, 2019; p. 2946. [Google Scholar]
- Whitfield, A.E.; Falk, B.W.; Rotenberg, D. Insect vector-mediated transmission of plant viruses. Virology 2015, 479, 278–289. [Google Scholar] [CrossRef] [PubMed]
- Hull, R. Plant. Virology, 5th ed.; Academic Press: New York, NY, USA, 2014; p. 1118. [Google Scholar]
- Jones, R.A.C. Plant and insect viruses in managed and natural environments: Novel and neglected transmission pathways. Adv. Virus Res. 2018, 101, 149–187. [Google Scholar] [PubMed]
- Lafforgue, G.; Tromas, N.; Elena, S.F.; Zwart, M.P. Dynamics of the establishment of systemic potyvirus infection: Independent yet cumulative action of primary infection sites. J. Virol. 2012, 86, 12912–12922. [Google Scholar] [CrossRef]
- Calil, I.P.; Fontes, E.P.B. Plant immunity against viruses: Antiviral immune receptors in focus. Ann. Bot. 2016, 119, 200–723. [Google Scholar] [CrossRef]
- Flores, R.; Hernández, C.; Martínez de Alba, A.E.; Daròs, J.A.; Di Serio, F. Viroids and viroid-host interactions. Annu. Rev. Phytopathol. 2005, 43, 117–139. [Google Scholar] [CrossRef]
- Flor, H.H. Current status of the gene-for-gene concept. Annu. Rev. Phytopathol. 1971, 9, 275–296. [Google Scholar] [CrossRef]
- Teixeira, P.J.P.; Colaianni, N.R.; Fitzpatrick, C.R.; Dangl, J.L. Beyond pathogens: Microbiota interactions with the plant immune system. Curr. Opin. Microbiol. 2019, 49, 7–17. [Google Scholar] [CrossRef]
- Mandadi, K.K.; Scholthof, K.B.G. Plant immune responses against viruses: How does a virus cause disease? Plant. Cell 2013, 25, 1489–1501. [Google Scholar] [CrossRef]
- Jones, J.; Dangl, J. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef]
- Soosaar, J.L.M.; Burch-Smith, T.M.; Dinesh-Kumar, S.P. Mechanisms of plant resistance to viruses. Nat. Rev. Microbiol. 2005, 3, 789–798. [Google Scholar] [CrossRef] [PubMed]
- Pallas, V.; García, J.A. How do plant viruses induce disease? Interactions and interference with host components. J. Gen. Virol. 2011, 92, 2691–2705. [Google Scholar] [CrossRef]
- Boualem, A.; Dogimont, C.; Bendahmane, A. The battle for survival between viruses and their host plants. Curr. Opin. Virol. 2016, 17, 32–38. [Google Scholar] [CrossRef]
- Pesti, R.; Kontra, L.; Paul, K.; Vass, I.; Csorba, T.; Havelda, Z.; Várallyay, É. Differential gene expression and physiological changes during acute or persistent plant virus interactions may contribute to viral symptom differences. PLoS ONE 2019, 14, e0216618. [Google Scholar] [CrossRef] [PubMed]
- Almási, A.; Harsányi, A.; Gáborjányi, R. Photosynthetic alterations of virus infected plants. Acta Phytopathol. Entomol. Hung. 2001, 36, 15–29. [Google Scholar] [CrossRef]
- Beimalt, S.; Sonnwald, U. Plant-microbe interactions to probe regulation of plant carbon metabolism. J. Plant. Physiol. 2006, 163, 307–318. [Google Scholar]
- Berger, S.; Sinha, A.K.; Roitsch, T. Plant physiology meets phytopathology: Plant primary metabolism and plant-pathogen interactions. J. Exp. Bot. 2007, 58, 4019–4026. [Google Scholar] [CrossRef] [PubMed]
- Rojas, C.M.; Senthil-Kumar, M.T.; Zin, V.; Mysore, K.S. Regulation of primary plant metabolism during plant-pathogen interactions and its contribution to plant defense. Front. Plant. Sci. 2014, 5, 17. [Google Scholar] [CrossRef]
- Fernández-Calvino, L.; Osorio, S.; Hernández, M.L.; Hamada, I.B.; del Toro, F.J.; Donaire, L.; Yu, A.; Bustos, R.; Fernie, A.R.; Martínez-Rivas, J.M.; et al. Virus-induced alterations in primary metabolism modulate susceptibility to Tobacco rattle virus in Arabidopsis. Plant. Physiol. 2014, 166, 1821–1838. [Google Scholar] [CrossRef]
- Sade, D.; Sade, N.; Shriki, O.; Lerner, S.; Gebremedhin, A.; Karavani, A.; Brotman, Y.; Osorio, S.; Fernie, A.R.; Willmitzer, L.; et al. Water balance, hormone homeostasis, and sugar signaling are all involved in tomato resistance to Tomato yellow leaf curl virus. Plant. Physiol. 2014, 165, 1684–1697. [Google Scholar] [CrossRef]
- Zhao, S.; Li, Y. Current understanding of the interplays between host hormones and plant viral infections. PLoS Pathog. 2021, 17, e1009242. [Google Scholar] [CrossRef]
- Palukaitis, P.; Carr, J.P.; Schoelz, J.E. Plant-virus interactions. In Plant Virology Protocols. Methods in Molecular Biology, 2nd ed.; Foster, G.D., Johansen, I.E., Hong, Y., Nagy, P.D., Eds.; Humana Press: New Jersey, NJ, USA, 2008; pp. 3–19. [Google Scholar]
- Alexander, H.M.; Mauck, K.E.; Whitfield, A.E.; Garrett, K.A.; Malmstrom, C.M. Plant-virus interactions and the agroecological interface. Eur. J. Plant. Pathol. 2014, 138, 529–547. [Google Scholar] [CrossRef]
- Fauquet, C.M. Taxonomy, classification and nomenclature of viruses. In Encyclopedia of Virology, 2nd ed.; Granoff, A., Webster, R.G., Eds.; Academic Press: Burlington, MA, USA, 1999; pp. 1730–1756. [Google Scholar]
- Koonin, E.V.; Dolja, V.V. A virocentric perspective on the evolution of life. Curr. Opin. Virol. 2013, 3, 546–557. [Google Scholar] [CrossRef] [PubMed]
- Lefkowitz, E.J.; Dempsey, D.M.; Hendrickson, R.C. Virus taxonomy: The database of the international committee on taxonomy of viruses (ICTV). Nucleic Acids Res. 2018, 46, D708–D717. [Google Scholar] [CrossRef]
- International Committee on Taxonomy of Viruses Executive Committee. The new scope of virus taxonomy: Partitioning the virosphere into 15 hierarchical ranks. Nat. Microbiol. 2020, 5, 668–674. [Google Scholar] [CrossRef] [PubMed]
- International Committee on Taxonomy of Viruses. Virus Metadata Repository: Version May 18. 2021; MSL36. Available online: https://talk.ictvonline.org/taxonomy/vmr/m/vmr-file-repository/12323 (accessed on 18 September 2021).
- Scholthof, K.B.G.; Adkins, S.; Czosnek, H.; Palukaitis, P.; Jacquot, E.; Hohn, T.; Hohn, B.; Saunders, K.; Candresse, T.; Ahlquist, P.; et al. Top 10 plant viruses in molecular plant pathology. Mol. Plant. Pathol. 2011, 12, 938–954. [Google Scholar] [CrossRef]
- Rybicki, E.P. A top ten list for economically important plant viruses. Arch. Virol. 2015, 160, 17–20. [Google Scholar] [CrossRef] [PubMed]
- Annual Report. Virus diseases of fruit trees. In The Kent Incorporated Society for Promoting Experiments in Horticulture; East Mailing Research Station Report; University of California: Oakland, CA, USA, 1962; p. 127.
- Sweet, J.B. Fruit tree virus infections of woody exotic and indigenous plants in Britain. Acta Phytopathol. 1980, 15, 231–238. [Google Scholar] [CrossRef]
- Martelli, G.P.; Uyemoto, J.K. Plant virus diseases: Fruit trees and grapevine. In Encyclopedia of Virology, 3rd ed.; Mahy, B.W.J., Van Regenmortel, M.H.V., Eds.; Academic Press: Burlington, MA, USA, 2008; pp. 201–207. [Google Scholar]
- Umer, M.; Liu, J.; You, H.; Xu, C.; Dong, K.; Luo, N.; Kong, L.; Li, X.; Hong, N.; Wang, G.; et al. Genomic, morphological and biological traits of the viruses infecting major fruit trees. Viruses 2019, 11, 515. [Google Scholar] [CrossRef]
- Cole, T.C.H.; Hilger, H.H.; Stevens, P.F. Angiosperm phylogeny poster 2019: Flowering plant systematics. Available online: http://www.mobot.org/MOBOT/research/APweb/ (accessed on 25 June 2021).
- Arcady, R.M.; Elena, S.F. Evolution of plant virus movement proteins from the 30K superfamily and of their homologs integrated in plant genomes. Virology 2015, 476, 304–315. [Google Scholar]
- Diop, S.I.; Geering, A.; Alfama-Depauw, F.; Loaec, M.; Teycheney, P.Y.; Maumus, F. Tracheophyte genomes keep track of the deep evolution of the Caulimoviridae. Sci. Rep. 2018, 8, 572. [Google Scholar] [CrossRef]
- Morris, C.E.; Moury, B. Revisiting the concept of host range of plant pathogens. Annu. Rev. Phytopathol. 2019, 57, 63–90. [Google Scholar] [CrossRef]
- Kumar, V.; Baweja, M.; Singh, P.K.; Shukla, P. Recent developments in systems biology and metabolic engineering of plant-microbe interactions. Front. Plant. Sci. 2016, 7, 1421. [Google Scholar] [CrossRef]
- Barba, M.; Ilardi, V.; Pasquini, G. Control of pome and stone fruit virus diseases. In Advances in Virus Research; Loebenstein, G., Katis, N.I., Eds.; Academic Press: Burlington, MA, USA, 2015; pp. 47–83. [Google Scholar]
- Roossinck, M.J.; Martin, D.P.; Roumagnac, P. Plant virus metagenomics: Advances in virus discovery. Phytopathology 2015, 105, 716–727. [Google Scholar] [CrossRef] [PubMed]
- Maliogka, V.; Minafra, A.; Saldarelli, P.; Ruiz-García, A.; Glasa, M.; Katis, N.; Olmos, A. Recent advances on detection and characterization of fruit tree viruses using high-throughput sequencing technologies. Viruses 2018, 10, 436. [Google Scholar] [CrossRef]
- Xu, Y.; Li, S.; Na, C.; Yang, L.; Lu, M. Analyses of virus/viroid communities in nectarine trees by next-generation sequencing and insight into viral synergisms implication in host disease symptoms. Sci. Rep. 2019, 9, 12261. [Google Scholar] [CrossRef] [PubMed]
- McLeish, M.J.; Fraile, A.; Garcia-Arenal, F. Evolution of plant-virus interactions: Host range and virus emergence. Curr. Opin. Virol. 2019, 34, 50–55. [Google Scholar] [CrossRef] [PubMed]
- Elena, S.F.; Fraile, A.; García-Arenal, F. Evolution and emergence of plant viruses. Adv. Virus Res. 2014, 88, 161–191. [Google Scholar]
- Jones, R.A.C. Disease pandemics and major epidemics arising from new encounters between indigenous viruses and introduced crops. Viruses 2020, 12, 1388. [Google Scholar] [CrossRef] [PubMed]
- Maclot, F.; Candresse, T.; Filloux, D.; Malmstrom, C.M.; Roumagnac, P.; van der Vlugt, R.; Massart, S. Illuminating an ecological blackbox: Using high throughput sequencing to characterize the plant virome across scales. Front. Microbiol. 2020, 11, 578064. [Google Scholar] [CrossRef] [PubMed]
- McLeish, M.J.; Fraile, A.; García-Arenal, F. Population genomics of plant viruses: The ecology and evolution of virus emergence. Phytopathology 2021, 111, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Mascia, T.; Gallitelli, D. Synergies and antagonisms in virus interactions. Plant. Sci. 2016, 252, 176–192. [Google Scholar] [CrossRef]
- Naidu, R.A.; Maree, H.J.; Burger, J.T. Grapevine leafroll disease and associated viruses: A unique pathosystem. Annu. Rev. Phytopathol. 2015, 53, 613–634. [Google Scholar] [CrossRef] [PubMed]
- Crnogorac, A.; Panno, S.; Mandić, A.; Gašpar, M.; Caruso, A.G.; Noris, E.; Davino, S.; Matić, S. Survey of five major grapevine viruses infecting Blatina and Žilavka cultivars in Bosnia and Herzegovina. PLoS ONE 2021, 16, e0245959. [Google Scholar] [CrossRef] [PubMed]
- Hanssen, I.M.; Paeleman, A.; Wittemans, L.; Goen, K.; Lievens, B.; Bragard, C.; Vanachter, A.C.R.C.; Thomma, B.P.H.J. Genetic characterization of Pepino mosaic virus isolates from Belgian greenhouse tomatoes reveals genetic recombination. Eur. J. Plant. Pathol. 2008, 121, 131–146. [Google Scholar] [CrossRef]
- Brioso, P.S.T.; Cordeiro, Z.J.M.; Rezende, J.A.M.; Kitajima, E.W.; Pimentel, J.P.; Figueiredo, A.R. Mixed infection by cucumber mosaic (CMV) and banana streak (BSV) viruses in banana in Brazil. Summa Phytopathol. 2000, 26, 254–257. [Google Scholar]
- Carnelossi, P.R.; Bijora, T.; Facco, C.U.; Silva, J.M.; Picoli, M.H.S.; Souto, E.R.; Oliveira, F.T. Episomal detection of Banana streak OL virus in single and mixed infection with Cucumber mosaic virus in Banana “Nanicão Jangada”. Trop. Plant. Pathol. 2014, 39, 342–346. [Google Scholar] [CrossRef]
- Lucía-Sanz, A.; Manrubia, S. Multipartite viruses: Adaptive trick or evolutionary treat? Syst. Biol. Appl. 2017, 3, 34. [Google Scholar] [CrossRef]
- Kappagantu, M.; Collum, T.D.; Dardick, C.; Culver, J.N. Viral Hacks of the Plant Vasculature: The Role of phloem alterations in systemic virus infection. Annu. Rev. Virol. 2020, 7, 10.2–10.20. [Google Scholar] [CrossRef]
- Navarro, J.A.; Serra-Soriano, M.; Corachán-Valencia, L.; Pallás, V. A conserved motif in three viral movement proteins from diferent genera is required for host factor recruitment and cell-to-cell movement. Sci. Rep. 2020, 10, 47–58. [Google Scholar] [CrossRef]
- Munir, N.; Hameed, A.A.; Haq, R.; Naz, S. Biochemical changes in cultivars of sweet oranges infected with citrus tristeza virus. Braz. J. Biol. 2019, 79, 742–748. [Google Scholar] [CrossRef]
- Moriones, E.; Navas-Castillo, J. Tomato yellow leaf curl virus, an emerging virus complex causing epidemics worldwide. Virus Res. 2000, 71, 123–134. [Google Scholar] [CrossRef]
- Andret-Link, P.; Laporte, C.; Valat, L.; Ritzenthaler, C.; Demangeat, G.; Vigne, E.; Laval, V.; Pfeiffer, P.; Stussi-Garaud, C.; Fuchs, M. Grapevine fanleaf virus: Still a major threat to the grapevine industry. J. Plant. Pathol. 2004, 86, 183–195. [Google Scholar]
- Morilla, G.; Janssen, D.; García-Andrés, S.; Moriones, E.; Cuadrado, I.M.; Bejarano, E.R. Pepper (Capsicum annuum) is a dead-end host for Tomato yellow leaf curl virus. Phytopathology 2005, 95, 1089–1097. [Google Scholar] [CrossRef] [PubMed]
- Bai, M.; Chen, W.; Xie, B.; Yang, G. A novel strategy to enhance resistance to Cucumber mosaic virus in tomato by grafting to transgenic rootstocks. J. Integr. Agric. 2016, 15, 2040–2048. [Google Scholar] [CrossRef]
- Basso, M.F.; Fajardo, T.V.M.; Saldarelli, P. Grapevine virus diseases: Economic impact and current advances in viral prospection and management. Rev. Bras. Frutic. 2017, 39, 1–22. [Google Scholar] [CrossRef]
- Hemmer, C.; Djennane, S.; Ackerer, L.; Hleibieh, K.; Marmonier, A.; Gersch, S.; Shahinez Garcia, S.; Vigne, E.; Komar, V.; Perrin, M.; et al. Nanobody-mediated resistance to Grapevine fanleaf virus in plants. Plant. Biotechnol. J. 2017, 16, 660–671. [Google Scholar] [CrossRef] [PubMed]
- Kinoti, W.; Constable, F.; Nancarrow, N.; Plummer, K.; Rodoni, B. The incidence and genetic diversity of apple mosaic virus (ApMV) and Prune dwarf virus (PDV) in Prunus species in Australia. Viruses 2018, 10, 136. [Google Scholar] [CrossRef]
- Liu, M.; Liang, Z.; Aranda, M.A.; Hong, N.; Liu, L.; Kang, B.; Gu, Q. A cucumber green mottle mosaic virus vector for virus-induced gene silencing in cucurbit plants. Plant. Methods. 2020, 16, 1–13. [Google Scholar] [CrossRef]
- Ertunç, F. Physiology of virus-infected plants. In Applied Plant Virology, Awasthi, L.P., Ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2020; pp. 199–205. [Google Scholar]
- Hull, R. Matthews’ Plant. Virology; Elsevier Inc.: Amsterdam, The Netherlands, 2002; Chapter 9; pp. 373–436. [Google Scholar]
- Mandahar, C.L.; Garg, I.D. Effect of cucumber mosaic virus on chlorophyll content, photosynthesis, respiration and carbohydrates of infected Luffa aegyptiaca Mill. J. Phytopathol. 1972, 75, 181–186. [Google Scholar] [CrossRef]
- Milavec, M.; Kovac, M.; Ravnikar, M. Photosynthetic pigments in potato plants (Solarium tuberosum L.) cv. Igor after primary infection with potato virus YNTN. Phyton 1999, 39, 265–269. [Google Scholar]
- Liu, J.; Yang, J.; Bi, H.; Zhang, P. Why mosaic? Gene expression profiling of african cassava mosaic virus-infected cassava reveals the effect of chlorophyll degradation on symptom development. J. Integr. Plant. Biol. 2014, 56, 122–132. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Zhang, X.; Hong, Y.; Liu, Y. Chloroplast in plant-virus interaction. Front. Microbiol. 2016, 7, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Bailiss, K. Infection of cucumber cotyledons by cucumber mosaic virus and the participation of chlorophyllase in the development of chlorotic lesions. Ann. Bot. 1970, 34, 647–655. [Google Scholar] [CrossRef]
- Radwan, D.E.M.; Fayez, K.A.; Younis Mahmoud, S.; Hamad, A.; Lu, G. Physiological and metabolic changes of Cucurbita pepo leaves in response to zucchini yellow mosaic virus (ZYMV) infection and salicylic acid treatments. Plant. Physiol. Biochem. 2007, 45, 480–489. [Google Scholar] [CrossRef] [PubMed]
- Moreno, P.; Ambrós, S.; Albiach-Martí, M.R.; Guerri, J.; Peña, L. Citrus tristeza virus: A pathogen that changed the course of the citrus industry. Physiol. Mol. Plant. Pathol. 2008, 9, 251–268. [Google Scholar] [CrossRef]
- Glick, E.; Levy, Y.; Gafni, Y. The viral etiology of tomato yellow leaf curl disease—A review. Plant. Protect. Sci. 2009, 45, 81–97. [Google Scholar] [CrossRef]
- Prasad, A.; Sharma, N.; Hari-Gowthem, G.; Muthamilarasan, M.; Prasad, M. Tomato yellow leaf curl virus: Impact, challenges, and management. Trends Plant. Sci. 2020, 25, 897–911. [Google Scholar] [CrossRef]
- Svoboda, J.; Polak, J. Relative concentration of apple mosaic virus coat protein in different parts of apple tree. Hort. Sci. 2010, 37, 22–26. [Google Scholar] [CrossRef]
- European and Mediterranean Plant Protection Organization. Available online: https://gd.eppo.int (accessed on 24 June 2021).
- Jimenez, I.; Lopez, L.; Alamillo, J.M.; Valli, A.; Garcia, J.A. Identification of a plum pox virus CI-interacting protein from chloroplast that has a negative effect in virus infection. Mol. Plant. Microbe Interact. 2006, 19, 350–358. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Chen, J.; Hong, X.; Chen, J.; Adams, M.J. A potyvirus P1 protein interacts with the Rieske Fe/S protein of its host. Mol. Plant. Pathol. 2007, 8, 785–790. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Y.; Li, H.F.; Wong, S.M.; Fan, Z.F. Plastocyanin transit peptide interacts with potato virus X coat protein, while silencing of plastocyanin reduces coat protein accumulation in chloroplasts and symptom severity in host plants. Mol. Plant. Microbe Interac. 2009, 22, 1523–1534. [Google Scholar] [CrossRef]
- Jang, C.; Seo, E.Y.; Nam, J.; Bae, H.; Gim, Y.G.; Kim, H.G.; Cho, S.; Lee, Z.W.; Bauchan, G.R.; Hammond, J.; et al. Insights into alternanthera mosaic virus TGB3 functions: Interactions with Nicotiana benthamiana PsbO correlate with chloroplast vesiculation and veinal necrosis caused by TGB3 over-expression. Front. Plant. Sci. 2013, 4, 1–15. [Google Scholar] [CrossRef]
- Sun, X.; Li, Y.; Shi, M.; Zhang, N.; Wu, G.; Li, T.; Qing, L.; Zhou, C. In vitro binding and bimolecular fluorescence complementation assays suggest an interaction between tomato mosaic virus coat protein and tobacco chloroplast ferredoxin I. Arch. Virol. 2013, 158, 2611–2615. [Google Scholar] [CrossRef]
- Ma, Y.X.; Zhou, T.; Hong, Y.G.; Fan, Z.; Li, H. Decreased level of ferredoxin I in tobacco mosaic virus-infected tobacco is associated with development of the mosaic symptom. Physiol. Mol. Plant. Pathol. 2008, 72, 39–45. [Google Scholar] [CrossRef]
- Feki, S.; Loukili, M.J.; Triki-Marrakchi, R.; Karimova, G.; Old, I.; Ounouna, H.; Nato, A.; Nato, F.; Guesdon, J.L.; Lafaye, P.; et al. Interaction between tobacco Ribulose-l,5-biphosphate Carboxylase/Oxygenase large subunit (RubisCO-LSU) and the PVY Coat Protein (PVY-CP). Eur. J. Plant. Pathol. 2005, 112, 221–234. [Google Scholar] [CrossRef]
- Rahoutei, J.; Garcia-Luque, I.; Baron, M. Inhibition of photosynthesis by viral infection: Effect on PSII structure and function. Physiol. Plant. 2000, 110, 286–292. [Google Scholar] [CrossRef]
- Sampol, B.; Bota, J.; Riera, D.; Medrano, H.; Flexas, J. Analysis of the virus-induced inhibition of photosynthesis in malmsey grapevines. New Phytol. 2003, 160, 403–412. [Google Scholar] [CrossRef]
- Pineda, M.; Sajnani, C.; Barón, M. Changes induced by the Pepper mild mottle tobamovirus on the chloroplast proteome of Nicotiana benthamiana. Photosynth. Res. 2009, 103, 31–45. [Google Scholar] [CrossRef]
- Kyseláková, H.; Prokopová, J.; Nauš, J.; Novák, O.; Navrátil, M.; Šafářová, D.; Spundová, M.; Ilík, P. Photosynthetic alterations of pea leaves infected systemically by pea enation mosaic virus: A coordinated decrease in efficiencies of CO2 assimilation and photosystem II photochemistry. Plant. Physiol. Biochem. 2011, 49, 1279–1289. [Google Scholar] [CrossRef]
- Shahrukh, S.; Um, E.; Khan, S.; Parveen, N.; Fatima, M.; Dahot, M.U. Certain growth-related attributes of bunchy top virus infected banana under ex-vitro conditions. Afr. J. Biotechnol. 2014, 13, 1876–1882. [Google Scholar] [CrossRef]
- Kumar, P.L.; Selvarajan, R.; Iskra-Caruana, M.L.; Chabannes, M.; Hanna, R. Biology, Etiology, and Control of Virus Diseases of Banana and Plantain. In Control of Plant Virus Diseases—Vegetatively-Propagated Crops; Loebenstein, N., Ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2015; Volume 91, pp. 229–269. [Google Scholar]
- Christov, I.; Stefanov, D.; Velinov, T.; Goltsev, V.; Georgieva, K.; Abracheva, P.; Genova, Y.; Christov, N. The symptomless leaf infection with grapevine leafroll associated virus 3 in grown in vitro plants as a simple model system for investigation of viral effects on photosynthesis. J. Plant. Physiol. 2007, 164, 1124–1133. [Google Scholar] [CrossRef] [PubMed]
- Palanisamy, P.; Michael, P.I.; Krishnaswamy, M. Physiological response of yellow vein mosaic virus-infected bhendi [Abelmoschus esculentus] leaves. Physiol. Mol. Plant. Pathol. 2009, 74, 129–133. [Google Scholar] [CrossRef]
- Halldorson, M.M.; Keller, M. Grapevine leafroll disease alters leaf physiology but has little effect on plant cold hardiness. Planta 2018, 248, 1201–1211. [Google Scholar] [CrossRef] [PubMed]
- Basso, M.F.; Fajardo, T.V.M.; Santos, H.P.; Guerra, C.C.; Ayub, R.A.; Nickel, O. Fisiologia foliar e qualidade enológica da uva em videiras infectadas por vírus. Trop. Plant Pathol. 2010, 35, 351–359. [Google Scholar] [CrossRef]
- Alabi, O.J.; Casassa, L.F.; Gutha, L.R.; Larsen, R.C.; Henick-Kling, T.; Harbertson, J.F.; Naidu, R.A. Impacts of grapevine leafroll disease on fruit yield and grape and wine chemistry in a wine grape (Vitis vinifera L.) cultivar. PLoS ONE 2016, 11, e0149666. [Google Scholar] [CrossRef]
- Shalitin, D.; Wolf, S. Cucumber mosaic virus infection affects sugar transport in melon plants. Plant. Physiol. 2000, 123, 597–604. [Google Scholar] [CrossRef]
- Reddy, G.S.; Murti, V.D. Citrus Disease and Their Control; Indian Council of Agricultural Research Publication: New Delhi, India, 1985; p. 79. [Google Scholar]
- Adkins, S.; McCollum, T.G.; Albano, J.P.; Kousik, C.S.; Baker, C.A.; Webster, C.G.; Roberts, P.D.; Webb, S.E.; Turechek, W.W. Physiological effects of squash vein yellowing virus infection on watermelon. Plant. Dis. 2013, 97, 1137–1148. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dória, M.S.; Sousa, A.O.; Barbosa, C.D.J.; Costa, M.G.C.; Gesteira, A.D.S.; Souza, R.M.; Freitas, A.C.O.; Pirovani, C.P. Citrus tristeza virus (CTV) causing proteomic and enzymatic changes in sweet orange variety “Westin.”. PLoS ONE 2015, 10, e0130950. [Google Scholar]
- Pazarlar, S.; Gümüş, M.; Öztekin, G.B. The Effects of Tobacco mosaic virus infection on growth and physiological parameters in some pepper varieties (Capsicum annuum L.). Not. Bot. Hortic. Agrobot. 2013, 41, 427–433. [Google Scholar] [CrossRef]
- Alazem, M.; Lin, N.S. Roles of plant hormones in the regulation of host-virus interactions. Mol. Plant. Pathol. 2015, 16, 529–540. [Google Scholar] [CrossRef]
- Pieterse, C.M.J.; Van der Does, D.; Zamioudis, C.; Leon-Reyes, A.; Van Wees, S.C.M. Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 2012, 28, 489–521. [Google Scholar] [CrossRef]
- Chivasa, S. Salicylic acid interferes with tobacco mosaic virus replication via a novel salicylhydroxamic acid-sensitive mechanism. The Plant. Cell. 1997, 9, 547–557. [Google Scholar] [CrossRef]
- Rodriguez, M.C.; Conti, G.; Zavallo, D.; Manacorda, C.A.; Asurmendi, S. TMV-Cg Coat Protein stabilizes DELLA proteins and in turn negatively modulates salicylic acid-mediated defense pathway during Arabidopsis thaliana viral infection. BMC Plant. Biol. 2014, 14, 1–17. [Google Scholar] [CrossRef]
- Huang, L.; Ren, Q.; Sun, Y.; Ye, L.; Cao, H.; Ge, F. Lower incidence and severity of tomato virus in elevated CO2 is accompanied by modulated plant induced defence in tomato. Plant. Biol. 2012, 14, 905–913. [Google Scholar] [CrossRef]
- Clarke, S.F.; McKenzie, M.J.; Burritt, D.J.; Guy, P.L.; Jameson, P.E. Influence of white clover mosaic potexvirus infection on the endogenous cytokinin content of bean . Plant Physiol. 1999, 120, 547–552. [Google Scholar]
- Synková, H.; Semorádová, Š.; Schnablová, R.; Müller, K.; Pospíšilová, J.; Ryšlavá, H.; Malbeck, J.; Čeřovská, N. Effects of biotic stress caused by Potato virus Y on photosynthesis in ipt transgenic and control Nicotiana tabacum L. Plant. Sci. 2006, 171, 607–616. [Google Scholar] [CrossRef]
- Bolouri Moghaddam, M.R.; Van den Ende, W. Sugars and plant innate immunity. J. Exp. Bot. 2012, 63, 3989–3998. [Google Scholar] [CrossRef]
- Kang, B.C.; Yeam, I.; Jahn, M.M. Genetics of plant virus resistance. Annu. Rev. Phytopathol. 2005, 43, 581–621. [Google Scholar] [CrossRef] [PubMed]
- Mauck, K.E.; De Moraes, C.M.; Mescher, M.C. Deceptive chemical signals induced by a plant virus attract insect vectors to inferior hosts. Proc. Natl. Acad. Sci. USA 2010, 107, 3600–3605. [Google Scholar] [CrossRef]
- Chen, G.; Pan, H.; Xie, W.; Wang, S.; Wu, Q.; Fang, Y.; Shi, X.; Zhang, Y. Virus infection of a weed increases vector attraction to and vector fitness on the weed. Sci. Rep. 2013, 3, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Feng, C.; Wu, K.; Chen, W.; Chen, Y.; Hao, X.; Wu, Y. Advances and prospects in biogenic substances against plant virus: A review. Pestic Biochem. Physiol. 2017, 135, 15–26. [Google Scholar] [CrossRef]
- Rendina, N.; Nuzzaci, M.; Scopa, A.; Cuypers, A.; Sofo, A. Chitosan-elicited defense responses in cucumber mosaic virus (CMV)-infected tomato plants. J. Plant. Physiol. 2019, 234–235, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Waterworth, H.E.; Hadidi, A. Economic losses due to plant viruses. In Plant Virus Disease Control; Hadidi, A., Khetarpal, R.K., Koganezawa, H., Eds.; APS Press: St. Paul, MN, USA, 1988; pp. 1–13. [Google Scholar]
- Anderson, P.K.; Cunningham, A.A.; Patel, N.G.; Morales, F.J.; Epstein, P.R.; Daszak, P. Emerging infectious diseases of plants: Pathogen pollution, climate change and agrotechnology drivers. Trends Ecol. Evol. 2004, 19, 535–544. [Google Scholar] [CrossRef]
- Sastry, K.S.; Zitter, T.A. Plant. Virus and Viroid Diseases in the Tropics; Springer: Berlin/Heidelberg, Germany, 2014; Volume 2, pp. 373–436. [Google Scholar]
- Jones, R.A.C. Future scenarios for plant virus pathogens as climate change progresses. In Advances in Virus Research; Kielian, M., Maramorosch, K., Mettenleitter, T.C., Eds.; Elsevier Inc.: Amsterdam, The Netherlands, 2016; Volume 95, pp. 87–147. [Google Scholar]
- Ranawaka, B.; Hayashi, S.; Waterhouse, P.M.; de Felippes, F.F. Homo sapiens: The superspreader of plant viral diseases. Viruses 2020, 12, 1462. [Google Scholar] [CrossRef]
- Tenllado, F.; Canto, T. Effects of a changing environment on the defenses of plants to viruses. Curr. Opin. Virol. 2020, 42, 40–46. [Google Scholar] [CrossRef]
- Trebicki, P. Climate change and plant virus epidemiology. Virus Res. 2020, 286, 1–25. [Google Scholar] [CrossRef]
- Van Munster, M. Impact of abiotic stresses on plant virus transmission by Aphids. Viruses 2020, 12, 216. [Google Scholar] [CrossRef]
- Legg, J. Emergence, spread and strategies for controlling the pandemic of cassava mosaic virus disease in east and central Africa. Crop. Pro. 1999, 18, 627–637. [Google Scholar] [CrossRef]
- Legg, J.P.; Thresh, J.M. Cassava mosaic virus disease in East Africa: A dynamic disease in a changing environment. Virus Res. 2000, 71, 135–149. [Google Scholar] [CrossRef]
- Calvert, L.A.; Tresh, J.M. The Viruses and Virus diseases of Cassava. In Cassava: Biology, Production and Utilization; Hillocks, R.J., Tresh, J.M., Bellotti, A.C., Eds.; CAB International: Wallingford, UK, 2002; pp. 237–257. [Google Scholar]
- Otim-Nape, G.W.; Thresh, J.M.; Shaw, M.W. The incidence and severity of cassava mosaic virus disease in Uganda: 1990–1922. Trop. Sci. 1998, 38, 25–37. [Google Scholar]
- Jeger, M.; Beresford, R.; Bock, C.; Brown, N.; Fox, A.; Newton, A.; Vicent, A.; Xu, X.; Yuen, J. Global challenges facing plant pathology: Multidisciplinary approaches to meet the food security and environmental challenges in the mid-twenty-first century. CABI Agric. Biosci. 2021, 2, 20. [Google Scholar] [CrossRef]
- Rao, G.P.; Reedy, M.G. Overview of yield losses due to plant viruses. In Applied Plant Virology. Advances, Detection, and Antiviral Strategies; Awasthi, L.P., Ed.; Elservier Inc.: Amsterdam, The Netherlands, 2020; pp. 531–562. [Google Scholar]
- Hanssen, I.M.; Lapidot, M. Major tomato viruses in the Mediterranean basin. In Viruses and Virus Diseases of Vegetables in the Mediterranean Basin, 1st ed.; Loebenstein, G., Lecoq, H., Eds.; Elservier Inc.: London, UK, 2012; Volume 84, pp. 31–66. [Google Scholar]
- Antignus. Y. Tomato. In Virus and Virus-Like Diseases of Major Crops in Developing Countries, 1st ed.; Loebenstein, G., Thottappilly, G., Eds.; Springer Science-Business Media: Dordrecht, The Netherlands, 2003; Volume 1, pp. 641–663. [Google Scholar]
- Levy, D.; Lapidot, M. Effect of plant age at inoculation on expression of genetic resistance to tomato yellow leaf curl virus. Arch. Virol. 2007, 153, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Varsani, A.; Shepherd, D.N.; Monjane, A.L.; Owor, B.E.; Erdmann, J.B.; Rybicki, E.P.; Peterschmitt, M.; Briddon, R.W.; Markham, P.G.; Oluwafemi, S.; et al. Recombination, decreased host specificity and increased mobility may have driven the emergence of maize streak virus as an agricultural pathogen. J. Gen. Virol. 2008, 89, 2063–2074. [Google Scholar] [CrossRef]
- García-Andrés, S.; Accotto, G.P.; Navas-Castillo, J.; Moriones, E. Founder effect, plant host, and recombination shape the emergent population of begomoviruses that cause the tomato yellow leaf curl disease in the Mediterranean basin. Virology 2007, 359, 302–312. [Google Scholar] [CrossRef]
- Lefeuvre, P.; Martin, D.P.; Harkins, G.; Lemey, P.; Gray, A.J.A.; Meredith, S.; Lakay, F.; Monjane, A.; Lett, J.M.; Varsani, A.; et al. The spread of tomato yellow leaf curl virus from the Middle East to the World. PLoS Pathog. 2010, 6, 1–13. [Google Scholar] [CrossRef]
- Patil, B.L.; Fauquet, C.M. Cassava mosaic geminiviruses: Actual kwoledge and perspectives. Mol. Plant. Patholog. 2009, 10, 685–707. [Google Scholar] [CrossRef]
- Lefeuvre, P.; Moriones, E. Recombination as a motor of host switches and virus emergence: Geminiviruses as case studies. Curr. Opin. Virol. 2015, 10, 14–19. [Google Scholar] [CrossRef]
- Pietersen, G.; Smith, M.F. Tomate yellow leaf curl virus resistant tomatoes show resistance to Tomato curly stunt virus. Plant. Dis. 2002, 86, 528–534. [Google Scholar] [CrossRef] [PubMed]
- Soler, S.; Cebolla-Cornejo, J.; Prohens, J.; Nuez, F. El pepino mosaic virus (PepMV), una nueva amenaza para el cultivo del tomate II. Vida Rural 2000, 119, 48–52. [Google Scholar]
- Verhoeven, J.T.J.; van der Vlugt, R.A.A.; Roenhorst, J.W. High similarity between tomato isolates of pepino mosaic virus suggests a common origin. Eur. J. Plant. Pathol. 2003, 109, 419–425. [Google Scholar] [CrossRef]
- Fakhro, A.; von Bargen, S.; Bandte, M.; Büttner, C.; Franken, P.; Schwarz, D. Susceptibility of different plant species and tomato cultivars to two isolates of pepino mosaic virus. Eur. J. Plant. Pathol. 2011, 129, 579–590. [Google Scholar] [CrossRef]
- Klap, C.; Luria, N.; Smith, E.; Hadad, L.; Bakelman, E.; Sela, N.; Belausov, E.; Lachman, O.; Leibman, D.; Dombrovsky, A. Tomato brown rugose fruit virus contributes to enhanced pepino mosaic virus titers in tomato plants. Viruses 2020, 12, 879. [Google Scholar] [CrossRef]
- Zaidi, S.S.E.A.; Martin, D.P.; Amin, I.; Farooq, M.; Mansoor, S. Tomato leaf curl New Delhi virus: A widespread bipartite begomovirus in the territory of monopartite begomoviruses. Mol. Plant. Patholog. 2016, 18, 901–911. [Google Scholar] [CrossRef] [PubMed]
- Juárez, M.; Tovar, R.; Fiallo-Olivé, E.; Aranda, M.A.; Gosálvez, B.; Castillo, P.; Moriones, E.; Navas-Castillo, J. First detection of Tomato leaf curl New Delhi virus infecting zucchini in Spain. Plant. Dis. 2014, 98, 857. [Google Scholar] [CrossRef]
- Tahir, M.; Haider, M.S. First report of Tomato leaf curl New Delhi virus infecting bitter gourd in Pakistan. Plant. Pathol. 2005, 54, 807. [Google Scholar] [CrossRef]
- Mizutani, T.; Daryono, B.S.; Ikegami, M.; Natsuaki, K.T. First report of tomato leaf curl New Delhi virus infecting cucumber in Central Java, Indonesia. Plant. Dis. 2011, 95, 1485. [Google Scholar] [CrossRef]
- Moriones, E.; Praveen, S.; Chakraborty, S. Tomato leaf curl New Delhi virus: An emerging virus complex threatening vegetable and fiber crops. Viruses 2017, 9, 264. [Google Scholar] [CrossRef]
- Kil, E.J.; Vo, T.T.B.; Fadhila, C.; Ho, P.T.; Lal, A.; Troiano, E.; Parrella, G.; Lee, S. Seed transmission of tomato leaf curl New Delhi virus from zucchini squash in Italy. Plants 2020, 9, 563. [Google Scholar] [CrossRef]
- Panno, S.; Caruso, A.G.; Barone, S.; Lo Bosco, G.; Rangel, E.A.; Davino, S. Spread of tomato brown rugose fruit virus in Sicily and evaluation of the spatiotemporal dispersion in experimental conditions. Agronomy 2020, 10, 834. [Google Scholar] [CrossRef]
- Kumar, P.L.; Hanna, R.; Alabi, O.J.; Soko, M.M.; Oben, T.T.; Vangu, G.H.P.; Naidu, R.A. Banana bunchy top virus in sub-Saharan Africa: Investigations on virus distribution and diversity. Virus Res. 2011, 159, 171–182. [Google Scholar] [CrossRef]
- Thomas, J.E.; Geering, A.D.W.; Dahaf, G.; Lockhart, B.E.L.; Thottappilly, G. Banana and Plantain. In Virus and Virus-Like Diseases of Major Crops in Developing Countries., 1st ed.; Loebenstein, G., Thottappilly, G., Eds.; Springer Science-Business Media: Dordrecht, The Netherlands, 2003; Volume 1, pp. 477–496. [Google Scholar]
- Niyongere, C.; Losenge, T.; Ateka, E.M.; Ntukamazina, N.; Ndayiragije, P.; Simbare, A.; Cimpaye, P.; Ninteje, P.; Lepoint, P.; Blomme, G. Understanding banana bunchy top disease epidemiology in Burundi for an enhanced and integrated management approach. Plant. Pathol. 2012, 62, 562–570. [Google Scholar] [CrossRef]
- Magee, C.J. Some aspects of the bunchy top disease of banana and other Musa spp. J. Proc. R. Soc. 1953, 87, 18. [Google Scholar]
- Magee, C.J. Investigation on the Bunchy Top Disease of the Banana; Bulletin of the Council for Scientific and Industrial Research (AUS): Melbourne, Australia, 1927; Bulletin 30; pp. 1–64. [Google Scholar]
- Lecoq, H. Cucubirts. In Virus and Virus-Like Diseases of Major Crops in Developing Countries, 1st ed.; Loebenstein, G., Thottappilly, G., Eds.; Springer Science-Business Media: Dordrecht, The Netherlands, 2003; Volume 1, pp. 665–668. [Google Scholar]
- Jacquemond, M. Cucumber Mosaic Virus. In Viruses and Virus Diseases of Vegetables in the Mediterranean Basin, 1st ed.; Loebenstein, G., Lecoq, H., Eds.; Elservier Inc.: London, UK, 2012; Volume 84, pp. 439–504. [Google Scholar]
- Nováková, S.; Šubr, Z.; Kováč, A.; Fialová, I.; Beke, G.; Danchenko, M. Corrigendum to “Cucumber mosaic virus resistance: Comparative proteomics of contrasting Cucumis sativus cultivars after long-term infection”. J. Proteomics. 2020, 222, 103–674. [Google Scholar] [CrossRef] [PubMed]
- Lovisolo, O. Virus and viroid diseases of cucurbits. Acta Hortic. 1981, 88, 33–82. [Google Scholar] [CrossRef]
- Desbiez, C.; Lecoq, H. Zucchini yellow mosaic virus. Plant. Pathol. 1997, 46, 809–829. [Google Scholar] [CrossRef]
- Lecoq, H.; Desbiez, C. Watermelon mosaic virus and zucchini yellow mosaic virus. In Encyclopedia of Virology, 4th ed.; Bamford, D.H., Zuckerman, M., Eds.; Elsevier Ltd.: Amsterdam, The Netherlands, 2021; Volume 3, pp. 862–870. [Google Scholar]
- Lecoq, H.; Fabre, F.; Joannon, B.; Wipf-Scheibel, C.; Chandeysson, C.; Schoeny, A.; Desbiez, C. Search for factors involved in the rapid shift in Watermelon mosaic virus (WMV) populations in South-eastern France. Virus Res. 2011, 159, 115–123. [Google Scholar] [CrossRef]
- Lecoq, H.; Cohen, S.; Pitrat, M.; Labonne, G. Resistance to cucumber mosaic virus transmission by Aphids in Cucumis melo. Phytopathology 1979, 69, 1223–1225. [Google Scholar] [CrossRef]
- Dombrovsky, A.; Tran-Nguyen, L.T.T.; Jones, R.A.C. Cucumber green mottle mosaic virus: Rapidly increasing global distribution, etiology, epidemiology, and management. Annu. Rev. Phytopathol. 2017, 55, 231–256. [Google Scholar] [CrossRef]
- Fletcher, J.T.; George, A.J.; Green, D.E. Cucumber green mottle mosaic virus, its effect on yield and its control in the Lea Valley, England. Plant. Pathol. 1969, 18, 16–22. [Google Scholar] [CrossRef]
- Reingold, V.; Lachman, O.; Koren, A.; Dombrovsky, A. First report of cucumber green mottle mosaic virus (CGMMV) symptoms in watermelon used for the discrimination of non-marketable fruits in Israeli commercial fields. Plant. Pathol. 2013, 28, 11. [Google Scholar]
- Noda, H.; Yamagishi, N.; Yaegashi, H.; Xing, F.; Xie, J.; Li, S.; Zhou, T.; Ito, T.; Yoshikawa, N. Apple necrotic mosaic virus, a novel ilarvirus from mosaic-diseased apple trees in Japan and China. J. Gen. Plant. Pathol. 2017, 83, 83–90. [Google Scholar] [CrossRef]
- Shi, W.; Yao, R.; Sunwu, R.; Huang, K.; Liu, Z.; Li, X.; Yang, Y.; Wang, J. Incidence and Molecular Identification of Apple Necrotic Mosaic Virus (ApNMV) in Southwest China. Plants 2020, 9, 415. [Google Scholar] [CrossRef] [PubMed]
- Noorani, M.S.; Khan, J.A. Development of a novel polyprobe for simultaneous detection of six viruses infecting stone and pome fruits. 3 Biotech. 2020, 10, 389. [Google Scholar] [CrossRef] [PubMed]
- Thokchom, T.; Rana, T.; Hallan, V.; Ram, R.; Zaidi, A. Molecular characterization of the Indian strain of apple mosaic virus isolated from apple (Malus domestica). Phytoparasitica 2009, 37, 375–379. [Google Scholar] [CrossRef]
- Akbas, B.; Ilhan, D. Widespread Distribution of apple mosaic virus on apple in Turkey. Plant. Dis. 2005, 89, 1010. [Google Scholar] [CrossRef] [PubMed]
- Robertson, N.L. First Report of Apple mosaic virus in Alaska. Plant. Dis. 2012, 96, 463. [Google Scholar] [CrossRef]
- Naidu, R.; Rowhani, A.; Fuchs, M.; Golino, D.; Martelli, G.P. Grapevine Leafroll: A complex viral disease affecting a high-value fruit crop. Plant. Dis. 2014, 98, 1172–1185. [Google Scholar] [CrossRef]
- European and Mediterranean Plant Protection Organization. Certification schemes Virus-free or virus-tested fruit trees and rootstocks. EPPO Bulletin. 1991, 2, 267–277. [Google Scholar]
- Tsai, C.W.; Rowhani, A.; Golino, D.A.; Daane, K.M.; Almeida, R.P.P. Mealybug transmission of grapevine leafroll viruses: An analysis of virus-vector specificity. Phytopathology 2010, 100, 830–834. [Google Scholar] [CrossRef] [PubMed]
- Demangeat, G.; Voisin, R.; Minot, J.C.; Bosselut, N.; Fuchs, M.; Esmenjaud, D. Survival of Xiphinema index in vineyard soil and retention of grapevine fan leaf virus over extended time in the absence of host plants. Phytopathology 2005, 95, 1151–1156. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cambra, M.; Gorris, M.T.; Marroquín, C.; Román, M.P.; Olmos, A.; Martínez, M.C.; Hrermoso de Mendoza, A.; López, A.; Navarro, L. Incidence and epidemiology of Citrus tristeza virus in the valencian community of Spain. Virus Res. 2020, 71, 85–95. [Google Scholar] [CrossRef]
- Herbario Virtual. Cátedra Fitopatología-FAUBA. Available online: http://herbariofitopatologia.agro.uba.ar/?page_id=4063 (accessed on 18 September 2021).
- Bar-Joseph, M.; Marcus, R.; Lee, R.F. The continuous challenge of citrus tristeza virus control. Annu. Rev. Phytopathol. 1989, 27, 291–316. [Google Scholar] [CrossRef]
- Wallace, J.M. Tristeza disease investigations, an example of progress through cooperative international research. In Citrus Virus Diseases; Wallace, J.M., Ed.; University of California, Division of Agricultural Science: Berkley, CA, USA, 1959; pp. 29–33. [Google Scholar]
- Lee, R.F.; Baker, P.S.; Rocha-Peña, M.A. The Citrus Tristeza Virus (CTV): An Introduction to Current Priorities, with Special Reference to Worsening Situation in Central America and Caribbean; Oxford University Press; Oxford, UK, 1994. [Google Scholar]
- Baranwal, V.K.; Kapoor, R.; Kumar, S.; Srivastava, N. Recent advances of virus diagnostics in horticultural crops. In Applied Plant Virology; Awasthi, L.P., Ed.; Elsevier: London, UK, 2020; pp. 27–38. [Google Scholar]
- Riley, M.B.; Williamson, M.R.; Maloy, O. Plant disease diagnosis. Plant. Health Instr. 2002, 10. [Google Scholar] [CrossRef]
- Clark, M.F.; Adams, A.N. Characteristics of the microplate method of enzyme linked immunosorbent assay for the detection of plant viruses. J. Gen. Virol. 1977, 34, 475–483. [Google Scholar] [CrossRef]
- Candresse, T.; Hammond, R.W.; Hadidi, A. Detection and identification of plant viruses and viroids using polymerase chain reaction (PCR). In Control of Plant Virus Diseases; Hadidi, A., Khetarpal, R.K., Koganezawa, K., Eds.; APS Press: St. Paul, MN, USA, 1998; pp. 399–416. [Google Scholar]
- Jeong, J.J.; Ju, H.J.; Noh, J. A review of detection methods for the plant viruses. Res. Plant. Dis. 2014, 20, 173–181. [Google Scholar] [CrossRef]
- Kirankumar, K.C.; Priya, N.; Jayasudha, S.M.; Bhat, G. Advances in protein-based diagnostic tools of plant viruses. In Applied Plant Virology; Awasthi, L.P., Ed.; Elsevier: London, UK, 2020; pp. 93–99. [Google Scholar]
- Rubio, L.; Galipienso, L.; Ferriol, I. Detection of plant viruses and disease management: Relevance of genetic diversity and evolution. Front. Plant. Sci. 2020, 11, 1–23. [Google Scholar] [CrossRef]
- Varma, A.; Singh, M.K. Diagnosis of plant virus diseases. In Applied Plant Virology; Awasthi, L.P., Ed.; Elsevier: London, UK, 2020; pp. 79–92. [Google Scholar]
- Mehetre, G.T.; Leo, V.V.; Singh, G.; Sorokan, A.; Maksimov, I.; Yadav, M.K.; Upadhyaya, K.; Hashem, A.; Alsaleh, A.N.; Dawoud, T.M.; et al. Current developments and challenges in plant viral diagnostics: A systematic review. Viruses 2021, 13, 412. [Google Scholar] [CrossRef] [PubMed]
- Engvall, R.; Perlmann, P. Enzyme-linked immunosorbent assay (ELISA). Quantitative assay of immunoglobulin G. Immunochemistry 1971, 8, 871–874. [Google Scholar] [CrossRef]
- Engvall, E.; Perlmann, P. Enzyme-linked immunosorbent assay, ELISA. HI. Quantitation of specific antibodies by enzyme-labelled anti-immunoglobulin in antigen-coated tubes. J. Immunol. 1972, 109, 129–135. [Google Scholar] [PubMed]
- Clark, M.F.; Bar-Joseph, M. Enzyme immunosorbent assays in plant virology. In Methods in Virology; Maramorosch, K., Koprowski, H., Eds.; Academic Press: New York, NY, USA, 1984; pp. 51–85. [Google Scholar]
- Boonham, N.; Kreuze, J.; Winter, S.; Van der Vlugt, R.; Bergervoet, J.; Tomlinson, J.; Mumford, R. Methods in virus diagnostics: From ELISA to next generation sequencing. Virus Res. 2014, 186, 20–31. [Google Scholar] [CrossRef] [PubMed]
- Prosser, S.W.; Goszezynski, D.E.; Meng, B. Molecular analysis of double-stranded RNAs reveals complex infection of grapevines with multiple viruses. Virus Res. 2007, 124, 151–159. [Google Scholar] [CrossRef]
- Folimonova, S.Y.; Robertson, C.J.; Shilts, T.; Folimonov, A.S.; Hilf, M.E.; Garnsey, S.M.; Dawson, W.O. Infection with strains of Citrus tristeza virus does not exclude superinfection by other strains of the virus. J. Virol. 2010, 84, 1314–1325. [Google Scholar] [CrossRef] [PubMed]
- Van Regenmortel, M.H.V. Serology and Immunochemistry of Plant. Viruses, 1st ed.; Academic Press: New York, NY, USA, 1982; pp. 10–308. [Google Scholar]
- Fox, J.L.; Klass, M. Antigens produced by recombinant DNA technology. Clin. Chem. 1989, 35, 1838–1842. [Google Scholar] [CrossRef]
- Agarwal, S.; Krishnareddy, M.; Jain, R.K. Production of polyclonal antibodies using recombinant coat protein of Papaya ringspot virus and their use in immunodiagnosis. J. Plant. Biochem. Biotechnol. 2009, 18, 109–111. [Google Scholar] [CrossRef]
- Sharma, S.K.; Kumar, P.V.; Baranwal, V.K. Immunodiagnosis of episomal Banana streak MY virus using polyclonal antibodies to an expressed putative coat protein. J. Virol. Methods 2014, 207, 86–94. [Google Scholar] [CrossRef]
- Rai, R.; Khurana, S.M.P.; Kumar, S.; Gupta, N.; Baranwal, V.K. Serological detection of grapevine leafroll-associated virus 4 in grapevine growing areas of India using polyclonal antiserum raised against the recombinant coat protein. Crop. Prot. 2018, 109, 128–135. [Google Scholar] [CrossRef]
- Kapoor, R.; Mandal, B.; Paul, P.K.; Chigurupati, P.; Jain, R.K. Production of cocktail of polyclonal antibodies using bacterial expressed recombinant protein for multiple virus detection. J. Virol. Methods 2014, 196, 7–14. [Google Scholar] [CrossRef]
- De Boer, S.H.; Lopez, M.M. New grower-friendly methods for plant pathogen monitoring. Ann. Rev. Phytopathol. 2012, 50, 197–218. [Google Scholar] [CrossRef]
- Salomone, A.; Mongelli, M.; Roggero, P.; Boscia, D. Reliability of detection of Citrus tristeza virus by an immunochromatographic lateral flow assay in comparison with ELISA. J. Plant. Pathol. 2004, 86, 43–48. [Google Scholar]
- Koczula, K.M.; Gallotta, A. Lateral flow assays. Essays Biochem. 2016, 60, 111–120. [Google Scholar] [PubMed]
- Posthuma-Trumpie, G.A.; Korf, J.; Amerongen, A.V. Lateral flow (immuno)assay: Its strengths, weaknesses, opportunities and threats: A literature survey. Anal. Bioanal. Chem. 2009, 393, 569–582. [Google Scholar] [CrossRef] [PubMed]
- Kusano, N.; Hirashima, K.; Kuwahara, M.; Narahara, K.; Imamura, T.; Mimori, T.; Nakahira, K.; Torii, K. Immunochromatographic assay for simple and rapid detection of Satsuma dwarf virus and related viruses using monoclonal antibodies. J. Gen. Plant. Pathol. 2007, 73, 66–71. [Google Scholar] [CrossRef] [PubMed]
- Maejima, K.; Himeno, M.; Netsu, O.; Ishikawa, K.; Yoshida, T.; Fujita, N.; Hashimoto, M.; Komatsu, K.; Yamaji, Y.; Namba, S. Development of an on-site plum pox virus detection kit based on immunochromatography. J. Gen. Plant. Pathol. 2014, 80, 176–183. [Google Scholar] [CrossRef]
- Yoon, J.Y.; Choi, G.S.; Cho, I.S.; Choi, S.K. Development of rapid immune-gold strip kit for on-site diagnosis of cucumber mosaic virus. J. Korean Soc. Int. Agricult. 2014, 26, 62–67. [Google Scholar] [CrossRef]
- Yoon, J.Y.; Choi, G.S.; Cho, I.S.; Choi, S.K. Development of rapid immune-gold strip kit for on-site diagnosis of tomato spotted wilt virus. Res. Plant. Dis. 2014, 20, 15–20. [Google Scholar] [CrossRef]
- Salomone, A.; Roggero, P. Host range, seed transmission and detection by ELISA and lateral flow of an italian isolate of pepino mosaic virus. J. Plant. Pathol. 2002, 84, 65–68. [Google Scholar]
- Agdia. Available online: https://orders.agdia.com/pathogen-tests/immunostrip-tests (accessed on 18 September 2021).
- Abd El-Aziz, M.H. Three modern serological methods to detect plant viruses. J. Plant. Sci. Phytopathol. 2019, 3, 101–106. [Google Scholar] [CrossRef]
- Banttari, E.E.; Goodwin, P.H. Detection of potato viruses S, X, and Y by enzyme-linked immunosorbent assay on nitrocellulose membranes (dot-ELISA). Plant. Dis. 1985, 69, 202–205. [Google Scholar] [CrossRef]
- Rocha-Peña, M.A.; Lee, R.F.; Niblett, C.L. Development of a dot-immunobinding assay for detection of citrus tristeza virus. J. Virol. Methods 1991, 34, 297–309. [Google Scholar] [CrossRef]
- Wang, Q.; Shi, Y.; Yang, J.; Sun, Q. Detection of apple chlorotic leaf spot virus and apple stem grooving virus by Dot-immunobinding assay. Acta Hortic. 1998, 472, 51–54. [Google Scholar] [CrossRef]
- Hu, J.S.; Sether, D.M.; Liu, X.P.; Wang, M.; Zee, F.; Ullman, D.E. Use of a tissue blotting immunoassay to examine the distribution of pineapple closterovirus in Hawaii. Plant. Dis. 1997, 81, 1150–1154. [Google Scholar] [CrossRef]
- Abd El-Aziz, M.H.; Younes, H.A. Detection of cucumber mosaic cucumovirus in infected cowpea plants (Vigna unguiculata L.) from northern Egypt. Novel Res. Microbiol. J. 2019, 3, 326–340. [Google Scholar] [CrossRef]
- Vunsh, R.; Rosner, A.; Stein, A. The use of the polymerase chain reaction (PCR) for the detection of bean yellow mosaic virus in gladiolus. Ann. Appl. Biol. 1990, 117, 561–569. [Google Scholar] [CrossRef]
- Mullis, K.F.; Faloona, F.; Scharf, S.; Saiki, R.; Horn, G.; Erlich, H. Specific enzymatic amplification of DNA in vitro: The polymerase chain reaction. Cold Spring Harb. Symposia Quant. Biol. 1986, 51, 263–273. [Google Scholar] [CrossRef] [PubMed]
- Murray, M.G.; Thompson, W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4325. [Google Scholar] [CrossRef] [PubMed]
- Dellaporta, S.L.; Wood, J.; Hicks, J.B. A plant DNA minipreparation version II. Plant. Mol. Biol. Rep. 1983, 1, 19–21. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. Isolation of plant DNA from fresh tissue. Focus 1990, 12, 13–15. [Google Scholar]
- Mirmajlessi, S.M.; Loit, E.; Mänd, M.; Mansouripour, S.M. Real-time PCR applied to study on plant pathogens: Potential applications in diagnosis—a review. Plant. Protect. Sci. 2015, 51, 177–190. [Google Scholar] [CrossRef]
- Rački, N.; Dreo, T.; Gutierrez-Aguirre, I.; Blejec, A.; Ravnikar, M. Reverse transcriptase droplet digital PCR shows high resilience to PCR inhibitors from plant, soil and water samples. Plant. Methods 2014, 10, 42. [Google Scholar] [CrossRef] [PubMed]
- Baranwal, V.K.; Majumder, S.; Ahlawat, Y.S.; Singh, R.P. Sodium sulphite yields improved DNA of higher stability for PCR detection of citrus yellow mosaic virus from citrus leaves. J. Virol. Methods 2003, 112, 155–159. [Google Scholar] [CrossRef]
- Selvarajan, R.; Balasubramanian, V.; Kavitha, K.; Kavitha, K.S.; Sathiamoorthy, S.; Ahlawat, Y.S. Detection of Banana bunchy top virus and Banana streak Mysore virus by PCR: Impact of storing virus infected banana samples. Indian J. Virol. 2008, 19, 155–159. [Google Scholar]
- Mahadev, S.R.; Thamilarasan, S.K.; Kathithachalam, A. PCR detection of banana bunchy top virus (BBTV) at tissue culture level for the production of virus-free planting materials. Int. Res. J. Biol. Sci. 2013, 2, 22–26. [Google Scholar]
- Kumar, S.; Baranwal, V.K.; Singh, P.; Jain, R.K.; Sawant, S.D.; Singh, S.K. Characterization of a grapevine leafroll-associated virus 3 from India showing incongruence in its phylogeny. Virus Genes 2012, 45, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Chamberlain, J.S.; Gibbs, R.A.; Ranier, J.E.; Nguyen, P.N.; Caskey, C.T. Deletion screening of the Duchenne muscular dystrophy locus via multiplex DNA amplification. Nucleic Acids Res. 1988, 16, 11141–11156. [Google Scholar] [CrossRef]
- Chauhan, R.P.; Wijayasekara, D.; Webb, M.A.; Verchot, J. A reliable and rapid multiplex RT-PCR assay for detection of two Potyviruses and a Pararetrovirus infecting canna plants. Plant. Dis. 2015, 99, 1695–1703. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hyun, J.W.; Hwang, R.Y.; Jung, K.E. Development of multiplex PCR for simultaneous detection of citrus viruses and the incidence of citrus viral diseases in late-maturity citrus trees in Jeju Island. Plant. Pathol. J. 2017, 33, 307–317. [Google Scholar] [CrossRef]
- Pallás, V.; Sánchez-Navarro, J.A.; James, D. Recent advances on the multiplex molecular detection of plant viruses and viroids. Front. Microbiol. 2018, 9, 2087–2111. [Google Scholar] [CrossRef]
- Ito, T.; Ieki, H.; Ozaki, K. Simultaneous detection of six citrus viroids and apple stem grooving virus from citrus plant by multiplex reverse transcription polymerase chain reaction. J. Virol. Methods 2002, 106, 235–239. [Google Scholar] [CrossRef]
- Malandraki, I.; Beris, D.; Isaioglou, I.; Olmos, A.; Varveri, C.; Vassilakos, N. Simultaneous detection of three pome fruit tree viruses by one-step multiplex quantitative RT-PCR. PLoS ONE 2017, 12, e0180877. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.R.; Wetzel, S.; Klerks, M.M.; Vaskova, D.; Schoen, C.D.; Spak, J.; Jelkmann, W. Multiplex RT-PCR detection of four aphid borne viruses infecting strawberry viruses in Fragaria spp in combination with a plant mRNA specific internal control. J. Virol. Methods 2003, 111, 85–93. [Google Scholar] [CrossRef]
- Menzel, W.; Jelkmann, W.; Maiss, E. Detection of four apple viruses by multiplex RT-PCR assays with co-amplification of plant mRNA as internal control. J. Virol. Methods 2002, 99, 81–92. [Google Scholar] [CrossRef]
- Selvarajan, R.; Sheeba, M.M.; Balasubramanian, V. Simultaneous detection of episomal banana streak mysore virus and banana bunchy top virus using multiplex RT-PCR. Curr. Sci. 2011, 100, 31–34. [Google Scholar]
- Kumar, S.; Singh, L.; Ram, R.; Zaidi, A.A.; Hallan, V. Simultaneous detection of major pome fruit viruses and a viroid. Indian J. Microbiol. 2014, 54, 203–210. [Google Scholar] [CrossRef]
- Gambino, G. Multiplex RT-PCR method for the simultaneous detection of nine grapevine viruses. Methods Mol. Biol. 2015, 1236, 39–47. [Google Scholar]
- Meena, R.P.; Baranwal, V.K. Development of multiplex polymerase chain reaction assay for simultaneous detection of clostero-, badna- and mandari-viruses along with huanglongbing bacterium in citrus trees. J. Virol. Methods 2016, 235, 58–64. [Google Scholar] [CrossRef]
- Guzaev, M.; Li, X.; Park, C.; Leung, W.Y.; Roberts, L. Comparison of nucleic acid gel stains cell permeability, safety, and sensitivity of ethidium bromide alternatives. Biotium 2017, 1–4. [Google Scholar]
- Mackay, I.M.; Arden, K.E.; Nitsche, A. Real-time PCR in virology. Nucleic Acids Res. 2002, 30, 1292–1305. [Google Scholar] [CrossRef]
- Mackay, I.M. Real-time PCR in the microbiology laboratory. Clin. Microbiol. Infect. 2004, 10, 190–212. [Google Scholar] [CrossRef] [PubMed]
- Saponari, M.; Loconsoleb, G.; Liaoc, H.H.; Jiangd, B.; Savinob, V.; Yokomie, R.K. Validation of high-throughput real time polymerase chain reaction assays for simultaneous detection of invasive citrus pathogens. J. Virol. Methods 2013, 193, 478–486. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zhou, Y.; Wang, X.; Zhou, C.; Yang, X.; Li, Z. Detection of citrus yellow vein clearing virus by quantitative realtime RT-PCR. J. Horticult. Plant. 2016, 2, 188–192. [Google Scholar] [CrossRef]
- Poojari, S.; Alabi, O.J.; Okubara, P.A.; Naidu, R.A. SYBR(s) Green-based real-time quantitative reverse-transcription PCR for detection and discrimination of grapevine viruses. J. Virol. Methods 2016, 235, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Nolasco, G.; de Blas, C.; Torres, V.; Ponz, F. A method combining immunocapture and PCR amplification in a microtiter plate for the detection of plant viruses and subviral pathogens. J. Virol. Methods 1993, 45, 201–218. [Google Scholar] [CrossRef]
- Mulholland, V. Immunocapture-PCR for plant virus detection. Methods Mol. Biol. 2009, 508, 183–192. [Google Scholar] [PubMed]
- Sharma, S.K.; Kumar, P.V.; Poswal, R.; Rai, R.; Geetanjali, A.S.; Prabha, K.; Jain, R.K.; Baranwal, V.K. Occurrence and distribution of banana streak disease and standardization of a reliable detection procedure for routine indexing of banana streak viruses in India. Sci. Horticult. 2014, 179, 277–283. [Google Scholar] [CrossRef]
- Le Provost, G.; Iskra-Caruana, M.L.; Acina, I.; Teycheney, P.Y. Improved detection of episomal banana streak viruses by multiplex immunocapture PCR. J. Virol. Methods 2006, 137, 7–13. [Google Scholar] [CrossRef]
- Sreenivasulu, M.; Sai Gopal, D.V.R. Development of recombinant coat protein antibody based IC-RT-PCR and comparison of its sensitivity with other immunoassays for the detection of Papaya ringspot virus isolates from India. J. Plant. Pathol. 2010, 26, 25–31. [Google Scholar] [CrossRef]
- Kumar, S.; Rai, R.; Baranwal, V.K. Development of an immunocapture-reverse transcription-polymerase chain reaction (IC-RTPCR) using modified viral RNA release protocol for the detection of Grapevine leafroll-associated virus 3 (GLRaV-3). Phytoparasitica 2015, 43, 311–316. [Google Scholar] [CrossRef]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, i–vii. [Google Scholar] [CrossRef]
- Mori, Y.; Nagamine, K.; Tomita, N.; Notomi, T. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem. Biophys. Res. Commun. 2001, 289, 150–154. [Google Scholar] [CrossRef]
- Wong, Y.P.; Othman, S.; Lau, Y.L.; Radu, S.; Chee, H.Y. Loop-mediated isothermal amplification (LAMP): A versatile technique for detection of micro-organisms. J. Appl. Microbiol. 2018, 124, 626–643. [Google Scholar] [CrossRef]
- Peng, J.; Fan, Z.; Huang, J. Rapid detection of banana streak virus by loop-mediated isothermal amplification assay in South China. J. Phytopathol. 2012, 160, 248–250. [Google Scholar] [CrossRef]
- Peng, J.; Shi, M.; Xia, Z.; Huang, J.; Fan, Z. Detection of cucumber mosaic virus isolates from banana by one step reverse transcription loop-mediated isothermal amplification. Arch. Virol. 2012, 157, 2213–2217. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Zhang, J.; Xia, Z.; Li, Y.; Huang, J.; Fan, Z. Rapid and sensitive detection of banana bunchy top virus by loop-mediated isothermal amplification. J. Virol. Methods 2012, 185, 254–258. [Google Scholar] [CrossRef]
- Walsh, H.N.; Pietersen, G. Rapid detection of grapevine leafroll associated virus type 3 using a reverse transcription loop-mediated amplification method. J. Virol. Methods 2013, 194, 308–316. [Google Scholar] [CrossRef]
- Anthony Johnson, A.M.; Dasgupta, I.; Sai Gopal, D.V.R. Development of loop-mediated isothermal amplification and SYBR green real-time PCR methods for the detection of Citrus yellow mosaic badnavirus in citrus species. J. Virol. Methods 2014, 203, 1–6. [Google Scholar] [CrossRef]
- Siljo, A.; Bhat, A.I. Reverse transcription loop-mediated isothermal amplification assay for rapid and sensitive detection of Banana bract mosaic virus in cardamom (Elettaria cardamomum). Eur. J. Plant. Pathol. 2014, 138, 209–214. [Google Scholar] [CrossRef]
- Zhao, L.; Feng, C.H.; Li, B.Q.; Hao, X.A.; Liu, H.; Wu, Y.; Wang, Q. Rapid detection of apple stem grooving virus by reverse transcription loop-mediated isothermal amplification. J. Plant. Pathol. 2014, 96, 407–409. [Google Scholar] [CrossRef]
- Selvarajan, R.; Balasubramanian, V.; Sasireka, T. A simple, rapid and solvent free nucleic acid extraction protocol for detection of Banana bunchy top virus by polymerase chain reaction and loopmediated isothermal amplification. Eur. J. Plant. Pathol. 2015, 142, 389–396. [Google Scholar] [CrossRef]
- Eiken Chemical Co., Ltd. Available online: https://primerexplorer.jp/e/v5_manual/pdf/PrimerExplorerV5_Manual_1.pdf (accessed on 18 September 2021).
- Mekuria, T.A.; Zhangb, S.; Eastwella, K.C. Rapid and sensitive detection of Little cherry virus 2 using isothermal reverse transcription-recombinase polymerase amplification. J. Virol. Methods 2014, 205, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Ravelonandro, M.; Russell, P.; McOwen, N.; Briard, P.; Bohannon, S.; Vrient, A. Rapid diagnostic detection of Plum pox virus in Prunus plants by isothermal AmplifyRP using reverse transcription-recombinase polymerase amplification. J. Virol. Methods 2014, 207, 114–120. [Google Scholar] [CrossRef]
- Kapoor, R.; Srivastava, N.; Kumar, S.; Saritha, R.K.; Sharma, S.K.; Jain, R.K.; Baranwal, V.K. Development of a recombinase polymerase amplification assay for the diagnosis of Banana bunchy top virus in different banana cultivars. Arch. Virol. 2017, 162, 2791–2796. [Google Scholar] [CrossRef]
- Londono, M.A.; Harmon, C.L.; Polston, J.E. Evaluation of recombinase polymerase amplification for detection of begomoviruses by plant diagnostic clinics. Virol. J. 2016, 13, 1–9. [Google Scholar] [CrossRef]
- Johne, R.; Muller, H.; Rector, A.; van Ranst, M.; Stevens, H. Rolling-circle amplification of viral DNA genomes using phi29 polymerase. Trends Microbiol. 2009, 17, 205–211. [Google Scholar] [CrossRef]
- Rector, A.; Tachezy, R.; Van Ranst, M. A sequence-independent strategy for detection and cloning of circular DNA virus genomes by using multiply primed rolling-circle amplification. J. Virol. 2004, 78, 4993–4998. [Google Scholar] [CrossRef]
- Inoue-Nagata, A.K.; Albuquerque, L.C.; Rocha, W.B.; Nagata, T. A simple method for cloning the complete begomovirus genome using the bacteriophage phi29 DNA polymerase. J. Virol. Methods 2004, 116, 209–211. [Google Scholar] [CrossRef]
- Haible, D.; Kober, S.; Jeske, H. Rolling circle amplification revolutionizes diagnosis and genomics of geminiviruses. J. Virol. Methods 2006, 135, 9–16. [Google Scholar] [CrossRef] [PubMed]
- James, A.P.; Geijskes, R.J.; Dale, J.L.; Harding, R.M. Development of a novel rolling circle amplification technique to detect banana streak virus that also discriminates between integrated and episomal virus sequences. Plant. Dis. 2011, 95, 57–62. [Google Scholar] [CrossRef]
- James, A.P.; Geijskes, R.J.; Dale, J.L.; Harding, R.M. Molecular characterization of six badnavirus species associated with leaf streak disease of banana in East Africa. Ann. Appl. Biol. 2011, 158, 346–353. [Google Scholar] [CrossRef]
- Baranwal, V.K.; Sharma, S.K.; Khurana, D.; Varma, R. Sequence analysis of shorter than genome length episomal Banana streak OL virus like sequences isolated from banana in India. Virus Genes 2014, 48, 120–127. [Google Scholar] [CrossRef]
- Grigoras, I.; Timchenko, T.; Katul, L.; Grande-Pérez, A.; Vetten, H.-J.; Gronenborn, B. Reconstitution of authentic nanovirus from multiple cloned DNAs. J. Virol. 2009, 83, 10778–10787. [Google Scholar] [CrossRef]
- Schena, M.; Shalon, D.; Davis, R.W.; Brown, P.O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 1995, 270, 467–470. [Google Scholar] [CrossRef]
- Hadidi, A.; Czosnek, H.; Barba, M. DNA microarrays and their potential applications for the detection of plant viruses, viroids and phytoplasmas. J. Plant. Pathol. 2004, 86, 97–104. [Google Scholar]
- Leveque, N.; Renois, F.; Andréoletti, L. The microarray technology: Facts and controversies. Clin. Microbiol. Infect. 2013, 19, 10–14. [Google Scholar] [CrossRef]
- McLoughlin, K.S. Microarrays for pathogen detection and analysis. Brief. Funct. Genomics 2011, 10, 342–353. [Google Scholar] [CrossRef]
- Lee, G.P.; Min, B.E.; Kim, C.S.; Choi, S.H.; Harn, C.H.; Kim, S.U.; Ryu, K.H. Plant virus cDNA chip hybridization for detection and differentiation of four cucurbit-infecting Tobamoviruses. J. Virol. Methods 2003, 110, 19–24. [Google Scholar] [CrossRef]
- Bystricka, D.; Lenz, O.; Mraz, I.; Piherova, L.; Kmoch, S.; Sip, M. Oligonucleotide-based microarray: A new improvement in microarray detection of plant viruses. J. Virol. Methods 2005, 128, 176–182. [Google Scholar] [CrossRef] [PubMed]
- Agindotan, B.O.; Patrick, J.S.; Philip, H.B. Simultaneous detection of potato viruses, PLRV, PVA, PVX and PVY from dormant potato tubers by TaqMans real-time RT-PCR. J. Virol. Methods 2007, 142, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Pasquini, G.; Barba, M.; Hadidi, A.; Faggioli, F.; Negri, R.; Sobol, I.; Tiberini, A.; Caglayan, K.; Mazyad, H.; Anfoka, G.; et al. Oligonucleotide microarray-based detection and genotyping of Plum pox virus. J. Virol. Methods 2008, 147, 118–126. [Google Scholar] [CrossRef]
- Engel, E.A.; Escobara, P.F.; Rojasa, L.A.; Riveraa, P.A.; Fiore, N.; Valenzuela, P.D.T. A diagnostic oligonucleotide microarray for simultaneous detection of grapevine viruses. J. Virol. Methods 2010, 163, 445–451. [Google Scholar] [CrossRef]
- Deyong, Z.; Willingmann, P.; Heinze, C.; Adam, G.; Pfunder, M.; Frey, B.; Frey, J.E. Differentiation of cucumber mosaic virus isolates by hybridization to oligonucleotides in a microarray format. J. Virol. Methods 2005, 123, 101–108. [Google Scholar] [CrossRef]
- Nicolaisen, M. An oligonucleotide-based microarray for detection of plant RNA viruses. J. Virol. Methods 2011, 173, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Nam, M.; Kim, J.S.; Lim, S.; Park, C.Y.; Kim, J.G.; Choi, H.S.; Lim, H.S.; Moon, J.S.; Lee, S.H. Development of the large-scale oligonucleotide chip for the diagnosis of plant viruses and its practical use. J. Plant. Pathol. 2014, 30, 51–57. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Thompson, J.R.; Fuchs, M.; McLane, H.; Celebi-Toprak, F.; Fischer, K.F.; Potter, J.L.; Perry, K.L. Profiling viral infections in grapevine using a randomly primed reverse transcription-polymerase chain reaction/macroarray multiplex platform. Phytopathology 2014, 104, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Jan, A.T.; Azam, M.; Warsi, M.K.; Ali, A.; Haq, Q.M.R. Technical advancement in plant virus diagnosis-an appraisal. Arch. Phytopathol. Plant. Prot. 2012, 45, 909–921. [Google Scholar] [CrossRef]
- Meyer, M.; Kircher, M. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb. Protoc. 2010, 6, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Al Rwahnih, M.; Daubert, S.; Golino, D.; Rowhani, A. Deep sequencing analysis of RNAs from a grapevine showing Syrah decline symptoms reveals a multiple virus infection that includes a novel virus. Virology 2009, 387, 395–401. [Google Scholar] [CrossRef] [PubMed]
- Kreuze, J.F.; Perez, A.; Untiveros, M.; Quispe, D.; Fuentes, S.; Barker, I.; Simon, R. Complete viral genome sequence and discovery of novel viruses by deep sequencing of small RNAs: A generic method for diagnosis, discovery and sequencing of viruses. Virology 2009, 388, 1–7. [Google Scholar] [CrossRef]
- Prabha, K.; Baranwal, V.K.; Jain, R.K. Applications of next generation high throughput sequencing technologies in characterization, discovery and molecular interaction of plant viruses. Indian J. Virol. 2013, 24, 157–165. [Google Scholar] [CrossRef]
- Barba, M.; Czosnek, H.; Hadidi, A. Historical perspective, development and applications of next-generation sequencing in plant virology. Viruses 2014, 6, 106–136. [Google Scholar] [CrossRef]
- Wang, M.B.; Metzlaff, M. RNA silencing and antiviral defense in plants. Curr. Opin. Plant. Biol. 2005, 8, 216–222. [Google Scholar] [CrossRef]
- Massart, S.; Olmos, A.; Jijakli, H.; Candresse, T. Current impact and future directions of high throughput sequencing in plant virus diagnostics. Virus Res. 2014, 188, 90–96. [Google Scholar] [CrossRef]
- Pooggin, M.M. Small RNA-omics for plant virus identification, virome reconstruction, and antiviral defense characterization. Front. Microbiol. 2018, 9, 1–20. [Google Scholar] [CrossRef]
- James, D.; Varga, A.; Pallas, V.; Candresse, T. Strategies for simultaneous detection of multiple plant viruses. Can. J. Plant. Pathol. 2006, 28, 16–29. [Google Scholar] [CrossRef]
- Gaafar, Y.; Ziebell, H. Comparative study on three viral enrichment approaches based on RNA extraction for plant virus/viroid detection using high-throughput sequencing. PLoS ONE 2020, 15, e0237951. [Google Scholar] [CrossRef]
- Wu, Q.; Ding, S.W.; Zhang, Y.; Zhu, S. Identification of viruses and viroids by next-generation sequencing and homology-dependent and homology-independent algorithms. Annu. Rev. Phytopathol. 2015, 53, 425–444. [Google Scholar] [CrossRef]
- Rackus, D.G.; Shamsi, M.H.; Wheeler, A.R. Electrochemistry, biosensors and microfluidics: A convergence of fields. Chem. Soc. Rev. 2015, 44, 5320–5340. [Google Scholar] [CrossRef]
- Sharma, S.K.; Sehgal, N.; Kumar, A. Biomolecules for development of biosensors and their applications. Curr. Appl. Phys. 2003, 3, 307–316. [Google Scholar] [CrossRef]
- Frossyniotis, D.; Anthopoulos, Y.; Kintzios, S.; Moschopoulou, G.; Yialouris, C.P. Artificial neural network selection for the detection of plant viruses. World J. Agric. Res. 2008, 4, 114–120. [Google Scholar]
- Rettcher, S.; Jungk, F.; Kühn, C.; Krause, H.J.; Nölke, G.; Commandeur, U.; Fischer, R.; Schillberg, S.; Schröper, F. Simple and portable magnetic immunoassay for rapid detection and sensitive quantification of plant viruses. Appl. Environ. Microbiol. 2015, 81, 3039–3048. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Kaushal, A.; Kulshrestha, S. A nano-Au/C-MWCNT based label free amperometric immunosensor for the detection of Capsicum chlorosis virus in bell pepper. Arch. Virol. 2017, 162, 2047–2052. [Google Scholar] [CrossRef] [PubMed]
- Sauerbrey, G. Verwendung von Schwingquarzen zur Wägung dünner Schichten und zur Mikrowägung. Z. Physik 1959, 155, 206–222. [Google Scholar] [CrossRef]
- Hayden, O.; Lieberzeit, P.A.; Blaas, D.; Dickert, F.L. Artificial antibodies for bioanalyte detection-sensing viruses and proteins. Adv. Funct. Mater. 2006, 16, 1269–1278. [Google Scholar] [CrossRef]
- Huang, X.; Xu, J.; Ji, H.-F.; Li, G.; Chen, H. Quartz crystal microbalance based biosensor for rapid and sensitive detection of maize chlorotic mottle virus. Anal. Methods 2014, 6, 4530–4536. [Google Scholar] [CrossRef]
- Kosslinger, C.; Drost, S.; Albert, F.; Wolf, H.; Koch, S.; Woias, P. A quartz crystal biosensor for measurement in liquids. Biosensors Bioelectr. 1992, 7, 397–404. [Google Scholar] [CrossRef]
- Konig, B.; Gratzel, M. Development of piezoelectric immunosensors for the detection of human erythrocytes. Analyt. Chim. Acta 1993, 276, 329–333. [Google Scholar] [CrossRef]
- Eun, A.J.C.; Huang, L.Q.; Chew, F.T.; Li, S.F.Y.; Wong, S.M. Detection of two orchid viruses using quartz crystal microbalance (QCM) immunosensors. J. Virol. Meth. 2002, 99, 71–79. [Google Scholar] [CrossRef]
- Malecka, K.; Michalczuk, L.; Radecka, H.; Radecki, J. Ion-Channel genosensor for the detection of specific DNA sequences derived from Plum pox virus in plant extracts. Sensors 2014, 14, 18611–18624. [Google Scholar] [CrossRef]
- Wongkaew, P.; Poosittisak, S. Diagnosis of Sugarcane white leaf disease using the highly sensitive DNA based voltammetric electrochemical determination. Am. J. Plant. Sci. 2014, 5, 2256–2268. [Google Scholar] [CrossRef]
- Singh, S.J. (Ed.) Advances in Diseases of Fruit Crops in India; Kalyani Publisher: New Delhi, India, 1996; p. 492. [Google Scholar]
- Thind, S.K. Principles of disease management in fruit crops. Int. Clin. Pathol. J. 2017, 4, 123–137. [Google Scholar]
- Ventura, J.A.; de Melo Lima, I.; Martins, M.V.V.; Culik, M.P.; Costa, H. Impact and management of diseases in the propagation of fruit plants. Rev. Bras. Frutic. 2019, 41, 1–13. [Google Scholar] [CrossRef]
- Sastry, K.S. Plant Virus Transmission Through Vegetative Propagules (Asexual Reproduction). In Seed-Borne Plant Virus Diseases; Springer: New Delhi, India, 2013; pp. 285–305. [Google Scholar]
- Roistacher, C.N. Diagnosis and management of virus and virus like diseases of citrus. In Diseases of Fruits and Vegetables and Their Management, 1st ed.; Thind, T.S., Ed.; Kalyani Publisher, Ludhiana: New Delhi, India, 2001; pp. 109–189. [Google Scholar]
- Singh, S.J. Virus Diseases of Pineapple. In Diseases of fruits and vegetables and their management, 1st ed.; Thind, T.S., Ed.; Kalyani Publisher, Ludhiana: New Delhi, India, 2001; pp. 639–651. [Google Scholar]
- Thind, S.K.; Mahal, J.S. Package of Practices for Cultivation of Fruits; Additional Director of Communication for Punjab Agricultural University: Ludhiana, India, 2021. [Google Scholar]
- Summanwar, A.; Marathe, T. Diagnostic technique for the detection of bunchy top and infectious chlorosis in banana suckers. Curr. Sci. 1982, 51, 47–49. [Google Scholar]
- Singh, S.J. Diseases of banana and their management. In Diseases of Horticultural Crops: Fruits; Indus Publishing Company: New Delhi, India, 1999; p. 456. [Google Scholar]
- Vapnek, J. Legislatively Establishing a Health Certification Programme for Citrus; FAO Legal Papers Online #81; Legal Officer with the United Nations Food and Agriculture Organization, Caracalla: Rome, Italy, 2009. [Google Scholar]
- Usha, K.; Thakre, M.; Goswami, A.K.; Nayan, D.G. Fundamental of Fruit Production; Division of Fruits and Horticultural Technology, Indian Agricultural Research Institute: New Delhi, India, 2015; p. 245. [Google Scholar]
- Weinbaum, S.A.; Johnson, R.S.; DeJong, T.M. Causes and consequences of overfertilization in orchards. HortTechnology 1992, 2, 112–121. [Google Scholar] [CrossRef]
- Marschner, H. Mineral. Nutrition of Higher Plants, 2nd ed.; Academic Press: London, UK, 1995; p. 892. [Google Scholar]
- Peterson, B.; Stevens, R. Tree Fruit Nutrition: A Comprehensive Manual of Deciduous Tree Fruit Nutrient Needs; Good Fruit Grower: Yakima, WA, USA, 1994; p. 211. [Google Scholar]
- Shear, C.B.; Faust, M. Nutritional ranges in deciduous tree fruits and nuts. Hortic. Rev. 1980, 2, 142–163. [Google Scholar]
- Dordas, C. Role of nutrients in controlling plant diseases in sustainable agriculture. A review. Agron. Sustain. Dev. 2008, 28, 33–46. [Google Scholar] [CrossRef]
- Singh, S.J.; Sastry, K.S.M. Effect of fertilizers and spacing on the incidence of pineapple wilt virus. Indian J. Mycol. and Plant. Pathol. 1975, 5, 156–160. [Google Scholar]
- Oladokun, J.O.; Halabi, M.H.; Barua, P.; Nath, P.D. Tomato brown rugose fruit disease: Current distribution, knowledge and future prospects. Plant. Pathol. 2019, 68, 1579–1586. [Google Scholar] [CrossRef]
- Jones, J.P.; Alexander, L.J. Relation of certain environmental factors and tobacco mosaic virus to blotchy ripening of tomatoes. Phytopathology 1962, 52, 524–528. [Google Scholar]
- Rich, S. Fetilizers influence the incidence of tomato internal browning in the field. Phytopathology 1958, 48, 448–450. [Google Scholar]
- Baghel, P.P.S.; Gupta, D.C. Effect of inter-cropping on root knot nematodes infesting grapevine variety Perlette. Indian Nematol. 1986, 16, 283–284. [Google Scholar]
- Manandhar, R.; Hooks, C.R.R. Using Protector Plants to Reduce the Incidence of Papaya Ringspot Virus-Watermelon Strain in Zucchini. Environ. Entomol. 2011, 40, 391–398. [Google Scholar] [CrossRef]
- Estelitta, S.; Radhakrishnan, T.; Paul, T.S. Infectious chlorosis disease of banana in Kerala. Infomusa 1996, 5, 25–26. [Google Scholar]
- Mansilla, P.J.; Moreira, A.G.; Mello, A.P.O.A.; Rezende, J.A.M.; Ventura, J.A.; Yuki, V.A.; Levatti, F.J. Importance of cucurbits in the epidemiology of Papaya ringspot virus type P. Plant. Pathol. 2013, 62, 571–577. [Google Scholar] [CrossRef]
- De Jesus Bermúdez-Guzmán, M.; Guzmán-González, S.; Lara-Reyna, J.; Palmeros-Suárez, P.A.; López-Muraira, I.G.; Gómez-Leyva, J.F. Presence of Papaya ringspot virus (PRSV) in weed associated with Carica papaya in Colima, Mexico. Rev. Mex. Fitopatol. 2018, 36, 1–15. [Google Scholar]
- Cameron, J.W. Sexual and nucellar embryony in F1 hybrids and advanced crosses of Citrus and Poncirus. J. Am. Soc. Hort. Sci. 1979, 104, 408–410. [Google Scholar]
- Frost, H.B.; Soost, R.K. Seed reproduction: Development of gametes and embryos. In The Citrus Industry; Reuther, W., Batchelor, L.D., Webber, H.J., Eds.; University of California Press: Berkeley, CA, USA, 1967; pp. 290–324. [Google Scholar]
- Aleza, P.; Juarez, J.; Cuenca, J.; Ollitrault, P.; Navarro, L. Recovery of citrus triploid hybrids by embryo rescue and flow cytometry from 2x 3 2x sexual hybridisation and its application to extensive breeding programs. Plant. Cell Rep. 2010, 29, 1023–1034. [Google Scholar] [CrossRef]
- Dhillon, R.S.; Kaundal, G.S.; Cheema, S.S. Nucellar embryony for propagating citrus. Indian Hort. 1993, 38, 44–45. [Google Scholar]
- Cameron, J.W.; Johnston, J.C. Nucellar seedlings: May permit development of disease-free citrus varieties. Calif. Agr. 1949, 3, 9–12. [Google Scholar]
- Litz, R.E. In vitro somatic embryogenesis from nucellar callus of monoembryonic Mangifera indica L. HortScience 1984, 19, 715–717. [Google Scholar]
- Krishna, H.; Singh, S.K. Biotechnological advances in mango (Mangifera indica L.) and their future implication in crop improvement: A review. Biotechnol. Adv. 2007, 25, 223–243. [Google Scholar] [CrossRef] [PubMed]
- James, D.J.; Passey, A.J.; Deeming, D.C. Adventitious Embryogenesis and the in vitro culture of Apple Seed Parts. J. Plant. Physiol. 1984, 115, 217–229. [Google Scholar] [CrossRef]
- Moore, S.; Kirkman, W. Citrus orchard sanitation with emphasis on false codling moth control. SA Fruit J. 2008, 7, 57–60. [Google Scholar]
- Trapman, M.; Jansonius, P.J. Disease management in organic apple orchards is more than applying the right product at the correct time. In Proceedings of the Ecofruit, 13th International Conference on Cultivation Technique and Phytopathological Problems in Organic Fruit-Growing, Weinsberg, Germany, 18–20 February 2008; pp. 16–22. [Google Scholar]
- Thind, S.K.; Cheema, S.S.; Arora, J.K. Recent approaches in managing the viral diseases of citrus. In Management of Plant Diseases; Madhu Publication: Bikaner, India, 2004; pp. 121–148. [Google Scholar]
- Ram, V.; Bhardwaj, L.N. Stone fruit diseases and their management. In Diseases of Fruits and Vegetables; Naqvi, S.A.M.H., Ed.; Kluwer Academic Publishers: Amsterdam, The Netherlands, 2004; pp. 485–510. [Google Scholar]
- Raut, S.P.; Ranade, S. Diseases of banana and their management. In Diseases of Fruits and Vegetables; Naqvi, S.A.M.H., Ed.; Kluwer Academic Publishers: Amsterdam, The Netherlands, 2004; pp. 37–52. [Google Scholar]
- Sholberg, P.L. Management of grape diseases in arid climates. In Diseases of Fruits and Vegetables; Naqvi, S.A.M.H., Ed.; Management Kluwer Academic Publishers: Amsterdam, The Netherlands, 2004; pp. 53–80. [Google Scholar]
- Ventura, J.A.; Costa, H.; da Silva Tatagiba, J. Papaya diseases and integrated control. In Diseases of Fruits and Vegetables; Naqvi, S.A.M.H., Ed.; Management Kluwer Academic Publishers: Amsterdam, The Netherlands, 2004; pp. 201–268. [Google Scholar]
- Gupta, V.K.; Sharma, S.K. Diseases of pome fruits and their management. In Diseases of Fruits and Vegetables and Their Management; Thind, T.S., Ed.; Kalyani Publishers: Hyderabad, India, 2001; pp. 1–24. [Google Scholar]
- Michigan State University. Available online: https://www.canr.msu.edu/news/sanitation_is_critical_to_prevent_plant_diseases_part_2_field_sanitation (accessed on 18 September 2021).
- USDA-CSREES, National Integrated Food Safety Initiative. Available online: https://www.kyagr.com/marketing/documents/GAP_tomato.pdf (accessed on 18 September 2021).
- Chatterji, A.; Fauquet, C.M. Chapter 8-Ecology of plant viruses, with special reference to whitefly-transmitted geminiviruses (WTGs). In Viral Ecology; Hurst, C.J., Ed.; Academic Press: San Diego, CA, USA, 2000; pp. 321–351. [Google Scholar]
- Meneghini, M. Experiences of Transmission of the Disease “Tristeza” of Citrus by the Black Orange Aphid (Experiências de Transmissão da Doença. “Tristeza” Dos Citrus Pelo Pulgão Preto da Laranjeira). Biológico 1948, 14, 115–118. [Google Scholar]
- Olson, E.O.; Sleeth, B. Tristeza virus carried by some Meyer lemon trees in Texas. Proc. Rio Gd. Val. Hortic. Inst. 1954, 8, 84–88. [Google Scholar]
- Michaud, J. A Review of the Literature on Toxoptera citricida (Kirkaldy) (Homoptera: Aphididae). Fla. Entomol. 1998, 81, 37–61. [Google Scholar] [CrossRef]
- Hansen, A.J. Origin, cause, host range and spread of cherry rasp leaf disease in North America. Phytopathology 1973, 64, 721–727. [Google Scholar] [CrossRef]
- U.S. Department of Agriculture (USDA), Virus Diseases and Noninfectious Disorders of Stone Fruits in North. America; Agriculture Handbook No. 437; Gilmer, R.M., Moore, J.D., Nyland, G., Welsh, M.F., Pine, T.S., Eds.; U.S. Department of Agriculture (USDA): Washington, DC, USA, 1975; p. 442.
- Wang, K.H.; Uyeda, J.; Sugano, J. Chapter IV: IPM Strategies against BBTV. In Banana Pest and Disease Management in the Tropical Pacific: A Guidebook for Banana Growers; University of Hawaii: Manoa, HI, USA, 2017; pp. 26–31. [Google Scholar]
- McCollum, D. Strategic Agrichemical Review Process (SARP)-Updates; Horticulture Innovation Australia (Hort Innovation): Sydney, NSW, Australia, 2020; pp. 1–16. [Google Scholar]
- Damsteegt, V.D.; Scorza, R.; Stone, A.L.; Schneider, W.L.; Webb, K.; Demuth, M.; Gildow, F.E. Prunus host range of Plum pox virus (PPV) in the United States by aphid and graft inoculation. Plant. Dis. 2007, 91, 18–23. [Google Scholar] [CrossRef]
- University of California, Integrated Pest Management (UC IPM). Pest Management Guidelines: Citrus; UC ANR Publication: Los Angeles, CA, USA, 2020; p. 3441. [Google Scholar]
- University of California, Integrated Pest Management (UC IPM). Pest Management Guidelines: Cherry; UC ANR Publication: Los Angeles, CA, USA, 2009; p. 3440. [Google Scholar]
- European and Mediterranean Plant Protection Organization (EPPO). Cherry Rasp Leaf Virus. EPPO Datasheets on Pests Recommended for Regulation. Available online: https://gd.eppo.int/taxon/CRLV00/documents (accessed on 29 July 2021).
- European and Mediterranean Plant Protection Organization (EPPO); CAB International. Peach rosette Mosaic Nepovirus. Data Sheets on Quarantine Pests. Available online: https://www.cabi.org/isc/datasheet/39224 (accessed on 29 July 2021).
- Ogawa, J.M.; Zehr, E.I.; Bird, G.B.; Ritchie, D.F.; Uriu, K.; Uyemoto, J.K. Compendium of Stone Fruit Diseases; APS Press, the American Phytopathological Society: Saint Paul, MN, USA, 1995; pp. 1–84. [Google Scholar]
- Sharma, P.D. Plant. Pathology: Methods of Control. of Plant. Diseases; Rastogi Publication: Meerut, India, 2001; pp. 106–144. [Google Scholar]
- European and Mediterranean Plant Protection Organization (EPPO); CAB International. Cherry Little Cherry ‘Virus’. Data Sheets on Quarantine Pests. Available online: https://www.cabi.org/isc/datasheet/16197 (accessed on 29 July 2021).
- Stapleton, J.J.; DeVay, J.E. Soil solarization: A non-chemical approach for management of plant pathogens and pests. Crop. Prot. 1986, 5, 190–198. [Google Scholar] [CrossRef]
- Luvisi, A.; Panattoni, A.; Materazzi, A. Heat treatments for sustainable control of soil viruses. Agron. Sustain. Dev. 2015, 35, 657–666. [Google Scholar] [CrossRef]
- Baker, K.F. The U.C. System for Producing Healthy Container Grown Plants; Manual 23; University of California: Los Angeles, CA, USA, 1957; p. 332. [Google Scholar]
- Wang, L.; Wang, G.; Hong, N.; Tang, R.; Deng, X. Effect of thermotherapy on elimination of apple stem grooving virus and apple chlorotic leaf spot virus for in vitro-cultured pear shoot tips. HortScience 2006, 41, 729–732. [Google Scholar] [CrossRef]
- Pandey, B.N.; Sindhan, G.S.; Singh, B.E. Note on the effect of heat treatment on the inactivation of some temperate-fruit viruses (hot water treatment). Progress. Agric. 1972, 4, 53–58. [Google Scholar]
- Lizárraga, A.; Ascasíbar, J.; González, M. Fast and effective thermotherapy treatment for in vitro virus eradication in apple and pear trees. Am. J. Plant. Sci. 2017, 8, 2474–2482. [Google Scholar] [CrossRef]
- Gupta, P.P. Eradication of mosaic disease and rapid clonal multiplication of bananas and plantains through meristem tip culture. Plant. Cell Tissue Org. 1986, 6, 33–39. [Google Scholar] [CrossRef]
- Reddy, P.P. Bacterial and Viral Disease and Their Management in Horticultural Crops; Scientific Publishers: Rajasthan, India, 2010; pp. 5–63. [Google Scholar]
- Sharma, S.; Singh, B.; Rani, G.; Zaidi, A.; Hallan, V.; Nagpal, A.; Virk, G. In vitro production of Indian citrus ringspot virus (ICRSV) free Kinnow plants employing phytotherapy coupled with shoot tip grafting. Vitro Cell. Dev. Biol. Plant 2007, 43, 254–259. [Google Scholar] [CrossRef]
- Dassanayake, E.M.; Perera, W.G.S. Effect of heat therapy to control pineapple wilt virus in pineapple (Ananas comosos). Vidyodaya J. Sci. 2001, 10, 135–150. [Google Scholar]
- Waage, J.; Greathead, D.; Brown, R.; Paterson, R.; Haskell, P.; Cook, R.; Krishnaiah, K. Biological Control: Challenges and opportunities [and discussion]. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1988, 318, 111–128. [Google Scholar]
- Bale, J.S.; van Lenteren, J.C.; Bigler, F. Biological control and sustainable food production. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2008, 363, 761–776. [Google Scholar] [CrossRef]
- Pal, K.K.; McSpadden Gardener, B. Biological control of plant pathogens. Plant. Health Instr. 2006, 1–25. [Google Scholar] [CrossRef]
- Pijnakker, J.; Vangansbeke, D.; Duarte, M.; Moerkens, R.; Wäckers, F.L. Predators and parasitoids-in-first: From inundative releases to preventative biological control in greenhouse crops. Front. Sustain. Food Syst. 2020, 4, 1–38. [Google Scholar] [CrossRef]
- Legaspi, J.C.; Correa, J.A.; Carruthers, R.I.; Legaspi Jr., B. C.; Nordlund, D.A. Effect of short-term releases of Chrysoperla rufilabris (Neuroptera: Chrysopidae) against silverleaf whitefly (Homoptera: Aleyrodidae) in field cages. J. Entomol. Sci. 1996, 31, 102–111. [Google Scholar] [CrossRef]
- Wisler, G.C.; Duffus, J.E.; Liu, H.-Y.; Li, R.H. Ecology and epidemiology of whitefly-transmitted closteroviruses. Plant. Dis. 1998, 82, 270–280. [Google Scholar] [CrossRef] [PubMed]
- Hoogerbrugge, H.; Houten, Y.V.; Knapp, M.; Bolckmans, K. Biological control of thrips and whitefly on strawberries with Amblydromalus limonicus and Amblyseius swirskii. IOBC/WPRS Bull. 2011, 68, 65–69. [Google Scholar]
- Ziebell, H.; Carr, J.P. Cross protection: A century of mystery. Adv. Virus Res. 2010, 76, 211–264. [Google Scholar]
- Roistacher, C.N.; da Graca, J.V.; Müller, G.W. Cross protection against Citrus tristeza virus—A review. In Proceedings of the 17th Conference of the International Organization of Citrus Virologists, Adana, Turkey, 22–26 October 2007; Duran-Vila, N., Milne, R.G., da Graca, J.V., Eds.; International Organization of Citrus Virologists Conference Proceedings: Riverside, CA, USA, 2010; pp. 1–27. [Google Scholar]
- Ahlawat, Y.S. Role of cross protection in virus diseases of citrus. In Management of Viral Diseases in Citrus Proceeding Group Discussion on Role of Cross Protection in Viral Diseases of Citrus and Production of Virus Free Planting Material; Indian Institute of Horticultural Research: Bengaluru Karnataka, India, 1994; pp. 30–31. [Google Scholar]
- Singh, S.; Kapur, S.P.; Cheema, S.S. Management of greening disease through cross protection. Indian J. Virol. 1994, 10, 113–121. [Google Scholar]
- Bederski, K.; Roistacher, C.N.; Silvestre, O.P.; Müller, G.W. Long-term cross protection of severe stem pitting citrus tristeza virus in Peru. In Proceedings of the 17th Conference of the International Organization of Citrus Virologists, Adana, Turkey, 22–26 October 2007; Hilf, M.E., Timmer, L.W., Milne, R.G., da Graca, J.V., Eds.; International Organization of Citrus Virologists Conference Proceedings: Riverside, CA, USA, 2010; pp. 67–79. [Google Scholar]
- Von Broembsen, L.A.; Lee, A.T. South Africa’s Citrus improvement programm International Organization of Citrus Virologists Conference Proceedings (1957–2010). Int. Organ. Citrus Virol. Conf. Proc. 1988, 10, 407–416. [Google Scholar]
- Simmonds, N.W. Bananas. In Tropical Agriculture Series; Longman Group Ltd.: London, UK, 1994; pp. 370–375. [Google Scholar]
- Saha, L.R. Handbook of Plant. Protection; Kalyani Publishers: Ludhiana, India, 1992; p. 900. [Google Scholar]
- Hernández Pérez, R.; Guillen Sanchez, D.; Pérez López, M.; Casanova Casio, E. Viral inhibitors to control the papaya ringspot virus on Carica papaya. Cienc. Investig. Agrar. 2017, 44, 312–319. [Google Scholar] [CrossRef]
- Chanda, B.; Shamimuzzaman, M.; Gilliard, A.; Ling, K.S. Effectiveness of disinfectants against the spread of tobamoviruses: Tomato brown rugose fruit virus and cucumber green mottle mosaic virus. Virol J. 2021, 18, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Davino, S.; Caruso, A.G.; Bertacca, S.; Barone, S.; Panno, S. Tomato brown rugose fruit virus: Seed transmission rate and efficacy of different seed disinfection treatments. Plants 2020, 9, 1615. [Google Scholar] [CrossRef]
- Paylan, I.C.; Erkan, S.; Cetinkaya, N.; Ergun, M.; Pazarlar, S. Effects of different treatments on the inactivation of various seedborne viruses in some vegetables. Ozone Sci. Eng. 2014, 36, 422–426. [Google Scholar] [CrossRef]
- Betti, L.; Trebbi, G.; Majewsky, V.; Scherr, C.; Shah-Rossi, D.; Jäger, T.; Baumgartner, S. Use of homeopathic preparations in phytopathological models and in field trials: A critical review. Homeopathy 2009, 98, 244–266. [Google Scholar] [CrossRef] [PubMed]
- Prasad, V.; Srivastava, S. Phytoproteins and Induced Antiviral Defense in Susceptible Plants: The Indian Context. In A Century of Plant Virology in India; Springer Nature: London, UK, 2017; pp. 689–728. [Google Scholar]
- Van Esse, H.P.; Reuber, T.L.; van der Does, D. Genetic modification to improve disease resistance in crops. New Phytol. 2020, 225, 70–86. [Google Scholar] [CrossRef] [PubMed]
- Paudel, D.B.; Sanfaçon, H. Exploring the diversity of mechanisms associated with plant tolerance to virus infection. Front. Plant. Sci. 2018, 9, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Vidhysekaran, P. Molecular biology of pathogens and induced systemic resistance. Indian Phytopath. 1998, 51, 111–121. [Google Scholar]
- Marais, L.J. Citrus tristeza virus and its effect on Southern African citrus industry. Citrus Industry 1994, 75, 58–60. [Google Scholar]
- Dhatt, A.S.; Dhiman, J.S. Citrus Decline Indian. Indian J. Hort. 2000, 58, 91–111. [Google Scholar]
- Fitch, M.M.M.; Manshardt, R.M.; Gonsalves, D.; Slightom, J.L.; Sanford, J.C. Virus resistant papaya plant derived from tissue bombarded with the Coat Protein gene of papaya ring spot virus. BioTechnology 1998, 10, 1166–1472. [Google Scholar]
- Zamir, D.; Ekstein-Michelson, I.; Zakay, Y. Mapping and introgression of a tomato yellow leaf curl virus tolerance gene, TY-1. Theor. Appl. Genet. 1994, 88, 141–146. [Google Scholar] [CrossRef]
- Vidavsky, F.; Czosnek, H. Tomato breeding lines resistant and tolerant to tomato yellow leaf curl virus issued from Lycopersicon hirsutum. Phytopathology 1998, 88, 910–914. [Google Scholar] [CrossRef]
- Cirilli, M.; Rossini, L.; Geuna, F. Genetic dissection of Sharka disease tolerance in peach (P. persica L. Batsch). BMC Plant. Biol. 2017, 17, 1–15. [Google Scholar] [CrossRef]
- Cosson, P.; Schurdi-Levraud, V.; Le, Q.H.; Sicard, O.; Caballero, M.; Roux, F.; Le Gall, O.; Candresse, T.; Revers, F. The RTM resistance to potyviruses in Arabidopsis thaliana: Natural variation of the RTM genes and evidence for the implication of additional genes. PLoS ONE 2012, 7, e39169. [Google Scholar] [CrossRef]
- Ishibashi, K.; Ishikawa, M. The resistance protein Tm-1 inhibits formation of a tomato mosaic virus replication protein-host membrane protein complex. J. Virol. 2013, 87, 7933–7939. [Google Scholar] [CrossRef]
- Ferreira, S.A.; Pitz, K.Y.; Manshardt, R.; Zee, F.; Fitch, M.; Gonsalves, D. Virus coatprotein transgenic papaya provides practical control of papaya ringspot virus in Hawaii. Plant. Dis. 2002, 86, 101–105. [Google Scholar] [CrossRef]
- Ye, C.; Li, H. 2010. 20 years of transgenic research in China for resistance to papaya ringspot virus. Transgenic Plant. J. 2010, 4, 58–63. [Google Scholar]
- Scorza, R.; Callahan, A.; Dardick, C.; Ravelonandro, M.; Polak, J.; Malinowski, T.; Zagrai, I.; Cambra, M.; Kamenova, I. Genetic engineering of plum pox virus resistance: ‘HoneySweet’ plum-from concept to product. Plant. Cell Tissue Organ. Cult. 2013, 115, 1–12. [Google Scholar] [CrossRef]
- Tricoll, D.M.; Carney, K.J.; Russell, P.F.; McMaster, J.R.; Groff, D.W.; Hadden, K.C.; Himmel, P.T.; Hubbard, J.P.; Boeshore, M.L.; Quemada, H.D. Field evaluation of transgenic squash containing single or multiple virus coat protein gene constructs for resistance to cucumber mosaic virus, watermelon mosaic virus 2, and zucchini yellow mosaic virus. Nat. Biotechnol. 1995, 13, 1458–1465. [Google Scholar] [CrossRef]
- Bastet, A.; Robaglia, C.; Gallois, J.L. eIF4E Resistance: Natural variation should guide gene editing. Trends Plant. Sci. 2017, 22, 411–419. [Google Scholar] [CrossRef] [PubMed]
- Van Schie, C.C.; Takken, F.L. Susceptibility genes 101: How to be a good host. Annu. Rev. Phytopathol. 2014, 52, 551–581. [Google Scholar] [CrossRef]
- Anbinder, I.; Reuveni, M.; Azari, R.; Paran, I.; Nahon, S.; Shlomo, H.; Chen, L.; Lapidot, M.; Levin, I. Molecular dissection of Tomato leaf curl virus resistance in tomato line TY172 derived from Solanum peruvianum. Theor. Appl. Genet. 2009, 119, 519–530. [Google Scholar] [CrossRef]
- Gill, U.; Scott, J.W.; Shekasteband, R.; Ogundiwin, E.; Schuit, C.; Francis, D.M.; Sim, S.C.; Smith, H.; Hutton, S.F. Ty-6, a major begomovirus resistance gene on chromosome 10, is effective against tomato yellow leaf curl virus and tomato mottle virus. Theor. Appl. Genet. 2019, 132, 1543–1554. [Google Scholar] [CrossRef]
- Castonguay, S. Creating an agricultural world order: Regional plant protection problems and international phytopathology, 1878–1939. Agric. Hist. 2010, 84, 46–73. [Google Scholar] [CrossRef]
- Devorshak, C. (Ed.) History of plant quarantine and the use of risk analysis. In Plant Pest Risk Analysis: Concepts and Application; CAB International: London, UK, 2012; p. 19. [Google Scholar]
- U.S. Department of Agriculture (USDA), Plants for Planting Manual; 1st ed., 2017. Available online: https://www.aphis.usda.gov/import_export/plants/manuals/ports/downloads/plants_for_planting.pdf (accessed on 29 July 2021).
- Wardlawa, C.W. Banana Diseases, Including Plantains and Abaca (William Clowes and Sons Ltd. London, Beccles and Colchester); Longmans, Green & Co., Ltd.: London, UK, 1972; pp. 11–68. [Google Scholar]
- Ferreira, S.A.; Trujillo, E.E.; Ogata, D.Y. Bunchy top Disease of Bananas Commodity Fact. Sheet; College of Tropical Agriculture and Human Resources. University of Hawaii: Manoa, HI, USA, 1989. [Google Scholar]
- Ferreira, S.A.; Trujillo, E.E.; Ogata, D.Y. Banana Bunchy Top. Virus; College of Tropical Agriculture and Human Resources. University of Hawaii: Manoa, HI, USA, 1997. [Google Scholar]
- Eastwell, K.C.; Li, T.S.C. Status of the little cherry disease eradication program in the Kootenay Valley of British Columbia. Can. Plant. Dis. Surv. 1994, 74, 115–116. [Google Scholar]
- Eastwell, K.C.; Jesperson, G.D. Little cherry disease control survey in British Columbia in 1996. Can. Plant. Dis. Surv. 1997, 77, 91–92. [Google Scholar]
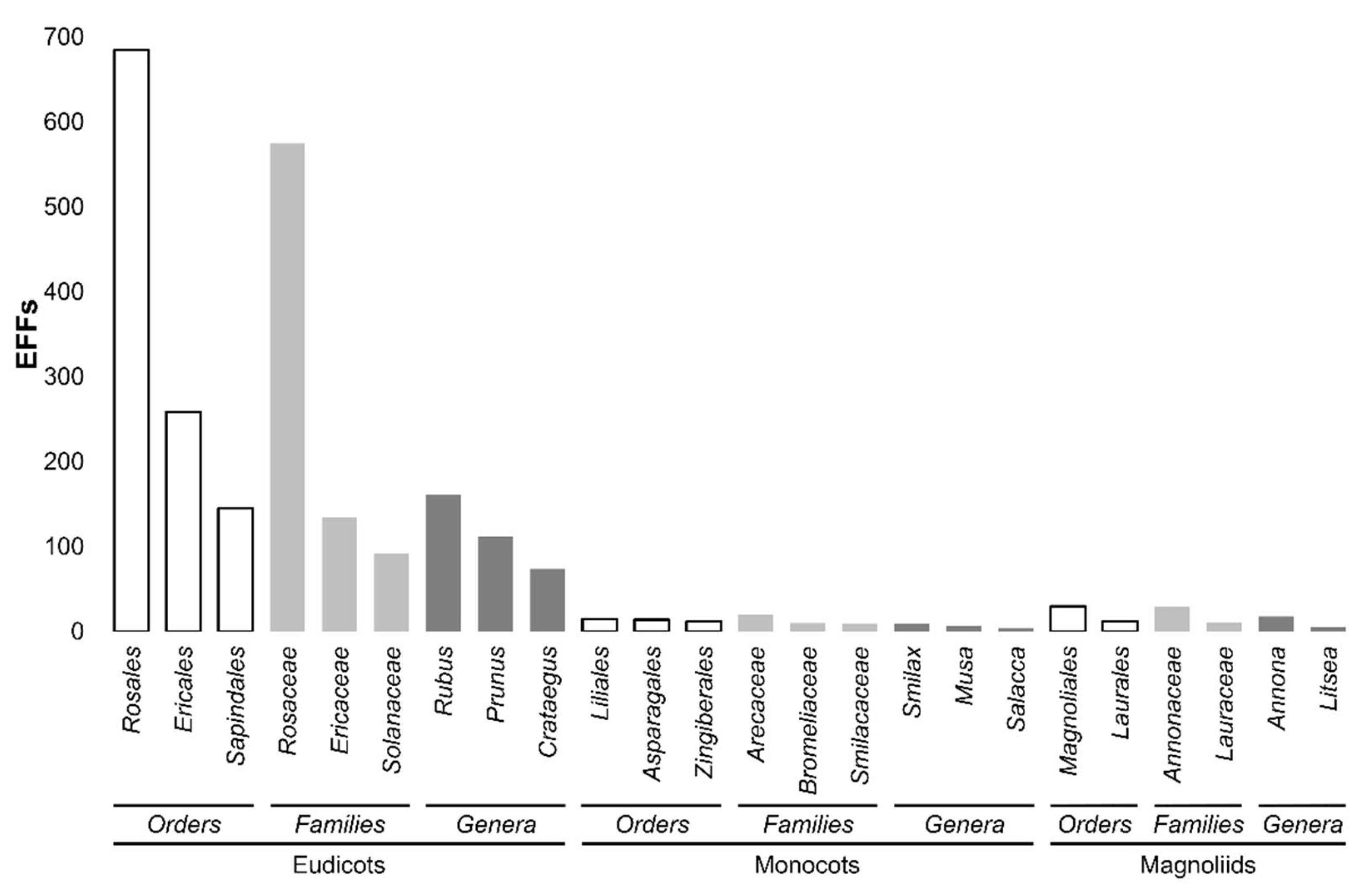

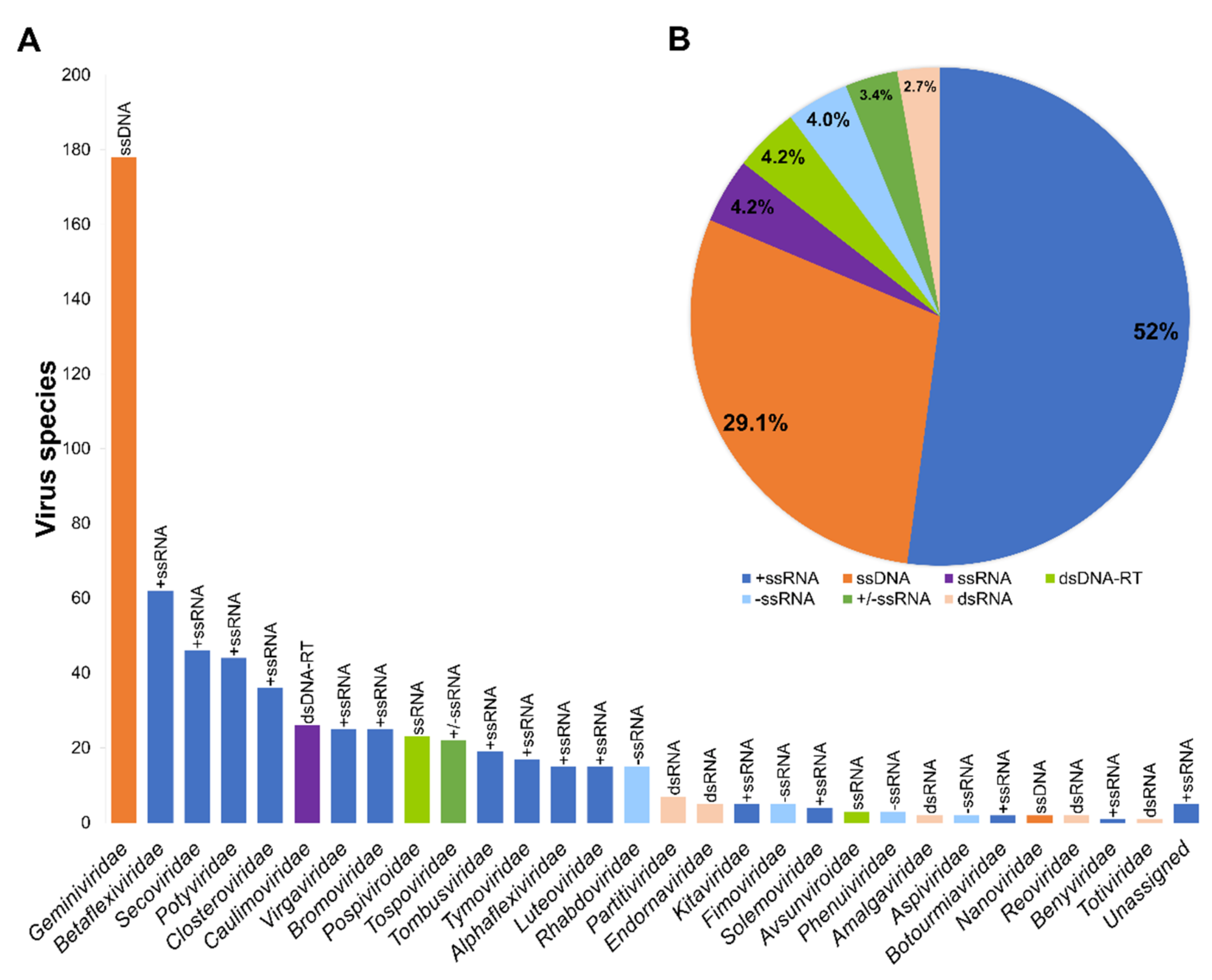
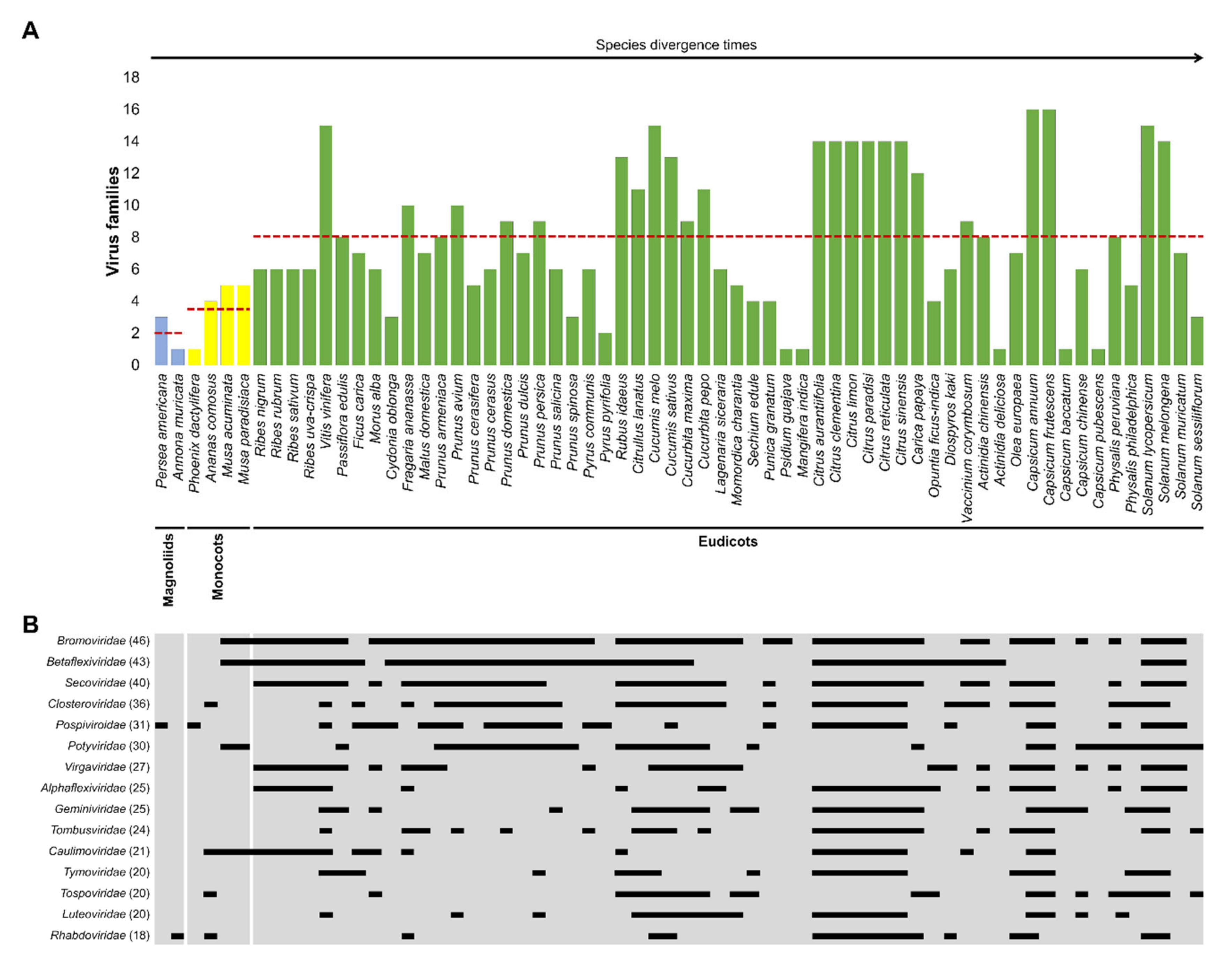
| Species | Genera | Families | Orders | Clade | % |
|---|---|---|---|---|---|
| 2119 | 438 | 119 | 38 | Eudicots | 94.8 |
| 74 | 32 | 16 | 7 | Monocots | 3.3 |
| 42 | 15 | 5 | 2 | Magnoliids | 1.8 |
| Fruit | Global Production (Million Tons per Year) | Market Worth (USD per Year) | References |
|---|---|---|---|
| Tomatoes | 186 | 9.4 billion | [45,46,47] |
| Bananas | 116 | 14.45 billion | [46,48,49] |
| Watermelons | 104 | 3.74 billion | [46,50] |
| Apples | 86 | 7.3 billion | [46,51] |
| Grapes | 79 | 10.9 billion | [46,51] |
| Oranges | 76 | 14.2 billion | [46,51] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodríguez-Verástegui, L.L.; Ramírez-Zavaleta, C.Y.; Capilla-Hernández, M.F.; Gregorio-Jorge, J. Viruses Infecting Trees and Herbs That Produce Edible Fleshy Fruits with a Prominent Value in the Global Market: An Evolutionary Perspective. Plants 2022, 11, 203. https://doi.org/10.3390/plants11020203
Rodríguez-Verástegui LL, Ramírez-Zavaleta CY, Capilla-Hernández MF, Gregorio-Jorge J. Viruses Infecting Trees and Herbs That Produce Edible Fleshy Fruits with a Prominent Value in the Global Market: An Evolutionary Perspective. Plants. 2022; 11(2):203. https://doi.org/10.3390/plants11020203
Chicago/Turabian StyleRodríguez-Verástegui, Lizette Liliana, Candy Yuriria Ramírez-Zavaleta, María Fernanda Capilla-Hernández, and Josefat Gregorio-Jorge. 2022. "Viruses Infecting Trees and Herbs That Produce Edible Fleshy Fruits with a Prominent Value in the Global Market: An Evolutionary Perspective" Plants 11, no. 2: 203. https://doi.org/10.3390/plants11020203
APA StyleRodríguez-Verástegui, L. L., Ramírez-Zavaleta, C. Y., Capilla-Hernández, M. F., & Gregorio-Jorge, J. (2022). Viruses Infecting Trees and Herbs That Produce Edible Fleshy Fruits with a Prominent Value in the Global Market: An Evolutionary Perspective. Plants, 11(2), 203. https://doi.org/10.3390/plants11020203





