Anticonvulsant Potential of 1-Aryl-6,7-dimethoxy-1,2,3,4-tetrahydroisoquinolines: Insights from Strychnine and Nicotine Models in In Vivo and In Silico Studies
Abstract
1. Introduction
2. Results
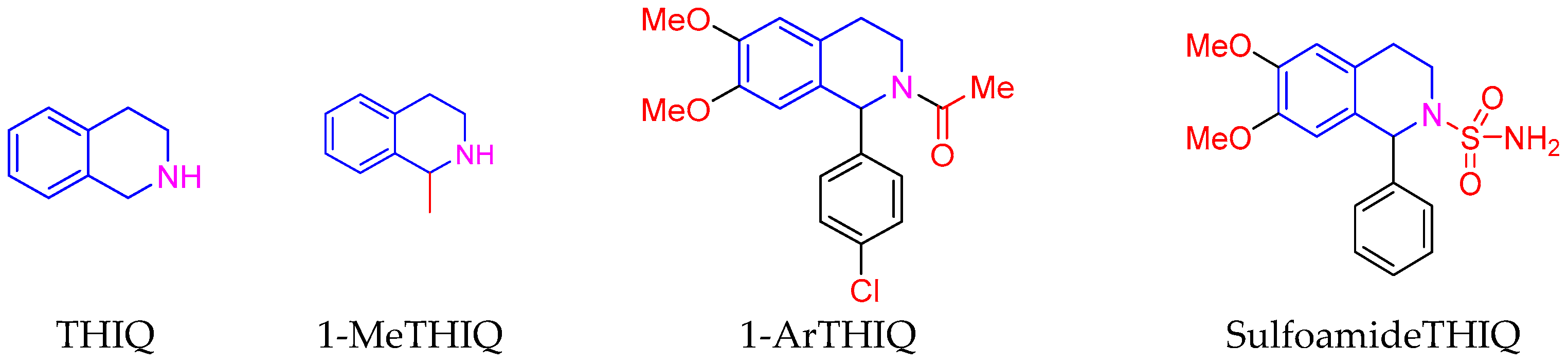
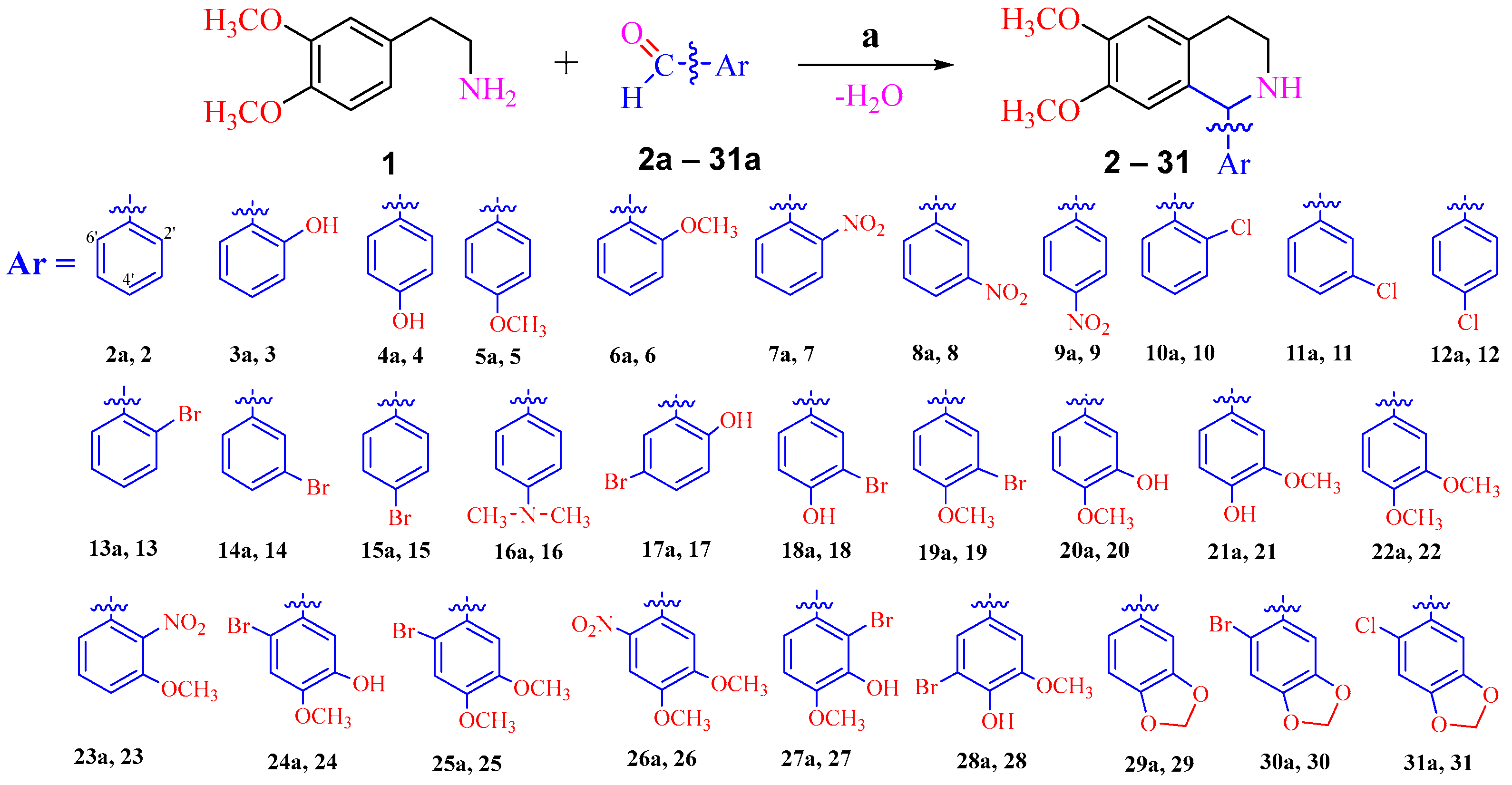
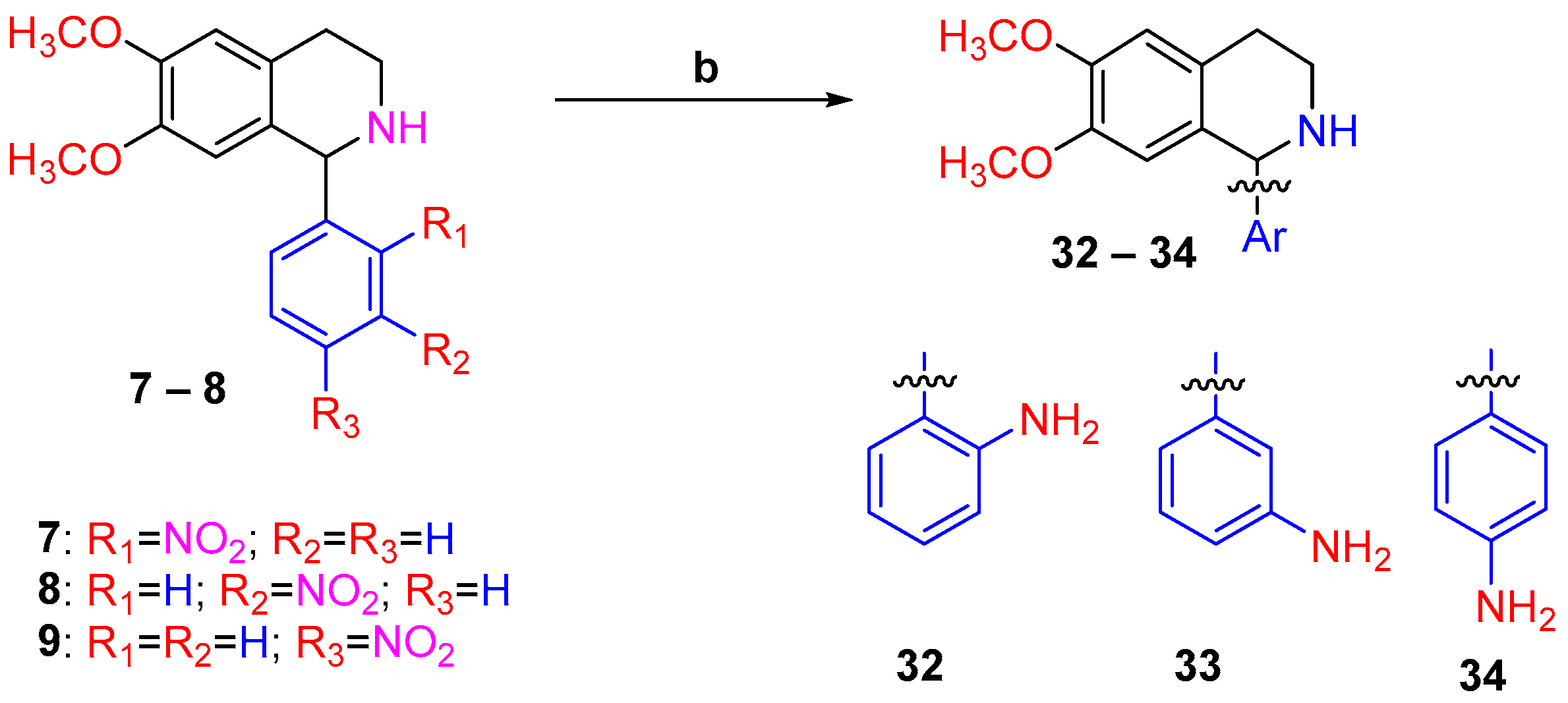
2.1. Synthesis of the 1-Aryl-1,2,3,4-tetrahydroisoquinoline Derivatives
2.2. Pharmacological Evaluation
2.2.1. Strychnine Seizure Model
2.2.2. Nicotine Seizure Model
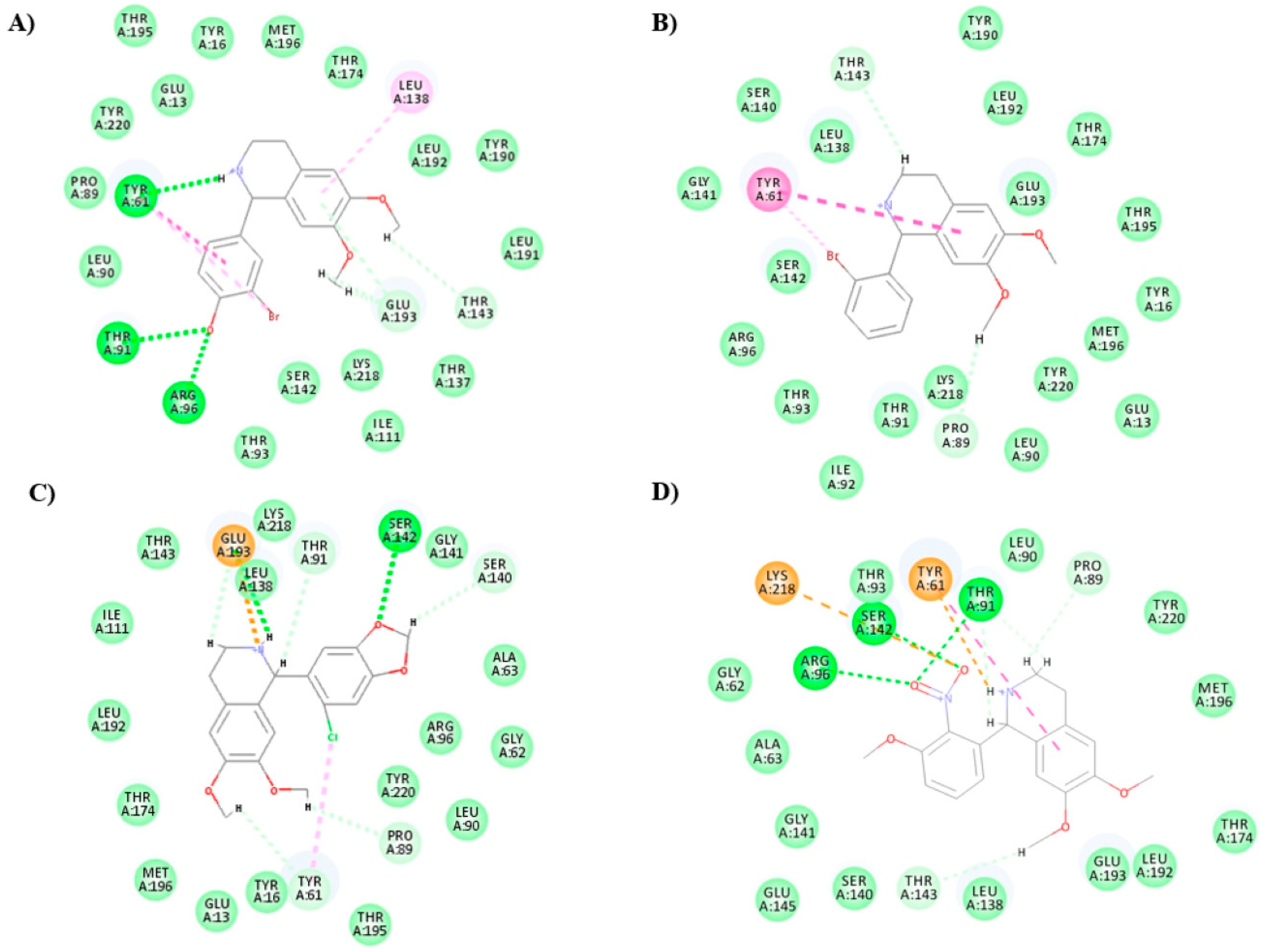
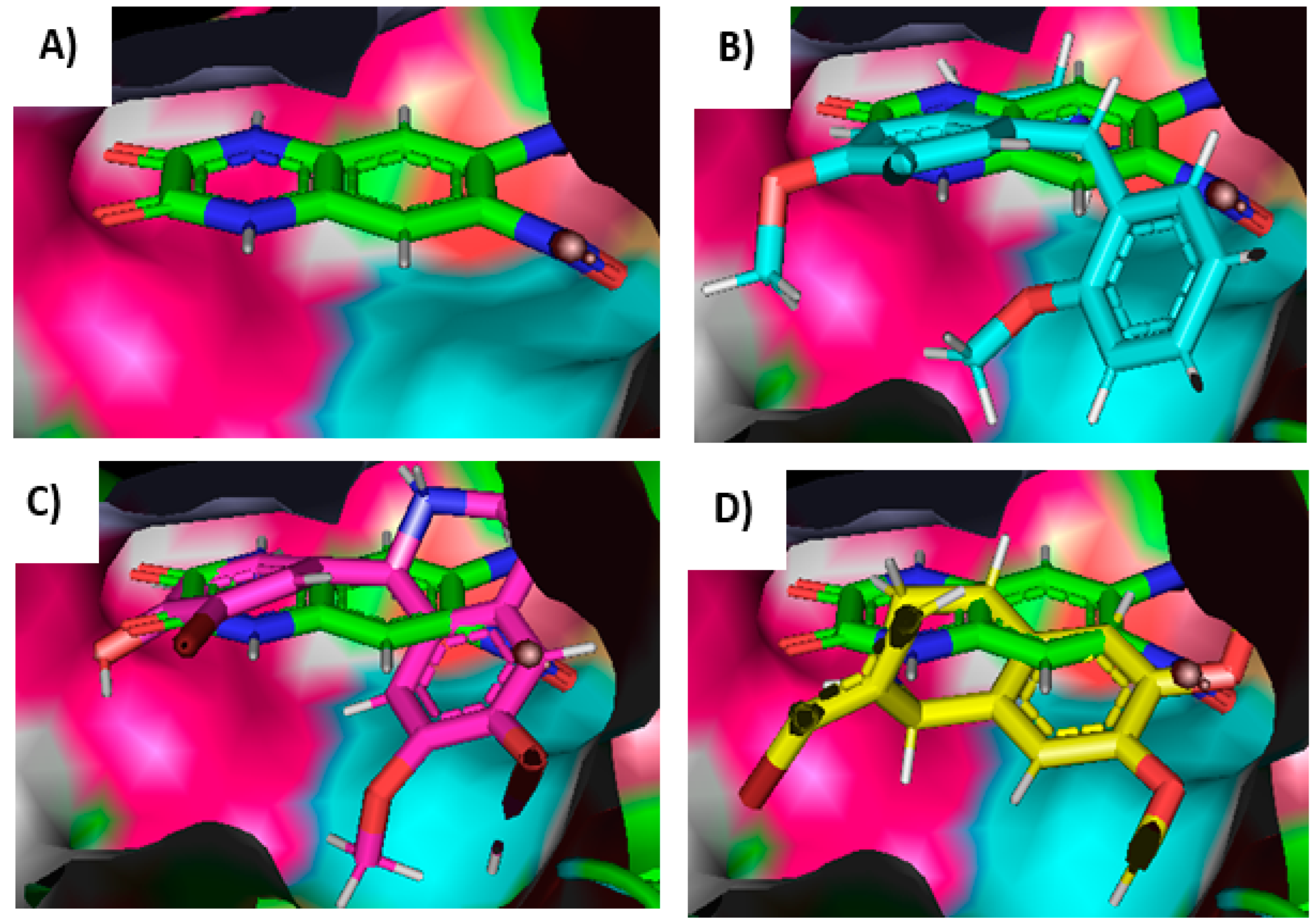
2.3. Molecular Docking
2.4. Structure–Activity Relationship
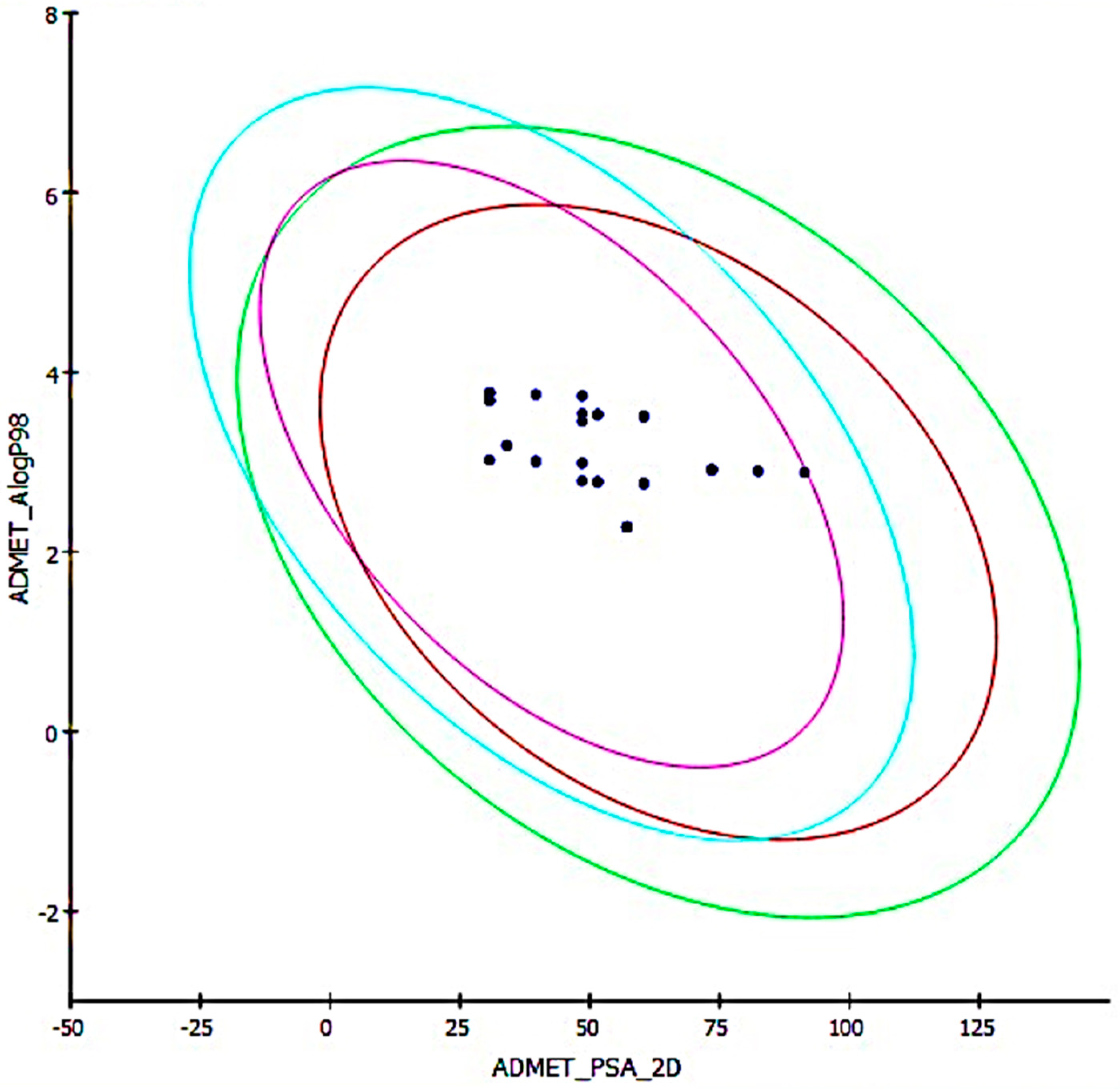
2.5. Predictive ADMET Study
3. Discussion
4. Materials and Methods
4.1. Synthesis of the Compounds
4.2. Biological Evaluation
4.3. Molecular Modeling
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sumadewi, K.T.; Harkitasari, S.; Tjandra, D.C. Biomolecular mechanisms of epileptic seizures and epilepsy: A review. Acta Epileptologica 2023, 5, 28. [Google Scholar] [CrossRef]
- Steinlein, O.K. Human disorders caused by the disruption of the regulation of excitatory neurotransmission. Results Probl. Cell Differ. 2008, 44, 223–242. [Google Scholar] [CrossRef] [PubMed]
- Teleanu, R.I.; Niculescu, A.-G.; Roza, E.; Vladâcenco, O.; Grumezescu, A.M.; Teleanu, D.M. Neurotransmitters—Key Factors in Neurological and Neurodegenerative Disorders of the Central Nervous System. Int. J. Mol. Sci. 2022, 23, 5954. [Google Scholar] [CrossRef]
- Reiner, A.; Levitz, J. Glutamatergic Signaling in the Central Nervous System: Ionotropic and Metabotropic Receptors in Concert. Neuron 2018, 98, 1080–1098. [Google Scholar] [CrossRef] [PubMed]
- De Sarro, G.; Gitto, R.; Russo, E.; Ibbadu, G.F.; Barreca, M.L.; Luca, L.D.; Chimirri, A. AMPA receptor antagonists as potential anticonvulsant drugs. Curr. Top. Med. Chem. 2005, 5, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Gitto, R.; De Luca, L.; Pagano, B.; Citraro, R.; De Sarro, G.; Costa, L.; Ciranna, L.; Chimirri, A. Synthesis and anticonvulsant evaluation of N-substituted isoquinoline AMPA receptor antagonists. Bioorg. Med. Chem. 2008, 16, 2379–2384. [Google Scholar] [CrossRef]
- Gupta, R.C.; Milatovic, D. Biomarkers in Toxicology; Academic Press: Cambridge, MA, USA, 2014; pp. 389–407. [Google Scholar] [CrossRef]
- Kuijpers, G.A.; Vergara, L.A.; Calvo, S.; Yadid, G. Inhibitory effect of strychnine on acetylcholine receptor activation in bovine adrenal medullary chromaffin cells. Br. J. Pharmacol. 1994, 113, 471–478. [Google Scholar] [CrossRef]
- Müller-Gärtner, H.W.; Mayberg, H.S.; Fisher, R.S.; Lesser, R.P.; Wilson, A.A.; Ravert, H.T.; Dannals, R.F.; Wagner, H.N., Jr.; Uematsu, S.; Frost, J.J. Decreased hippocampal muscarinic cholinergic receptor binding measured by 123I-iododexetimide and single-photon emission computed tomography in epilepsy. Ann. Neurol. 1993, 34, 235–238. [Google Scholar] [CrossRef]
- Albuquerque, E.X.; Pereira, E.F.; Alkondon, M.; Rogers, S.W. Mammalian nicotinic acetylcholine receptors: From structure to function. Physiol. Rev. 2009, 89, 73–120. [Google Scholar] [CrossRef]
- Berkovic, S.F.; Scheffer, I.E. Genetics of the epilepsies. Epilepsia 2001, 42, 16–23. [Google Scholar] [CrossRef]
- Chen, Y.; Wu, L.; Fang, Y.; He, Z.; Peng, B.; Shen, Y.; Xu, Q. A novel mutation of the nicotinic acetylcholine receptor gene CHRNA4 in sporadic nocturnal frontal lobe epilepsy. Epilepsy Res. 2009, 83, 152–156. [Google Scholar] [CrossRef]
- Luszczki, J.J.; Antkiewicz-Michaluk, L.; Czuczwar, S.J. 1-Methyl-1,2,3,4-tetrahydroisoquinoline enhances the anticonvulsant action of carbamazepine and valproate in the mouse maximal electroshock seizure model. Neuropharmacology 2006, 50, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Rogawski, M.A. Revisiting AMPA receptors as an antiepileptic drug target. Epilepsy Curr. 2011, 11, 56–63. [Google Scholar] [CrossRef]
- Rogawski, M.A.; Donevan, S.D. AMPA receptors in epilepsy and as targets for antiepileptic drugs. Adv. Neurol. 1999, 79, 947–963. [Google Scholar] [PubMed]
- De Luca, L.; Gitto, R.; Barreca, M.L.; Caruso, R.; Quartarone, S.; Citraro, R.; De Sarro, G.; Chimirri, A. 3D pharmacophore models for 1,2,3,4-tetrahydroisoquinoline derivatives acting as anticonvulsant agents. Arch. Pharm. 2006, 339, 388–400. [Google Scholar] [CrossRef] [PubMed]
- Gitto, R.; Ferro, S.; Agnello, S.; De Luca, L.; De Sarro, G.; Russo, E.; Vullo, D.; Supuran, C.T.; Chimirri, A. Synthesis and evaluation of pharmacological profile of 1-aryl-6,7-dimethoxy-3,4-dihydroisoquinoline-2(1H)-sulfonamides. Bioorg. Med. Chem. 2009, 17, 3659–3664. [Google Scholar] [CrossRef]
- Azamatov, A.A.; Zhurakulov, S.N.; Vinogradova, V.I.; Tursunkhodzhaeva, F.; Khinkar, R.M.; Malatani, R.T.; Aldurdunji, M.M.; Tiezzi, A.; Mamadalieva, N.Z. Evaluation of the Local Anesthetic Activity, Acute Toxicity, and Structure—Toxicity Relationship in Series of Synthesized 1-Aryltetrahydroisoquinoline Alkaloid Derivatives In Vivo and In Silico. Molecules 2023, 28, 477. [Google Scholar] [CrossRef]
- Gitto, R.; De Luca, L.; Ferro, S.; Agnello, S.; Russo, E.; De Sarro, G.; Chimirri, A. Synthesis and structure-active relationship of 1-aryl-6,7-dimethoxy-1,2,3,4-tetrahydroisoquinoline anticonvulsants. Chem. Pharm. Bull. 2010, 58, 1602–1605. [Google Scholar] [CrossRef][Green Version]
- Salaria, P.; Subrahmanyeswara Rao, N.N.; Dhameliya, T.M.; Amarendar Reddy, M. In silico investigation of potential phytoconstituents against ligand-and voltage-gated ion channels as antiepileptic agents. 3 Biotech 2024, 14, 99. [Google Scholar] [CrossRef]
- Azmy, E.M.; Nassar, I.F.; Hagras, M.; Fawzy, I.M.; Hegazy, M.; Mokhtar, M.M.; Yehia, A.M.; Ismail, N.S.; Lashin, W.H. New indole derivatives as multitarget anti-Alzheimer’s agents: Synthesis, biological evaluation and molecular dynamics. Future Med. Chem. 2023, 15, 473–495. [Google Scholar] [CrossRef]
- Babu, D.D.; Saranga Pani, A.; Joshi, S.D.; Naik, P.; Jayaprakash, G.K.; Al-Ghorbani, M.; Rodrigues, B.; Momidi, B.K. Computational and experimental insights into pharmacological potential: Synthesis, in vitro evaluation, and molecular docking analysis of bioactive urea and thiourea derivatives. Microb. Pathog. 2025, 200, 107209. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.B.; Hariprasad, V.; Joshi, S.D.; Jayaprakash, G.K.; Parashuram, L.; Pani, A.S.; Babu, D.D.; Naik, P. Bis(azolyl)pyridine-2,6-dicarboxamide Derivatives: Synthesis, Bioassay Analysis and Molecular Docking Studies. ChemistrySelect 2023, 8, e202204927. [Google Scholar] [CrossRef]
- Gupta, P.K. Chapter 11—Poisonous plants. In Illustrated Toxicology; Gupta, P.K., Ed.; Academic Press: Cambridge, MA, USA, 2018; pp. 309–329. [Google Scholar] [CrossRef]
- Iha, H.A.; Kunisawa, N.; Shimizu, S.; Tokudome, K.; Mukai, T.; Kinboshi, M.; Ikeda, A.; Ito, H.; Serikawa, T.; Ohno, Y. Nicotine Elicits Convulsive Seizures by Activating Amygdalar Neurons. Front. Pharmacol. 2017, 8, 57. [Google Scholar] [CrossRef]
- Protein Data Bank. Available online: https://www.rcsb.org (accessed on 11 November 2024).
- Mandour, A.A.; Elkaeed, E.B.; Hagras, M.; Refaat, H.M.; Ismail, N.S. Virtual screening approach for the discovery of selective 5α-reductase type II inhibitors for benign prostatic hyperplasia treatment. Future Med. Chem. 2023, 15, 2149–2163. [Google Scholar] [CrossRef]
| № | Tested Compound | Dose (mg/kg) | Latency to Tremor and Seizure Onset (min) | Duration of Tremor and Seizures (min) | Survival Rate (%) |
|---|---|---|---|---|---|
| 2 | 1-Phenyl-6,7-dimethoxy-1,2,3,4-tetrahydroisoquinoline | 1.0 | 14.1 ± 1.5 | 0.43 ± 0.9 | 60 |
| 3 | 1-(2′-Hydroxyphenyl)-6,7-dimethoxy-1,2,3,4-tetrahydroisoquinoline | 1.0 | 14.5 ± 1.7 | 0.29 ± 0.6 | 60 |
| 4 | 1-(4′-Hydroxyphenyl)-6,7-dimethoxy-1,2,3,4-tetrahydroisoquinoline | 1.0 | 14.2 ± 1.8 | 0.57 ± 0.7 | 70 |
| 5 | 1-(4′-Methoxyphenyl)-6,7-dimethoxy-1,2,3,4-tetrahydroisoquinoline | 1.0 | 16.5 ± 1.8 | 0.54 ± 0.7 | 60 |
| 6 | 1-(2′-Methoxyphenyl)-6,7-dimethoxy-1,2,3,4-tetrahydroisoquinoline | 1.0 | 15.7 ± 1.7 | 0.50 ± 0.4 | 30 |
| 7 | 1-(2′-Nitrophenyl)-6,7-dimethoxy-1,2,3,4-tetrahydroisoquinoline | 1.0 | 14.2 ± 1.7 | 0.32 ± 0.7 | 40 |
| 8 | 1-(3′-Nitrophenyl)-6,7-dimethoxy-1,2,3,4-tetrahydroisoquinoline | 1.0 | 15.9 ± 1.7 | 0.56 ± 0.4 | 40 |
| 9 | 1-(4′-Nitrophenyl)-6,7-dimethoxy-1,2,3,4-tetrahydroisoquinoline | 1.0 | 13.5 ± 1.9 | 0.51 ± 0.5 | 40 |
| 10 | 1-(2′-Chlorophenyl)-6,7-dimethoxy-1,2,3,4-tetrahydroisoquinoline | 1.0 | 17.2 ± 1.7 | 0.60 ± 0.4 | 30 |
| 11 | 1-(3′-Chlorophenyl)-6,7-dimethoxy-1,2,3,4-tetrahydroisoquinoline | 1.0 | 15.5 ± 1.2 | 0.46 ± 0.5 | 70 |
| 12 | 1-(4′-Chlorophenyl)-6,7-dimethoxy-1,2,3,4-tetrahydroisoquinoline | 1.0 | 18.5 ± 1.8 | 0.23 ± 0.6 | 20 |
| 13 | 1-(2′-Bromophenyl)-6,7-dimethoxy-1,2,3,4-tetrahydroisoquinoline | 1.0 | 10.5 ± 1.3 | 0.31 ± 0.7 | 40 |
| 14 | 1-(3′-Bromophenyl)-6,7-dimethoxy-1,2,3,4-tetrahydroisoquinoline | 1.0 | 18.5 ± 1.5 | 0.35 ± 0.8 | 60 |
| 15 | 1-(4′-Bromophenyl)-6,7-dimethoxy-1,2,3,4-tetrahydroisoquinoline | 1.0 | 12.5 ± 1.2 | 0.50 ± 0.5 | 40 |
| 16 | 1-(4′-Dimethylaminophenyl)-6,7-dimethoxy-1,2,3,4-tetrahydroisoquinoline | 1.0 | 13.1 ± 1.5 | 0.31 ± 0.6 | 50 |
| 17 | 1-(2′-Hydroxy-5′-bromophenyl)-6,7-dimethoxy-1,2,3,4-tetrahydroisoquinoline | 1.0 | 15.7 ± 1.6 | 0.61 ± 0.5 | 40 |
| 18 | 1-(3′-Bromo-4′-hydroxyphenyl)-6,7-dimethoxy-1,2,3,4-tetrahydroisoquinoline | 1.0 | 14.6 ± 1.5 | 0.56 ± 0.6 | 50 |
| 19 | 1-(3′-Bromo-4′-methoxyphenyl)-6,7-dimethoxy-1,2,3,4-tetrahydroisoquinoline | 1.0 | 16.0 ± 1.4 | 0.59 ± 0.9 | 40 |
| 20 | 1-(3′-Hydroxy-4′-methoxyphenyl)-6,7-dimethoxy-1,2,3,4-tetrahydroisoquinoline | 1.0 | 18.2 ± 1.4 | 0.63 ± 0.7 | 90 |
| 21 | 1-(4′-Hydroxy-3′-methoxyphenyl)-6,7-dimethoxy-1,2,3,4-tetrahydroisoquinoline | 1.0 | 15.4 ± 1.5 | 0.52 ± 0.4 | 40 |
| 22 | 1-(3′,4′-Dimethoxyphenyl)-6,7-dimethoxy-1,2,3,4-tetrahydroisoquinoline | 1.0 | 12.7 ± 1.3 | 0.71 ± 0.6 | 30 |
| 23 | 1-(3′-Methoxy-2′-nitrophenyl)-6,7-dimethoxy-1,2,3,4-tetrahydroisoquinoline | 1.0 | 14.6 ± 1.4 | 0.60 ± 0.4 | 30 |
| 24 | 1-(2′-Bromo-4′-methoxy-5′-hydroxyphenyl)-6,7-dimethoxy-1,2,3,4-tetrahydroisoquinoline | 1.0 | 10.2 ± 1.2 | 0.32 ± 0.4 | 30 |
| 25 | 1-(2′-Bromo-4′,5′-dimethoxyphenyl)-6,7-dimethoxy-1,2,3,4-tetrahydroisoquinoline | 1.0 | 16.5 ± 1.5 | 0.14 ± 0.6 | 90 |
| 26 | 1-(4′,5′-Dimethoxy-2′-nitrophenyl)-6,7-dimethoxy-1,2,3,4- tetrahydroisoquinoline | 1.0 | 16.7 ± 1.2 | 0.51 ± 0.8 | 40 |
| 27 | 1-(2′-Bromo-3′-hydroxy-4′-methoxyphenyl)-6,7-dimethoxy-1,2,3,4-tetrahydroisoquinoline | 1.0 | 10.5 ± 1.5 | 0.53 ± 0.3 | 40 |
| 28 | 1-(5′-Bromo-4′-hydroxy-3′-methoxyphenyl)-6,7- dimethoxy-1,2,3,4-tetrahydroisoquinoline | 1.0 | 15.5 ± 1.5 | 0.34 ± 0.8 | 20 |
| 29 | 1-(3′,4′-Methylenedioxyphenyl)-6,7-dimethoxy-1,2,3,4-tetrahydroisoquinoline | 1.0 | 12.5 ± 1.8 | 0.32 ± 0.9 | 50 |
| 30 | 1-(2′-Bromo-4′,5′-methylenedioxyphenyl)-6,7-dimethoxy-1,2,3,4-tetrahydroisoquinoline | 1.0 | 11.2 ± 1.2 | 0.41 ± 0.4 | 30 |
| 31 | 1-(4′,5′-Methylenedioxy-2′-chlorophenyl)-6,7-dimethoxy-1,2,3,4-tetrahydroisoquinoline | 1.0 | 13.6 ± 1.9 | 0.59 ± 0.5 | 30 |
| 32 | 1-(2′-Aminophenyl)-6,7-dimethoxy-1,2,3,4-tetrahydroisoquinoline | 1.0 | 18.2 ± 1.7 | 0.34 ± 0.4 | 50 |
| 33 | 1-(3′-Aminophenyl)-6,7-dimethoxy-1,2,3,4-tetrahydroisoquinoline | 1.0 | 16.7 ± 1.0 | 0.42 ± 0.8 | 50 |
| 34 | 1-(4′-Aminophenyl)-6,7-dimethoxy-1,2,3,4-tetrahydroisoquinoline | 1.0 | 15.5 ± 1.8 | 0.34 ± 0.7 | 40 |
| 35 | Control (Strychnine) | 1.5 | 9.8 ± 1.2 | 0.52 ± 0.7 | 0 |
| 36 | Carbamazepine | 5.0 | 14.6 ± 1.4 | 0.43 ± 0.5 | 40 |
| 10.0 | 12.6 ± 1.7 | 0.38 ± 0.8 | 50 | ||
| 37 | Valproate (Convulex) | 75.0 | 15.4 ± 1.5 | 0.34 ± 0.6 | 40 |
| 100.0 | 14.3 ± 1.3 | 0.32 ± 0.4 | 20 |
| Compound | Dose (mg/kg) | LD50 (mg/kg, per os) | Latency to Seizure Onset (min) | Duration of Seizures (min) | Survival Rate (%) | Compound | Dose (mg/kg) | LD50 (mg/kg, per os) | Latency to Seizure Onset (min) | Duration of Seizures (min) | Survival Rate (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 1.0 | 273.2 (249.7 ± 296.7) | 3.2 ± 0.07 | 3.2 ± 0.6 | 70 | 19 | 1.0 | 327.9 (287.1 ± 363.8) | 1.8 ± 0.07 | 19.0 ± 0.8 | 80 |
| 3 | 1.0 | 365.3 (326.1 ± 394.5) | 2.6 ± 0.08 | 9.0 ± 0.4 | 90 | 20 | 1.0 | 964.8 (916.5 ± 998.2) | 2.8 ± 0.07 | 11.0 ± 0.7 | 70 |
| 4 | 1.0 | 924.2 (893.9 ± 953.9) | 1.8 ± 0.05 | 10.5 ± 0.8 | 60 | 21 | 1.0 | 1663.6 (1623.1 ± 1629.1) | 1.8 ± 0.04 | 14.3 ± 0.6 | 60 |
| 5 | 1.0 | 1040.3 (986.0 ± 1079.8) | 2.1 ± 0.09 | 10.3 ± 0.6 | 80 | 22 | 1.0 | 666.6 (557.9 ± 760.2) | 2.2 ± 0.04 | 8.4 ± 0.50.5 | 80 |
| 6 | 1.0 | 286.7 (256.9 ± 318.7) | 2.4 ± 0.03 | 10.5 ± 0.5 | 100 | 23 | 1.0 | 315.4 (281.0 ± 344.2) | 1.5 ± 0.08 | 5.4 ± 0.9 | 20 |
| 7 | 1.0 | 948.9 (916.8 ± 975.8) | 2.3 ± 0.07 | 7.6 ± 0.8 | 70 | 24 | 1.0 | 546.9 (516.3 ± 571.6) | 2.1 ± 0.09 | 7.3 ± 0.3 | 70 |
| 8 | 1.0 | 531.4 (501.3 ± 554.1) | 2.3 ± 0.08 | 15.5 ± 0.3 | 90 | 25 | 1.0 | 272.1 (249.1 ± 295.6) | 2.2 ± 0.04 | 9.0 ± 0.9 | 90 |
| 9 | 1.0 | 1898.8 (703.5 ± 2273.4) | 2.5 ± 0.03 | 16.7 ± 0.4 | 90 | 26 | 1.0 | 524.5 (501.3 ± 547.7) | 2.6 ± 0.03 | 10.8 ± 0.4 | 80 |
| 10 | 1.0 | 264.2 (224.8 ± 293.9) | 2.2 ± 0.03 | 9.2 ± 0.9 | 70 | 27 | 1.0 | 4937.4 (4827.7 ± 5084.9) | 2.2 ± 0.02 | 9.7 ± 0.9 | 90 |
| 11 | 1.0 | 232.4 (211.8 ± 251.1) | 2.2 ± 0.07 | 8.3 ± 0.3 | 70 | 28 | 1.0 | 1532.7 (1471.5 ± 1579.5) | 2.7 ± 0.04 | 9.2 ± 0.4 | 70 |
| 12 | 1.0 | 730.4 (598.2 ± 883.7) | 2.1 ± 0.09 | 7.0 ± 0.4 | 70 | 29 | 1.0 | 267.2 (226.3 ± 301.1) | 2.5 ± 0.07 | 3.7 ± 0.9 | 90 |
| 13 | 1.0 | 315.8 (281.2 ± 345.2) | 2.2 ± 0.03 | 5.0 ± 0.7 | 50 | 30 | 1.0 | 711.7 (604.3 ± 792.9) | 2.5 ± 0.09 | 7.0 ± 0.2 | 90 |
| 14 | 1.0 | 446.7 (416.2 ± 471.3) | 2.8 ± 0.02 | 2.5 ± 0.4 | 90 | 31 | 1.0 | 520.8 (491.9 ± 545.4) | 2.3 ± 0.08 | 10.7 ± 0.9 | 90 |
| 15 | 1.0 | 422.0 (399.4 ± 445.3) | 2.2 ± 0.07 | 8.3 ± 0.3 | 70 | 32 | 1.0 | 366.3 (326.3 ± 395.6) | 1.8 ± 0.06 | 12.2 ± 0.8 | 60 |
| 16 | 1.0 | 498.3 (471.6 ± 524.6) | 2.7 ± 0.08 | 8.5 ± 0.7 | 90 | 33 | 1.0 | 732.0 (598.2 ± 885.4) | 2.2 ± 0.04 | 17.0 ± 0.6 | 80 |
| 17 | 1.0 | 297.3 (270.2 ± 323.7) | 2.1 ± 0.07 | 8.1 ± 0.8 | 70 | 34 | 1.0 | 1043.2 (993.3 ± 1082.6) | 2.2 ± 0.04 | 10.2 ± 0.7 | 90 |
| 18 | 1.0 | 3793.8 (3395.8 ± 4088.1) | 1.7 ± 0.04 | 8.9 ± 0.9 | 70 | Nicotine | 10.0 | 40sec ± 0.03 | 13.5 ± 0.8 | 30 | |
| Carbamazepine | 5.0 | 1.57 ± 0.07 | 15.2 ± 0.9 | 60 | |||||||
| 10.0 | 1.5 ± 0.02 | 4.7 ± 0.6 | 80 | ||||||||
| Valproate | 75.0 | 1.2 ± 0.04 | 6.2 ± 0.4 | 90 | |||||||
| 100.0 | 1.5 ± 0.06 | 10.8 ± 0.8 | 90 |
| Compound | Ligand/C-Docker Interaction Energy (Kcal/mol) | Amino Acid Residues Interaction |
|---|---|---|
| Ligand (6,7-dinitroquinoxaline-2,3-dione) | −41.60 | 2 HBAs with ARG96, 1 HBA with THR195, GLU193, and THR174, 1 HBD with PRO89, 2 pi–pi bonds with TYR 61, 1 carbon–hydrogen bond with LEU90 |
| 2 | −34.78 | 1 HBD and 1 carbon–hydrogen bond with THR91, 1 carbon–hydrogen bond with THR143, 1 pi–pi bond and 1 pi–carbon with TYR 61, 1 pi–alkyl with LEU138 |
| 3 | −39.20 | 2 HBAs with ARG96, 1 HBD with THR174, 1 carbon–hydrogen bond with GLY62, 2 pi–pi bonds with TYR 61, 1 pi–alkyl with MET196, 1 donor–donor with GLU193 |
| 4 | −36.33 | 2 HBAs with ARG96, 1 carbon–hydrogen bond with GLY62, 2 pi–pi bonds with TYR 61, 1 donor–donor with GLU193 |
| 5 | −38.24 | 1 HBD and 1 carbon–hydrogen bond with THR91, 1 carbon–hydrogen bond with THR143, 1 carbon–hydrogen bond with SER140, 1 pi–pi bond and 1 pi–carbon with TYR 61, 1 pi–alkyl with LEU138 |
| 6 | −37.21 | 1 HBA with ARG96, 1 carbon–hydrogen bond with THR91, 2 carbon–hydrogen bonds with GLU193, 1 pi–pi bond with TYR 61, 1 pi–sigma with LEU138 |
| 7 | −35.31 | 1 HBA with ARG96, THR91, and SER142, 2 pi–pi bonds with TYR 61, 1 carbon–hydrogen bond with PRO89 and TYR220, 1 attractive charge with LYS218 |
| 8 | −38.23 | 1 HBA with ARG96, 1 carbon–hydrogen bond with THR143, 2 pi–pi bonds with TYR 61, 1 pi–sigma with LEU138, 1 donor–donor with GLU193 and TYR220 |
| 9 | −38.09 | 1 HBD and 1 carbon–hydrogen bond with THR91, 1 carbon–hydrogen bond with THR143, 1 pi–pi bond and 1 pi–carbon with TYR 61, 1 pi–alkyl with LEU138 |
| 10 | −34.46 | 1 HBD with THR91, 1 carbon–hydrogen bond with THR143 and SER142 (halogen bond), 1 pi–pi bond and 1 pi–carbon with TYR 61, 2 pi–alkyl with LEU138 |
| 11 | −36.48 | 2 carbon–hydrogen bonds with THR91, 1 carbon–hydrogen bond with PRO89, 1 pi–pi bond and 1 pi–carbon with TYR 61 and 1 alkyl bond (halogen bond), 1 pi–alkyl with LEU138 and 1 alkyl bond (halogen bond) |
| 12 | −36.62 | 1 HBD and 1 carbon–hydrogen bond with THR91, 1 carbon–hydrogen bond with THR143, 1 pi–pi bond and 1 pi–carbon with TYR 61, 1 alkyl bond (halogen bond) with ALA63 |
| 13 | −33.82 | 1 carbon–hydrogen bond with PRO89 and THR143, 1 pi–pi bond with TYR 61 and 1 alkyl bond (halogen bond) |
| 14 | −38.00 | 1 carbon–hydrogen bond with PRO89 and THR143, 2 carbon–hydrogen bonds with THR91, 1 pi–pi bond and 1 alkyl bond (halogen bond) with TYR 61, 1 pi–alkyl with LEU138 and 1 alkyl bond (halogen bond) |
| 15 | −35.83 | 2 HBDs with THR91, 1 HBA with THR174 and 1 alkyl bond with ALA63 (halogen bond), 1 pi–carbon with GLU193, 1 pi–alkyl with LEU138 |
| 16 | −41.44 | 1 HBD with THR91, 1 carbon–hydrogen bond with THR91, PRO89, THR143, and SER140, 1 pi–carbon bond with TYR 61 |
| 17 | −38.28 | Halogen bonds with each of TYR16, PRO89, and TYR220, 1 carbon–hydrogen bond with GLY62, 1 donor–donor with GLU193 |
| 18 | −45.41 | 1 HBA with ARG96 and THR91, 1 HBD, 1 pi–pi bond, and 1 alkyl bond (halogen bond) with TYR 61, 1 carbon–hydrogen bond with THR143, 2 with GLU193, 1 pi–alkyl with LEU138 |
| 19 | −39.87 | 1 HBD with THR91, 1 carbon–hydrogen bond with THR143, 1 pi–pi bond, 1 pi–carbon, and 1 alkyl bond (halogen bond) with TYR 61, 1 pi–alkyl, and 1 alkyl bond (halogen bond) with LEU138 |
| 20 | −38.90 | 1 HBA with ARG96, 1 HBD with GLU13 and TYR61, 1 pi–pi bond with TYR 61, 1 carbon–hydrogen bond with PRO89and THR91, 1 pi–alkyl with LEU138 and MET196 |
| 21 | −38.50 | 1 HBA with ARG96, 1 HBD, 1 attractive charge, and 1 donor–donor with GLU193, 1 carbon–hydrogen bond with GLY62, PRO89, THR143, and TYR220, 1 pi–pi bond with TYR 61, 1 pi–alkyl with LEU138 |
| 22 | −41.19 | 1 HBD and 1 carbon–hydrogen bond with THR91, 1 carbon–hydrogen bond with THR143 and SER140, 1 pi–pi bond and 1 pi–carbon with TYR 61, 1 pi–alkyl with LEU138 |
| 23 | −44.10 | 3 HBAs with each of ARG96, THR91, and SER142, 1 pi–pi bond and 1 pi–carbon with TYR 61, 1 attractive charge with LYS218, 1 carbon–hydrogen bond with THR143 and PRO89, 2 carbon–hydrogen bonds with THR91 |
| 24 | −41.48 | 1 HBA with ARG96, 1 HBD and 1 attractive charge with GLU193, 1 alkyl bond (halogen bond) with TYR 61, 1 carbon–hydrogen bond with GLY62 and THR91 |
| 25 | −43.00 | 1 HBA with ARG96, THR91, and SER142, 1 pi–pi bond, 1 carbon–hydrogen bond, and 1 alkyl bond (halogen bond) with TYR 61, 1 carbon–hydrogen bond with THR91, SER140, GLY141, and GLY62, 1 attractive charge with GLU193 |
| 26 | −43.48 | 3 HBAs with each of ARG96, THR91, and SER142, 1 pi–pi bond and 1 pi–carbon with TYR 61, 1 pi–carbon with LYS218, 1 carbon–hydrogen bond with THR143 and PRO89, 2 carbon–hydrogen bonds with THR91, 1 attractive charge with LYS218, 1 pi–alkyl with LEU138 |
| 27 | −38.67 | 1 HBD with THR91, 1 pi–pi bond and 1 pi–carbon with TYR 61, 2 carbon–hydrogen bonds with SER140, 1 with THR143, 2 pi–alkyl with LEU138 |
| 28 | −39.89 | 1 HBA with ARG96, 2 pi–pi bonds with TYR 61, 1 carbon–hydrogen bond with THR91 and GLY62, 1 attractive charge with GLU193, 3 halogen bonds with each of TYR 16, TYR220, and PRO89 |
| 29 | −42.46 | 1 HBD with THR91, 1 HBA with SER142, 1 pi–pi bond, 1 carbon–hydrogen bond, and 1 alkyl bond (halogen bond) with TYR 61, 1 carbon–hydrogen bond with PRO89, LEU138, GLY141, and GLU145, 1 pi–carbon with TYR61 |
| 30 | −38.17 | 1 HBA with ARG96, 1 pi–pi bond with TYR 61, 1 carbon–hydrogen bond with GLY62, PRO89, and GLU193, 1 alkyl bond (halogen bond) with LEU138 and LEU192 |
| 31 | −40.36 | 1 HBA with SER142, 1 HBD, 1 attractive charge, and 1 carbon–hydrogen bond with GLU193, 1 carbon–hydrogen bond with TYR61, PRO89, THR91, and SER140, 1 alkyl bond (halogen bond) with TYR61 |
| 32 | −37.62 | 2 HBAs with ARG96, 1 HBD with THR174, 2 pi–pi bonds with TYR 61, 1 carbon–hydrogen bond with GLY62, 1 pi–alkyl with MET196 |
| 33 | −37.06 | 2 carbon–hydrogen bonds with THR91, 1 carbon–hydrogen bond with PRO89 and THR143 and 2 with THR91, 1 pi–pi bond and 1 pi–carbon with TYR61, 1 pi–alkyl with LEU138 |
| 34 | −34.41 | 1 HBD with PRO89, 1 donor–donor, 1 attractive charge, and 1 carbon–hydrogen bond with GLU193, 1 carbon–hydrogen bond with SER140 |
| Compound | ADMET Sol Level | ADMET A Log P98 | ADMET BBB Level | CYP2D6 Prediction (Non-Inhibitor) | Hepatotoxic Probability | Absorption Level | PPB Binding Prediction | Alog P98 | PSA 2D | TOPKAT Carcinogenic Potency TD 50 Rat | TOPKAT Ames Prediction |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 2 | 0 | 1 | FALSE | TRUE | 0 (good) | TRUE | 3.021 | 30.67 | 21.9697 | Non-Mutagen |
| 3 | 3 | 0 | 2 | FALSE | TRUE | 0 (good) | TRUE | 2.779 | 51.485 | 49.3691 | Non-Mutagen |
| 4 | 3 | 0 | 2 | FALSE | TRUE | 0 (good) | TRUE | 2.779 | 51.485 | 17.9936 | Non-Mutagen |
| 5 | 2 | 0 | 1 | FALSE | TRUE | 0 (good) | TRUE | 3.005 | 39.6 | 5.46952 | Non-Mutagen |
| 6 | 2 | 0 | 1 | FALSE | TRUE | 0 (good) | TRUE | 3.005 | 39.6 | 8.99628 | Non-Mutagen |
| 7 | 2 | 0 | 2 | FALSE | TRUE | 0 (good) | TRUE | 2.916 | 73.493 | 4.87626 | Non-Mutagen |
| 8 | 2 | 0 | 2 | FALSE | TRUE | 0 (good) | TRUE | 2.916 | 73.493 | 4.69132 | Non-Mutagen |
| 9 | 2 | 0 | 2 | FALSE | TRUE | 0 (good) | TRUE | 2.916 | 73.493 | 1.77725 | Non-Mutagen |
| 10 | 2 | 0 | 1 | FALSE | TRUE | 0 (good) | TRUE | 3.686 | 30.67 | 6.10272 | Non-Mutagen |
| 11 | 2 | 0 | 1 | FALSE | TRUE | 0 (good) | TRUE | 3.686 | 30.67 | 5.87126 | Non-Mutagen |
| 12 | 2 | 0 | 1 | FALSE | TRUE | 0 (good) | TRUE | 3.686 | 30.67 | 2.22426 | Non-Mutagen |
| 13 | 2 | 0 | 1 | FALSE | TRUE | 0 (good) | TRUE | 3.77 | 30.67 | 6.51486 | Non-Mutagen |
| 14 | 2 | 0 | 1 | FALSE | TRUE | 0 (good) | TRUE | 3.77 | 30.67 | 6.26777 | Non-Mutagen |
| 15 | 2 | 0 | 1 | FALSE | TRUE | 0 (good) | TRUE | 3.77 | 30.67 | 2.37448 | Non-Mutagen |
| 16 | 2 | 0 | 1 | FALSE | TRUE | 0 (good) | TRUE | 3.184 | 34.022 | 7.61886 | Non-Mutagen |
| 17 | 2 | 0 | 1 | FALSE | TRUE | 0 (good) | TRUE | 3.528 | 51.485 | 9.89607 | Non-Mutagen |
| 18 | 2 | 0 | 1 | FALSE | TRUE | 0 (good) | TRUE | 3.528 | 51.485 | 12.5742 | Non-Mutagen |
| 19 | 2 | 0 | 1 | FALSE | TRUE | 0 (good) | TRUE | 3.753 | 39.6 | 3.98643 | Non-Mutagen |
| 20 | 3 | 0 | 2 | FALSE | TRUE | 0 (good) | TRUE | 2.763 | 60.416 | 22.3668 | Non-Mutagen |
| 21 | 3 | 0 | 2 | FALSE | TRUE | 0 (good) | TRUE | 2.763 | 60.416 | 17.9743 | Non-Mutagen |
| 22 | 2 | 0 | 1 | FALSE | TRUE | 0 (good) | TRUE | 2.989 | 48.53 | 5.73009 | Non-Mutagen |
| 23 | 2 | 0 | 3 | FALSE | TRUE | 0 (good) | TRUE | 2.899 | 82.423 | 7.94352 | Non-Mutagen |
| 24 | 2 | 0 | 2 | FALSE | TRUE | 0 (good) | TRUE | 3.511 | 60.416 | 7.04772 | Non-Mutagen |
| 25 | 2 | 0 | 1 | FALSE | TRUE | 0 (good) | TRUE | 3.737 | 48.53 | 1.79015 | Non-Mutagen |
| 26 | 2 | 0 | 3 | FALSE | TRUE | 0 (good) | TRUE | 2.883 | 91.353 | 1.36115 | Non-Mutagen |
| 27 | 2 | 0 | 2 | FALSE | TRUE | 0 (good) | TRUE | 3.511 | 60.416 | 15.3108 | Non-Mutagen |
| 28 | 2 | 0 | 2 | FALSE | TRUE | 0 (good) | TRUE | 3.511 | 60.416 | 7.04772 | Non-Mutagen |
| 29 | 2 | 0 | 2 | FALSE | TRUE | 0 (good) | TRUE | 2.789 | 48.53 | 2.25834 | Non-Mutagen |
| 30 | 2 | 0 | 1 | FALSE | TRUE | 0 (good) | TRUE | 3.538 | 48.53 | 0.712376 | Non-Mutagen |
| 31 | 2 | 0 | 1 | FALSE | TRUE | 0 (good) | TRUE | 3.454 | 48.53 | 0.678266 | Non-Mutagen |
| 32 | 3 | 0 | 2 | FALSE | TRUE | 0 (good) | TRUE | 2.275 | 57.21 | 16.9758 | Non-Mutagen |
| 33 | 3 | 0 | 2 | FALSE | TRUE | 0 (good) | TRUE | 2.275 | 57.21 | 6.88315 | Non-Mutagen |
| 34 | 3 | 0 | 2 | FALSE | TRUE | 0 (good) | TRUE | 2.275 | 57.21 | 2.6076 | Non-Mutagen |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Azamatov, A.A.; Mamadalieva, N.Z.; Mandour, A.A.; Zhurakulov, S.N.; Aytmuratova, U.K.; Vinogradova, V.I.; Jalilov, F.S.; Tursunkhodzhaeva, F.M. Anticonvulsant Potential of 1-Aryl-6,7-dimethoxy-1,2,3,4-tetrahydroisoquinolines: Insights from Strychnine and Nicotine Models in In Vivo and In Silico Studies. Pharmaceuticals 2025, 18, 1350. https://doi.org/10.3390/ph18091350
Azamatov AA, Mamadalieva NZ, Mandour AA, Zhurakulov SN, Aytmuratova UK, Vinogradova VI, Jalilov FS, Tursunkhodzhaeva FM. Anticonvulsant Potential of 1-Aryl-6,7-dimethoxy-1,2,3,4-tetrahydroisoquinolines: Insights from Strychnine and Nicotine Models in In Vivo and In Silico Studies. Pharmaceuticals. 2025; 18(9):1350. https://doi.org/10.3390/ph18091350
Chicago/Turabian StyleAzamatov, Azizbek A., Nilufar Z. Mamadalieva, Asmaa A. Mandour, Sherzod N. Zhurakulov, Urkhiya K. Aytmuratova, Valentina I. Vinogradova, Fazliddin S. Jalilov, and Firuza M. Tursunkhodzhaeva. 2025. "Anticonvulsant Potential of 1-Aryl-6,7-dimethoxy-1,2,3,4-tetrahydroisoquinolines: Insights from Strychnine and Nicotine Models in In Vivo and In Silico Studies" Pharmaceuticals 18, no. 9: 1350. https://doi.org/10.3390/ph18091350
APA StyleAzamatov, A. A., Mamadalieva, N. Z., Mandour, A. A., Zhurakulov, S. N., Aytmuratova, U. K., Vinogradova, V. I., Jalilov, F. S., & Tursunkhodzhaeva, F. M. (2025). Anticonvulsant Potential of 1-Aryl-6,7-dimethoxy-1,2,3,4-tetrahydroisoquinolines: Insights from Strychnine and Nicotine Models in In Vivo and In Silico Studies. Pharmaceuticals, 18(9), 1350. https://doi.org/10.3390/ph18091350






