Heteroaryl-Capped Hydroxamic Acid Derivatives with Varied Linkers: Synthesis and Anticancer Evaluation with Various Apoptosis Analyses in Breast Cancer Cells, Including Docking, Simulation, DFT, and ADMET Studies
Abstract
1. Introduction
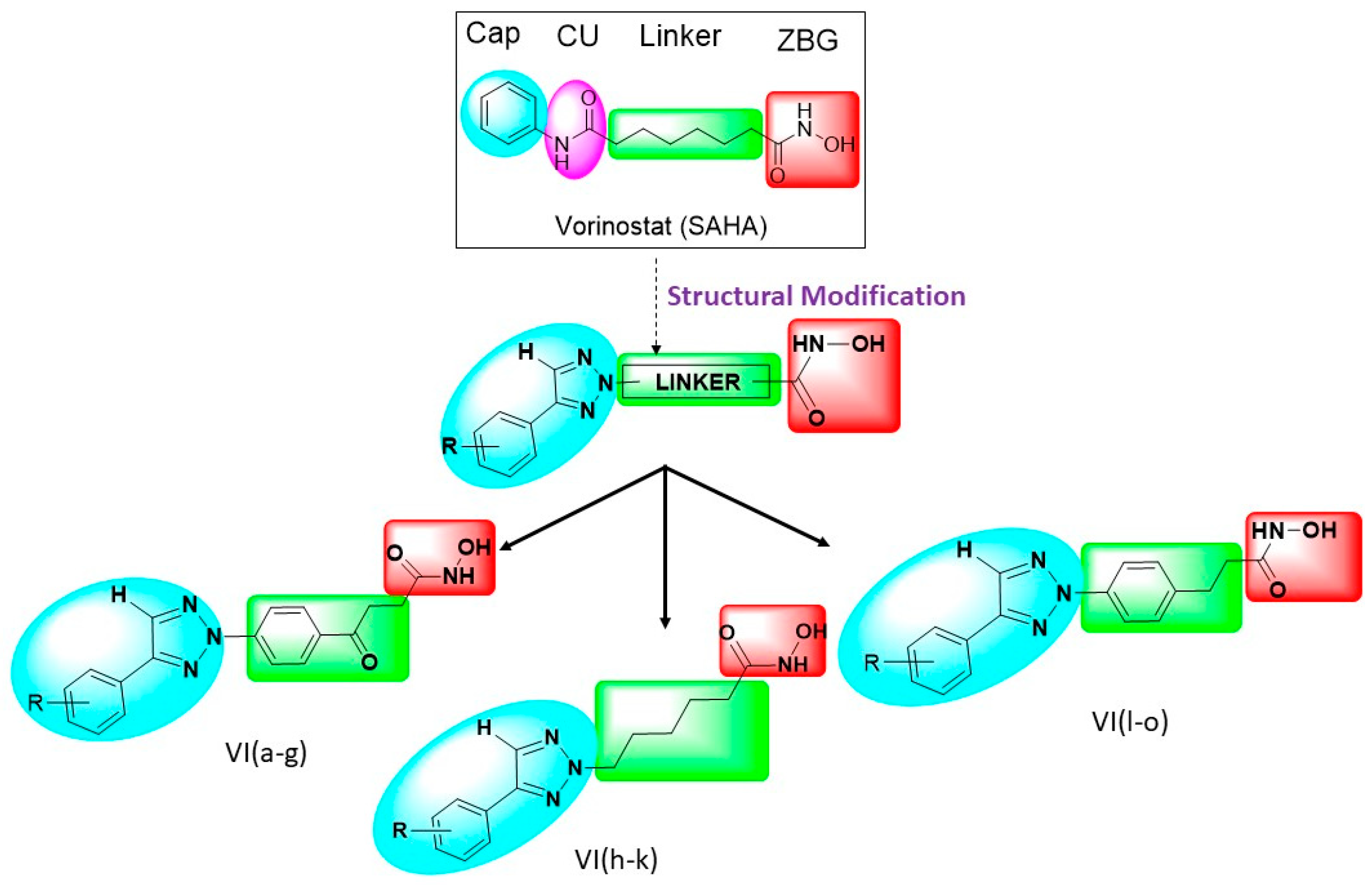
Rationale of This Research
2. Results and Discussion
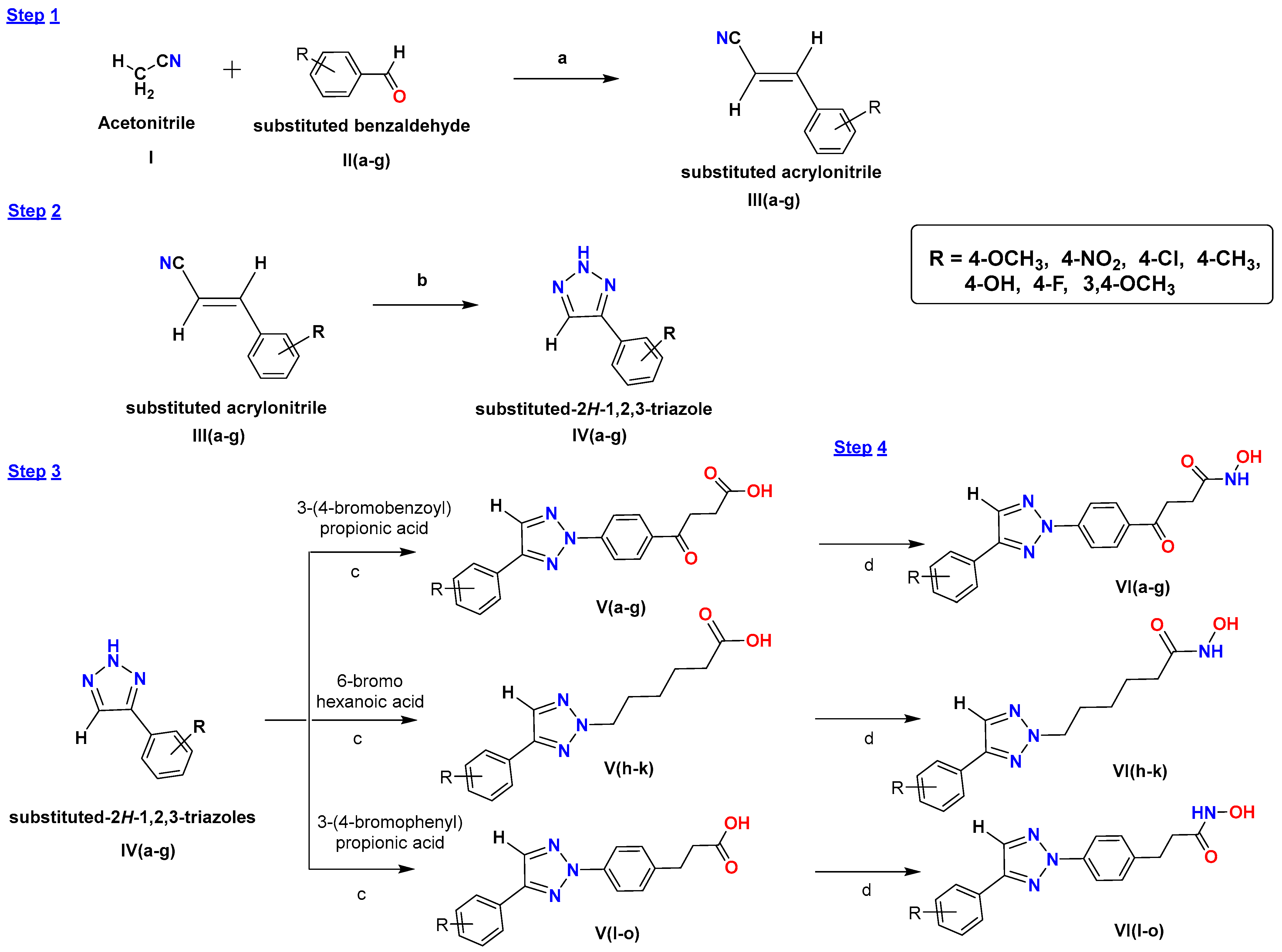
2.1. Chemistry of Synthesized Compounds
2.2. Description of FTIR Spectra
2.3. Description of NMR Spectra
2.4. Description of Physical and Chemical Properties
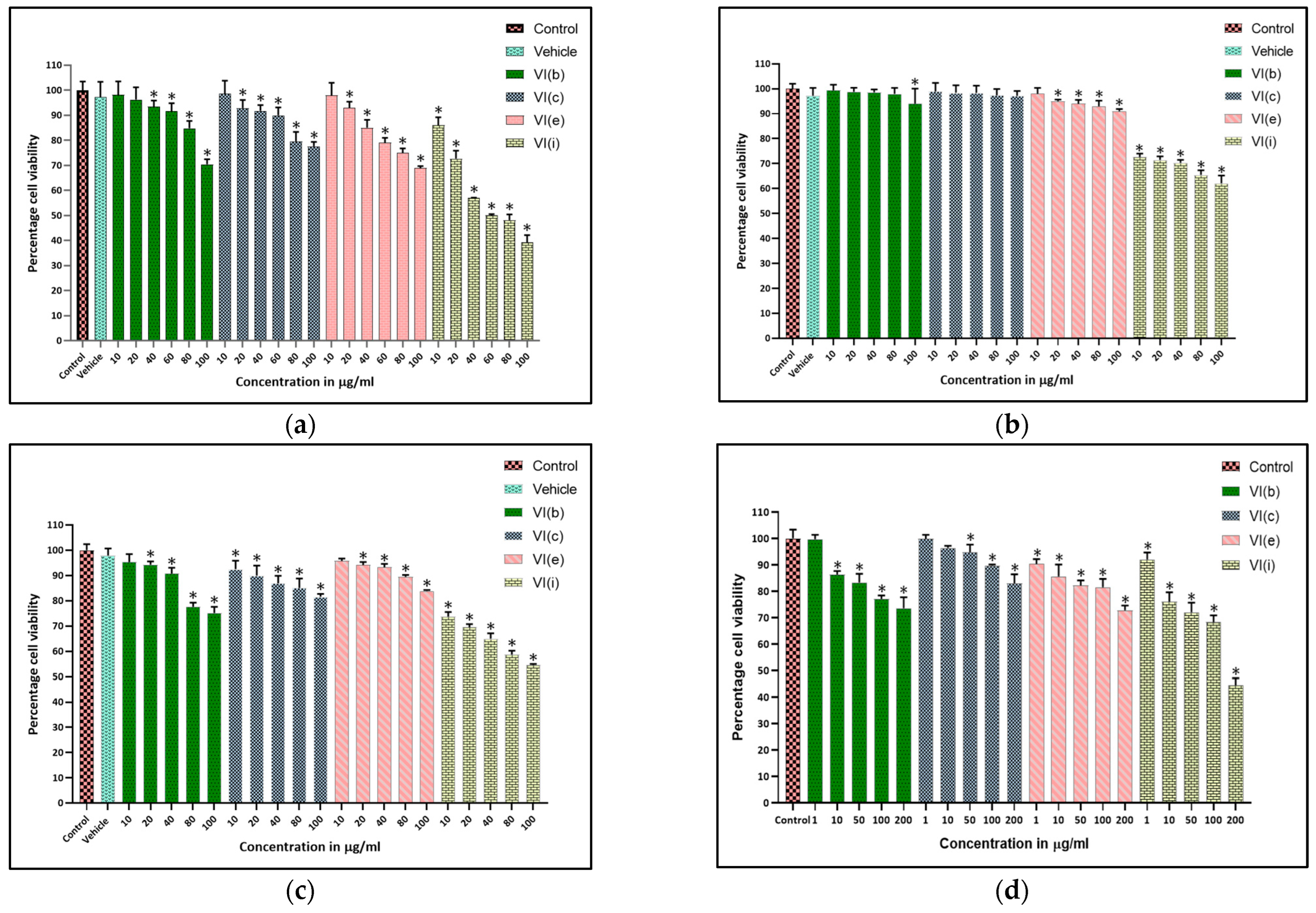
2.5. Antiproliferative Assay
2.6. MTT Assay
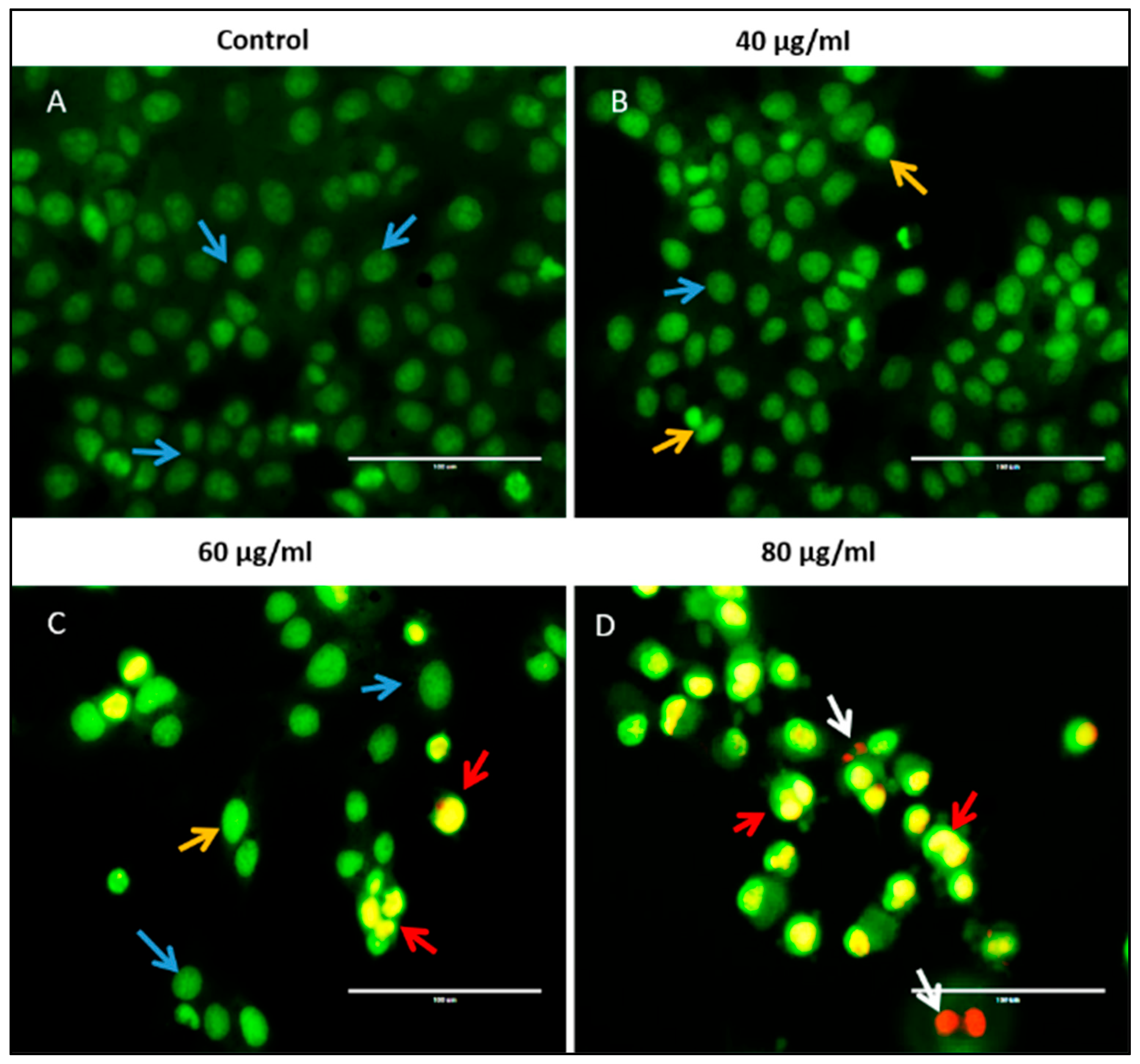
2.7. Apoptosis Study Using Acridine Orange/Ethidium Bromide Dual Staining Method
2.8. Enhanced Reactive Oxygen Species (ROS) Analysis
2.9. Cell Cycle Analysis
2.10. Apoptosis Assay Through AnnexinV/PI
2.11. HDAC–Inhibitory Assay
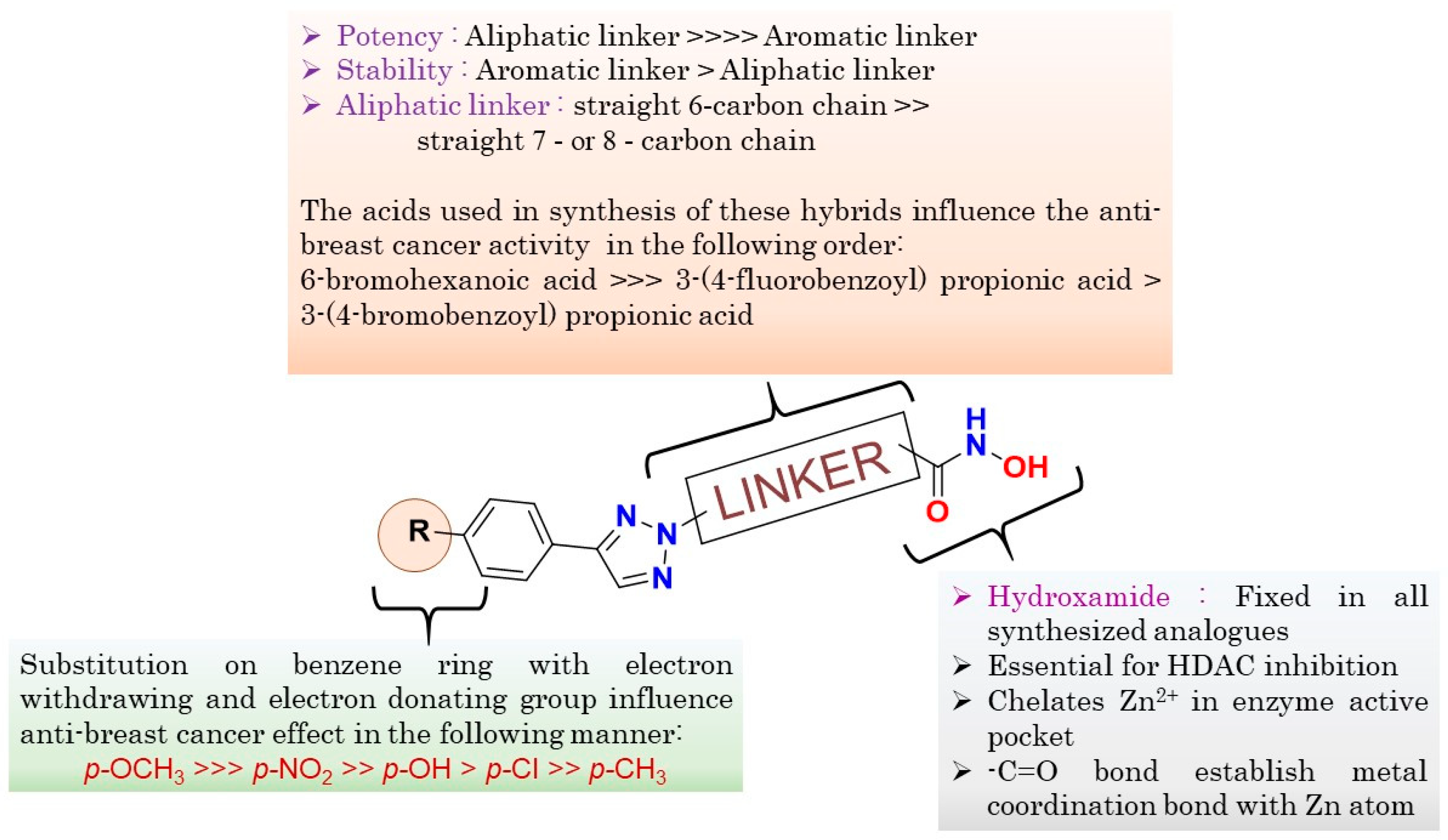
2.12. Structure-Activity Relationship (SAR) (Figure 7)
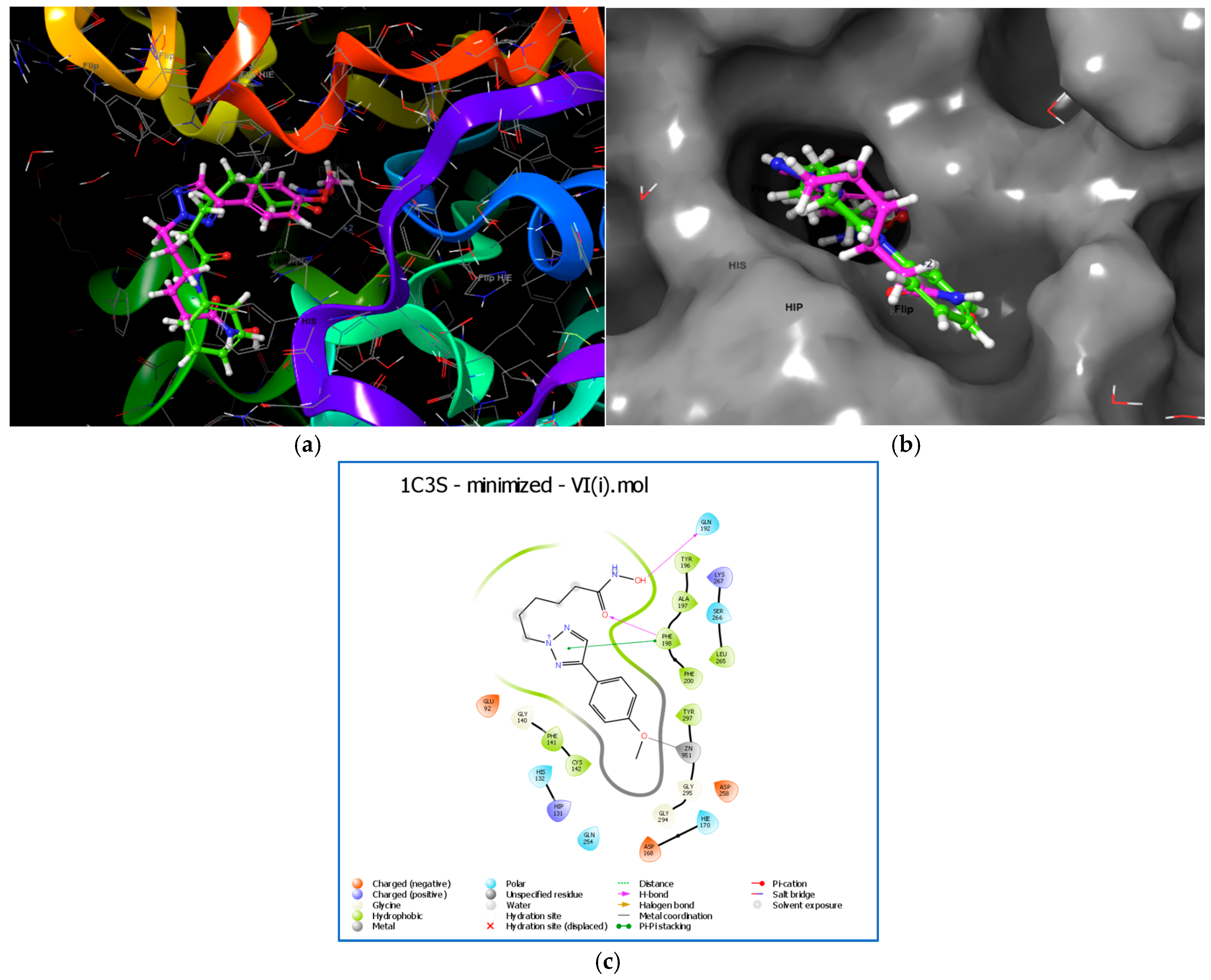
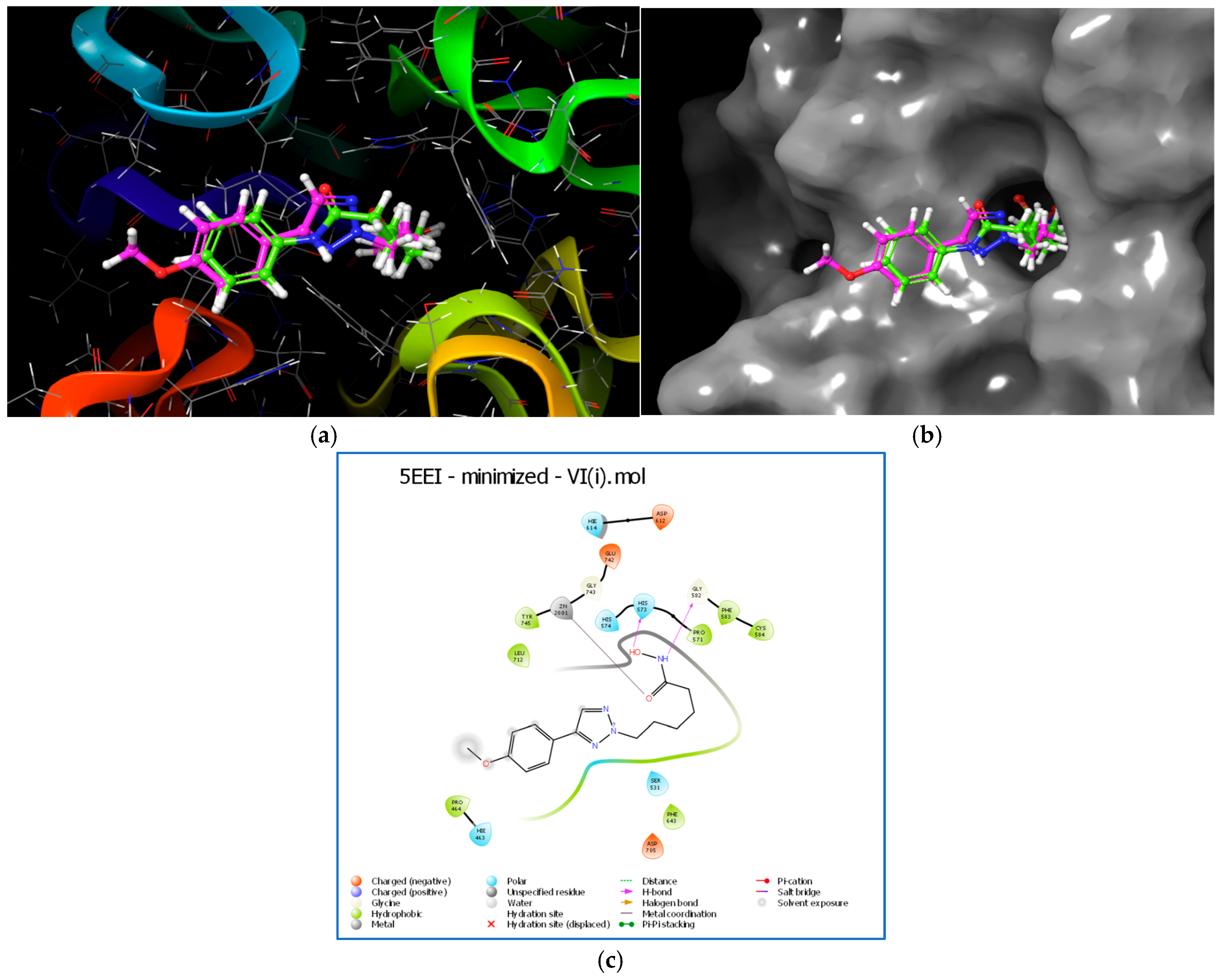
2.13. Molecular Docking
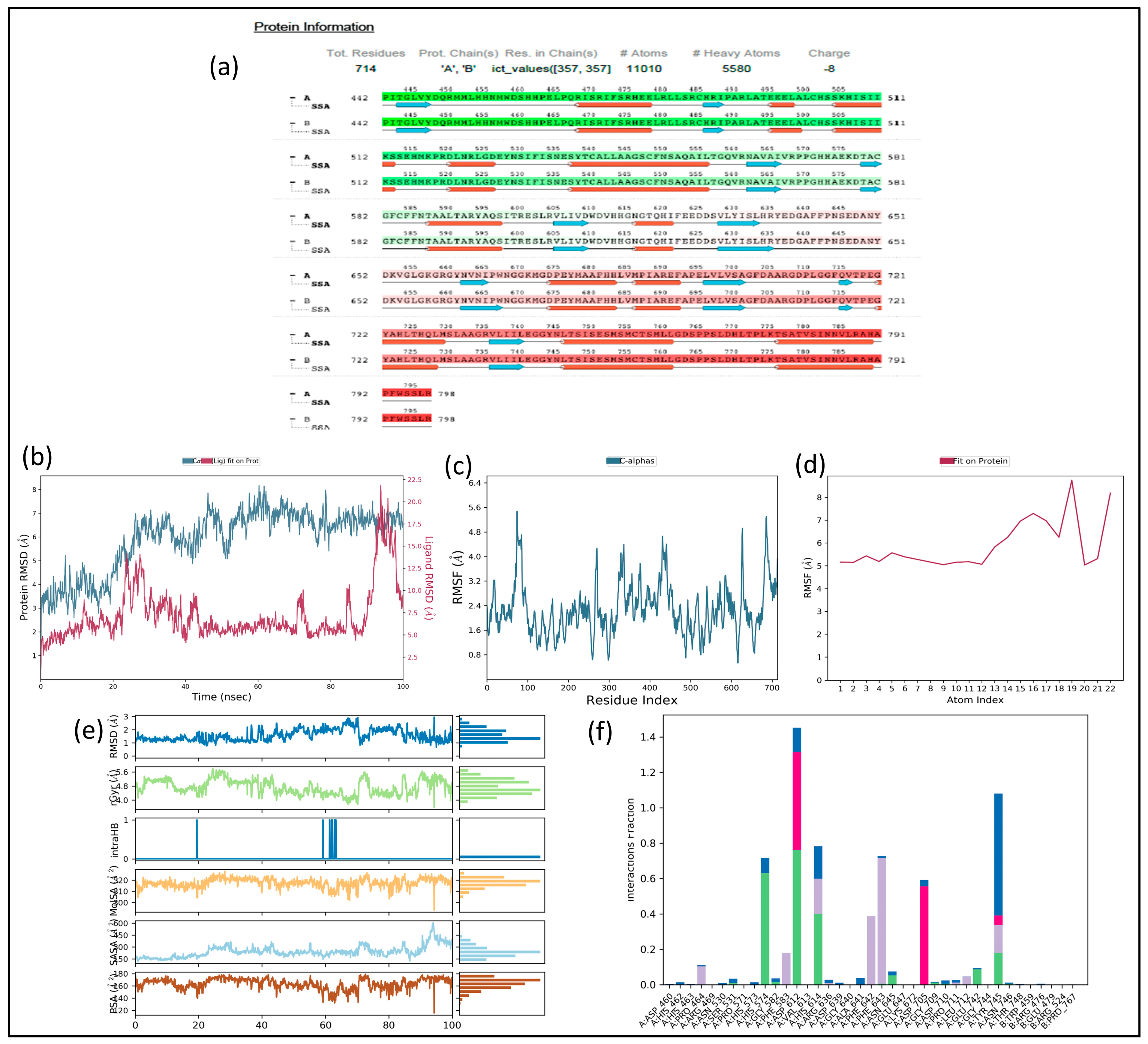
2.14. MD Simulation
2.15. Trajectory Analysis for VI(i)-1C3S Complex (Figure 10)
2.16. Trajectory Analysis for VI(i)-5EEI Complex (Figure 11)
2.17. MM-GBSA Analysis
2.18. ADME and Toxicity Evaluation
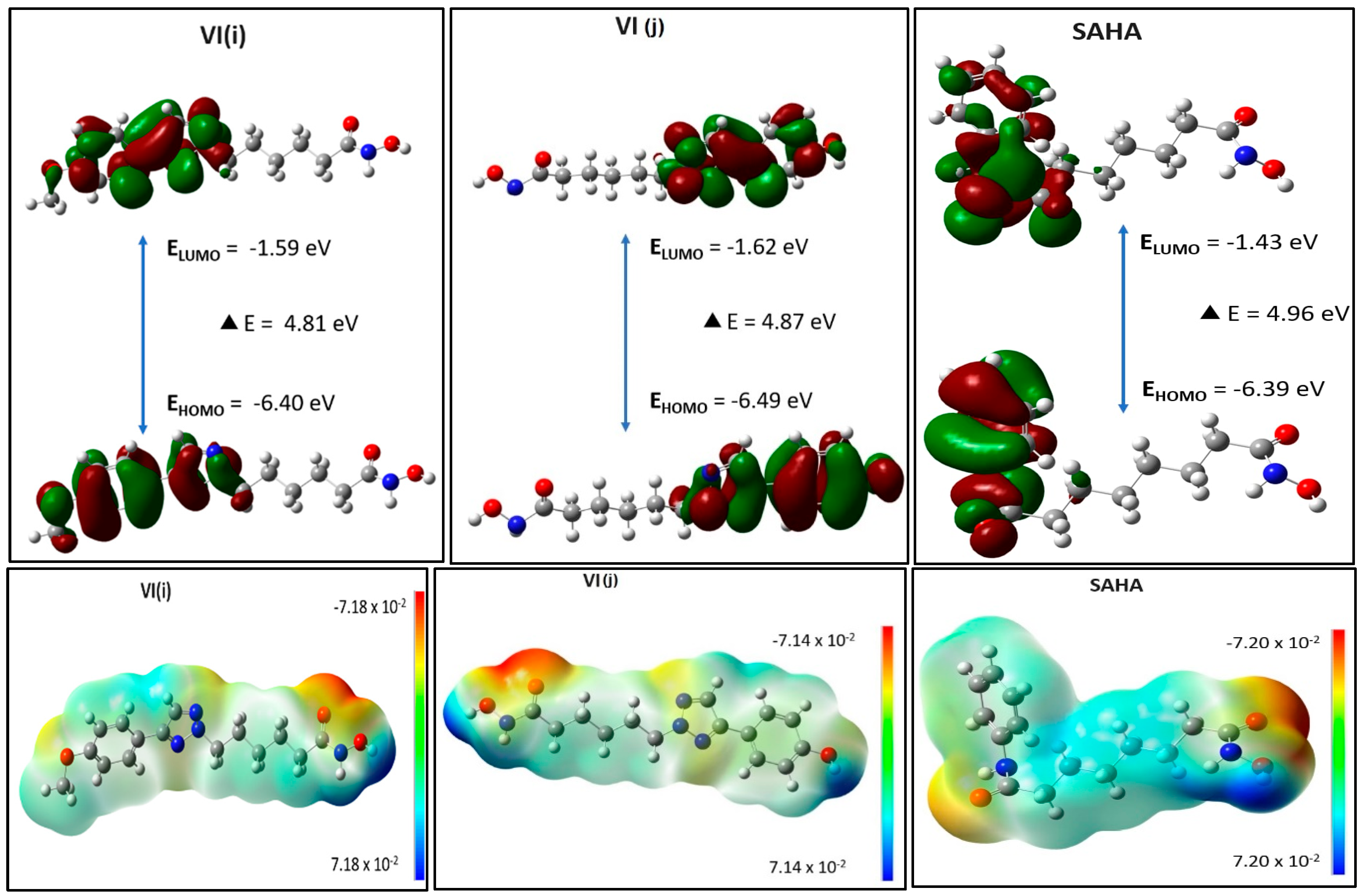
2.19. Energy Calculation of Molecular Orbitals Through DFT
3. Materials and Methods
3.1. Materials
3.2. Synthesis of Target Compounds
3.2.1. General Procedure for Synthesis of Substituted Acrylonitrile III(a–g)
3.2.2. General Procedure for Synthesis of Substituted 1,2,3-Triazole IV(a–g)
3.2.3. General Procedure for Synthesis of Substituted 1,2,3-Triazole-Based Hydroxamic Acid Analogs V(a–o)
3.2.4. General Procedure for Synthesis of Substituted 1,2,3-Triazole-Based Hydroxamide Analogs V(a–o)
3.3. Analytical Data for the Compounds
3.3.1. 3-(4-Methoxyphenyl)acrylonitrile, III(a)
3.3.2. 4-(4-Methoxyphenyl)-2H-1,2,3-triazole, IV(a)
3.3.3. 4-(4-(4-(4-Methoxyphenyl)-2H-1,2,3-triazol-2-yl)phenyl)-4-oxobutanoic Acid V(a)
3.3.4. N-Hydroxy-4-(4-(4-(4-methoxyphenyl)-2H-1,2,3-triazol-2-yl)phenyl)-4-oxobutanamide VI(a)
3.3.5. N-Hydroxy-4-(4-(4-(4-nitrophenyl)-2H-1,2,3-triazol-2-yl)phenyl)-4-oxobutanamide VI(b)
3.3.6. 4-(4-(4-(4-Chlorophenyl)-2H-1,2,3-triazol-2-yl)phenyl)-N-hydroxy-4-oxobutanamide VI(c)
3.3.7. N-Hydroxy-4-oxo-4-(4-(4-(p-tolyl)-2H-1,2,3-triazol-2-yl)phenyl)butanamide VI(d)
3.3.8. N-Hydroxy-4-(4-(4-(4-hydroxyphenyl)-2H-1,2,3-triazol-2-yl)phenyl)-4-oxobutanamide VI(e)
3.3.9. 4-(4-(4-(4-Fluorophenyl)-2H-1,2,3-triazol-2-yl)phenyl)-N-hydroxy-4-oxobutanamide VI(f)
3.3.10. 4-(4-(4-(3,4-Dimethoxyphenyl)-2H-1,2,3-triazol-2-yl)phenyl)-N-hydroxy-4-oxobutanamide VI(g)
3.3.11. 6-(4-(4-Chlorophenyl)-2H-1,2,3-triazol-2-yl)-N-hydroxyhexanamide VI(h)
3.3.12. N-Hydroxy-6-(4-(4-methoxyphenyl)-2H-1,2,3-triazol-2-yl)hexanamide VI(i)
3.3.13. N-Hydroxy-6-(4-(4-hydroxyphenyl)-2H-1,2,3-triazol-2-yl)hexanamide VI(j)
3.3.14. N-Hydroxy-6-(4-(p-tolyl)-2H-1,2,3-triazol-2-Yl)hexanamide VI(k)
3.3.15. 3-(4-(4-(4-Chlorophenyl)-2H-1,2,3-triazol-2-Yl)phenyl)-N-hydroxypropanamide VI(l)
3.3.16. N-Hydroxy-3-(4-(4-(p-tolyl)-2H-1,2,3-triazol-2-yl)phenyl)propanamide VI(m)
3.3.17. N-Hydroxy-3-(4-(4-(4-hydroxyphenyl)-2H-1,2,3-triazol-2-yl)phenyl)propanamide VI(n)
3.3.18. N-Hydroxy-3-(4-(4-(4-methoxyphenyl)-2H-1,2,3-triazol-2-Yl)phenyl)propanamide VI(o)
3.4. Biological Evaluation
3.4.1. Maintenance of Mammalian Cell Lines
3.4.2. Antiproliferative Assay
3.4.3. Cytotoxicity Assay
3.4.4. Apoptosis Study Using Acridine Orange/Ethidium Bromide (AO/EtBr) Dual-staining Technique
3.4.5. Detection of Enhanced Reactive Oxygen Species (ROS)
3.4.6. Analysis of Cell Cycle Distribution
3.4.7. Determination of Apoptosis Through Annexin V/Propidium Iodide (PI)
3.4.8. HDAC Inhibition Assay
3.5. Molecular Modeling Studies
3.5.1. Molecular Docking
3.5.2. Molecular Dynamic (MD) Simulations
3.5.3. MD Trajectory Analysis
3.5.4. Free Binding Energy Calculation (MM-GBSA)
3.5.5. ADME and Toxicity Predictability Studies
3.5.6. Density Functional Theory (DFT) Investigations
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Garg, P.; Malhotra, J.; Kulkarni, P.; Horne, D.; Salgia, R.; Singhal, S.S. Emerging therapeutic strategies to overcome drug resistance in cancer cells. Cancers 2024, 16, 2478. [Google Scholar] [CrossRef]
- Ingham, J.; Ruan, J.L.; Coelho, M.A. Breaking barriers: We need a multidisciplinary approach to tackle cancer drug resistance. BJC Rep. 2025, 3, 11. [Google Scholar] [CrossRef]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2020, 22, 96–118. [Google Scholar] [CrossRef]
- Sorrenti, V.; Buriani, A.; Fortinguerra, S.; Davinelli, S.; Scapagnini, G.; Cassidy, A.; Vivo, I.D. Cell survival, death, and proliferation in senescent and cancer cells: The role of (poly)phenols. Adv. Nutr. 2023, 14, 1111–1130. [Google Scholar] [CrossRef]
- Lu, Y.; Chan, Y.T.; Tan, H.Y.; Li, S.; Wang, N.; Feng, Y. Epigenetic regulation in human cancer: The potential role of epi-drug in cancer therapy. Mol. Cancer 2020, 19, 79. [Google Scholar] [CrossRef]
- Chen, H.P.; Zhao, Y.T.; Zhao, T.C. Histone deacetylases and mechanisms of regulation of gene expression. Crit. Rev. Oncog. 2015, 20, 35–47. [Google Scholar] [CrossRef] [PubMed]
- Routholla, G.; Pulya, S.; Patel, T.; Amin, S.A.; Adhikari, N.; Biswas, S.; Jha, T.; Ghosh, B. Synthesis, biological evaluation, and molecular docking analysis of novel linker-less benzamide-based potent and selective HDAC3 inhibitors. Bioorganic Chem. 2021, 114, 105050. [Google Scholar] [CrossRef]
- Li, G.; Tian, Y.; Zhu, W.-G. The roles of histone deacetylases and their inhibitors in cancer therapy. Front. Cell Dev. Biol. 2020, 8, 576946. [Google Scholar] [CrossRef] [PubMed]
- Hamm, C.A.; Costa, F.F. Epigenomes as therapeutic targets. Pharmacol. Ther. 2015, 151, 72–86. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.H.; Nosella, M.L.; Bolik-Coulon, N.; Harkness, R.W.; Huang, S.K.; Kay, L.E. Correlating histone acetylation with nucleosome core particle dynamics and function. Proc. Natl. Acad. Sci. USA 2023, 120, e2301063120. [Google Scholar] [CrossRef]
- Anh, D.T.; Hai, P.T.; Huy, L.D.; Ngoc, H.B.; Ngoc, T.T.M.; Dung, D.T.M.; Park, E.J.; Song, I.K.; Kang, J.S.; Kwon, J.-H.; et al. Novel 4-oxoquinazoline-based N-hydroxypropenamides as histone deacetylase inhibitors: Design, synthesis, and biological evaluation. ACS Omega 2021, 6, 4907–4920. [Google Scholar] [CrossRef]
- Chen, Y.C.; Koutelou, E.; Dent, S.Y.R. Now open: Evolving insights to the roles of lysine acetylation in chromatin organization and function. Mol. Cell 2022, 82, 716–727. [Google Scholar] [CrossRef]
- Nepali, K.; Liou, J.P. Recent developments in epigenetic cancer therapeutics: Clinical advancement and emerging trends. J. Biomed. Sci. 2021, 28, 27. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Wang, S.; Liu, X.; Lin, R.; Deng, F.; Jia, Z.; Zhang, C.; Li, Z.; Zhu, H.; Tang, L.; et al. Synthesis and biological evaluation of HDAC inhibitors with a novel zinc binding group. Front. Chem. 2020, 8, 256. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Seto, E. HDACs and HDAC inhibitors in cancer development and therapy. Cold Spring Harb. Perspect. Med. 2016, 6, a026831. [Google Scholar] [CrossRef]
- Sanaei, M.; Kavoosi, F. Histone deacetylases and histone deacetylase inhibitors: Molecular mechanisms of action in various cancers. Adv. Biomed. Res. 2019, 8, 63. [Google Scholar] [CrossRef]
- Luan, Y.; Li, J.; Bernatchez, J.A.; Li, R. Kinase and histone deacetylase hybrid inhibitors for cancer therapy. J. Med. Chem. 2019, 62, 3171–3183. [Google Scholar] [CrossRef]
- Zhang, Q.; Wang, S.; Chen, J.; Yu, Z. Histone deacetylases (HDACs) guided novel therapies for T-cell lymphomas. Int. J. Med. Sci. 2019, 16, 424–442. [Google Scholar] [CrossRef]
- Yang, F.; Zhao, N.; Ge, D.; Chen, Y. Next-generation of selective histone deacetylase inhibitors. RSC Adv. 2019, 9, 19571–19583. [Google Scholar] [CrossRef] [PubMed]
- Eckschlager, T.; Plch, J.; Stiborova, M.; Hrabeta, J. Histone deacetylase inhibitors as anticancer drugs. Int. J. Mol. Sci. 2017, 18, 1414. [Google Scholar] [CrossRef]
- Li, Y.; Wang, F.; Chen, X.; Wang, J.; Zhao, Y.; Li, Y.; He, B. Zinc-dependent deacetylase (HDAC) inhibitors with different zinc binding groups. Curr. Top. Med. Chem. 2019, 19, 223–241. [Google Scholar] [CrossRef]
- Curcio, A.; Rocca, R.; Alcaro, S.; Artese, A. The histone deacetylase family: Structural features and application of combined computational methods. Pharmaceuticals 2024, 17, 620. [Google Scholar] [CrossRef]
- Nasrollahzadeh, S.; Eskandarpour, V.; Maleki, M.F.; Eisvand, F.; Mashreghi, M.; Hadizadeh, F.; Tayarani-Najaran, Z.; Ghodsi, R. Design, synthesis and biological evaluation of novel imidazole-based benzamide and hydroxamic acid derivatives as potent histone deacetylase inhibitors and anticancer agents. J. Mol. Struct. 2024, 1297, 136951. [Google Scholar] [CrossRef]
- Sun, L.; Han, L.; Zhang, L.; Chen, C.; Zheng, C. Design, synthesis, and antitumor activity evaluation of carbazole derivatives with potent HDAC inhibitory activity. Med. Chem. Res. 2023, 32, 1677–1689. [Google Scholar] [CrossRef]
- Ajmal, M.; Mahato, A.K.; Khan, M.; Rawat, S.; Husain, A.; Almalki, E.B.; Alzahrani, M.A.; Haque, A.; Hakme, M.J.M.; Albalawi, A.S.; et al. Significance of Triazole in Medicinal chemistry: Advancement in drug design, reward and biological activity. Chem. Biodivers. 2024, 21, e202400637. [Google Scholar] [CrossRef]
- Çot, A.; Çeşme, M.; Onur, S.; Aksakal, E.; Şahin, I.; Tümer, F. Rational design of 1,2,3-triazole hybrid structures as novel anticancer agents: Synthesis, biological evaluation and molecular docking studies. J. Biomol. Struct. Dyn. 2023, 41, 6857–6865. [Google Scholar] [CrossRef]
- Bozorov, K.; Zhao, J.; Aisa, H.A. 1,2,3-Triazole-containing hybrids as leads in medicinal chemistry: A recent overview. Bioorg. Med. Chem. 2019, 27, 3511–3531. [Google Scholar] [CrossRef]
- Vala, D.P.; Vala, R.M.; Patel, H.M. Versatile synthetic platform for 1,2,3-triazole chemistry. ACS Omega 2022, 7, 36945–36987. [Google Scholar] [CrossRef] [PubMed]
- Matin, M.M.; Matin, P.; Rahman, M.R.; Hadda, T.B.; Almalki, F.A.; Mahmud, S.; Ghoneim, M.M.; Alruwaily, M.; Alshehri, S. Triazoles and their derivatives: Chemistry, synthesis, and therapeutic applications. Front. Mol. Biosci. 2022, 9, 864286. [Google Scholar] [CrossRef]
- Ashwini, N.; Garg, M.; Mohan, C.D.; Fuchs, J.; Rangappa, S.; Anusha, S.; Swaroop, T.R.; Rakesh, K.S.; Kanojia, D.; Madan, V.; et al. Synthesis of 1,2-benzisoxazole tethered 1,2,3-triazoles that exhibit anticancer activity in acute myeloid leukemia cell lines by inhibiting histone deacetylases, and inducing p21 and tubulin acetylation. Bioorg. Med. Chem. 2015, 23, 6157–6165. [Google Scholar] [CrossRef] [PubMed]
- Taddei, M.; Ferrini, S.; Giannotti, L.; Corsi, M.; Manetti, F.; Giannini, G.; Vesci, L.; Milazzo, F.M.; Alloatti, D.; Guglielmi, M.B.; et al. Synthesis and evaluation of new Hsp90 inhibitors based on a 1,4,5-trisubstituted 1,2,3-triazole scaffold. J. Med. Chem. 2014, 57, 2258–2274. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.C.; Patil, V.; Guerrant, W.; Green, P.; Oyelere, A.K. Synthesis and structure–activity relationship of histone deacetylase (HDAC) inhibitors with triazole-linked cap group. Bioorg. Med. Chem. 2008, 16, 4839–4853. [Google Scholar] [CrossRef]
- Kalinin, D.V.; Jana, S.K.; Pfafenrot, M.; Chakrabarti, A.; Melesina, J.; Shaik, T.B.; Lancelot, J.; Pierce, R.J.; Sippl, W.; Romier, C.; et al. Structure-based design, synthesis, and biological evaluation of triazole-based smHDAC8 inhibitors. ChemMedChem 2020, 15, 571–584. [Google Scholar] [CrossRef] [PubMed]
- Sun, N.; Yang, K.; Yan, W.; Yao, M.; Yu, C.; Duan, W.; Gu, X.; Jiang, H.; Xie, C.; Cheng, J. Design and synthesis of triazole-containing HDAC inhibitors that induce antitumor effects and immune response. J. Med. Chem. 2023, 66, 4802–4826. [Google Scholar] [CrossRef]
- Hassani, I.A.E.; Rouzi, K.; Hassani, A.A.E.; Karrouchi, K.; Ansar, M. Recent developments towards the synthesis of triazole derivatives: A review. Organics 2024, 5, 450–471. [Google Scholar] [CrossRef]
- Bonandi, E.; Christodoulou, M.S.; Fumagalli, G.; Perdicchia, D.; Rastelli, G.; Passarella, D. The 1,2,3-triazole ring as a bioisostere in medicinal chemistry. Drug Discov. Today 2017, 22, 1572–1581. [Google Scholar] [CrossRef]
- Madadi, N.R.; Penthala, N.R.; Howk, K.; Ketkar, A.; Eoff, R.L.; Borrelli, M.J.; Crooks, P.A. Synthesis and biological evaluation of novel 4,5-disubstituted 2H-1,2,3-triazoles as cis-constrained analogues of combretastatin A-4. Eur. J. Med. Chem. 2015, 103, 123–132. [Google Scholar] [CrossRef]
- Kode, J.; Kovvuri, J.; Nagaraju, B.; Jadhav, S.; Barkume, M.; Sen, S.; Kasinathan, N.K.; Chaudhari, P.; Mohanty, B.S.; Gour, J.; et al. Synthesis, biological evaluation and molecular docking analysis of phenstatin based indole linked chalcones as anticancer agents and tubulin polymerization inhibitors. Bioorg. Chem. 2020, 105, 104447. [Google Scholar] [CrossRef]
- Orellana, E.A.; Kasinski, A.L. Sulforhodamine B (SRB) assay in cell culture to investigate cell proliferation. Bio-Protocol 2016, 6, e1984. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Nagarajan, A.; Uchil, P.D. Analysis of cell viability by the MTT assay. Cold Spring Harb. Protoc. 2018, 2018, pdb-prot095505. [Google Scholar] [CrossRef]
- Sharma, G.; Rana, N.K.; Singh, P.; Dubey, P.; Pandey, D.S.; Koch, B. p53 dependent apoptosis and cell cycle delay induced by heteroleptic complexes in human cervical cancer cells. Biomed. Pharmacother. 2017, 88, 218–231. [Google Scholar] [CrossRef]
- Singh, M.; Rana, N.K.; Muthu, M.S.; Jha, A.; Baul, T.S.B.; Koch, B. Enhanced in vitro therapeutic efficacy of triphenyltin (IV) loaded vitamin E TPGS against breast cancer therapy. Mater. Today Commun. 2022, 31, 103256. [Google Scholar] [CrossRef]
- Bardaweel, S.K.; Gul, M.; Alzweiri, M.; Ishaqat, A.; ALSalamat, H.A.; Bashatwah, R.M. Reactive oxygen species: The dual role in physiological and pathological conditions of the human body. Eurasian J. Med. 2018, 50, 193–201. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, Y.; Wang, L.; Lee, S. Levistolide A induces apoptosis via ROS-mediated ER stress pathway in colon cancer cells. Cell. Physiol. Biochem. 2017, 42, 929–938. [Google Scholar] [CrossRef]
- Crowley, L.C.; Marfell, B.J.; Scott, A.P.; Waterhouse, N.J. Quantitation of apoptosis and necrosis by annexin V binding, propidium iodide uptake, and flow cytometry. Cold Spring Harb. Protoc. 2016, 2016, pdb.prot087288. [Google Scholar] [CrossRef]
- Aboeldahab, M.A.A.; Beshr, E.A.M.; Shoman, M.E.; Rabea, S.M.; Aly, O.M. Spirohydantoins and 1,2,4-triazole-3-carboxamide derivatives as inhibitors of histone deacetylase: Design, synthesis, and biological evaluation. Eur. J. Med. Chem. 2018, 146, 79–92. [Google Scholar] [CrossRef]
- Padige, G.; Negmeldin, A.T.; Pflum, M.K. Development of an ELISA-based HDAC activity assay for characterization of isoform-selective inhibitors. J. Biomol. Screen. 2015, 20, 1277–1285. [Google Scholar] [CrossRef] [PubMed]
- Sahayarayan, J.J.; Rajan, K.S.; Vidhyavathi, R.; Nachiappan, M.; Prabhu, D.; Alfarraj, R.; Arokiyaraj, S.; Daniel, A.N. In-silico protein-ligand docking studies against the estrogen protein of breast cancer using pharmacophore-based virtual screening approaches. Saudi J. Biol. Sci. 2021, 28, 400–407. [Google Scholar] [CrossRef]
- Drakontaeidi, A.; Pontiki, E. A review on molecular docking on HDAC isoforms: Novel tool for designing selective inhibitors. Pharmaceuticals 2023, 16, 1639. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, N.; Asari, A.; Yousaf, N.; Ahmad, M.; Ahmed, M.; Faisal, A.; Saleem, M.; Muddassar, M. Combined 3D-QSAR, molecular docking and dynamics simulations studies to model and design TTK inhibitors. Front. Chem. 2022, 10, 1003816. [Google Scholar] [CrossRef] [PubMed]
- Pai, P.; Kumar, A.; Shetty, M.G.; Kini, S.G.; Krishna, M.B.; Satyamoorthy, K.; Babitha, K.S. Identification of potent HDAC2 inhibitors using E-pharmacophore modeling, structure-based virtual screening and molecular dynamic simulation. J. Mol. Model. 2022, 28, 119. [Google Scholar] [CrossRef]
- Ramachandran, B.; Kesavan, S.; Rajkumar, T. Molecular modeling and docking of small molecule inhibitors against NEK2. Bioinformation 2016, 12, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Baselious, F.; Hilscher, S.; Robaa, D.; Barinka, C.; Schutkowski, M.; Sippl, W. Comparative structure-based virtual screening utilizing optimized AlphaFold model identifies selective HDAC11 inhibitor. Int. J. Mol. Sci. 2024, 25, 1358. [Google Scholar] [CrossRef]
- Patel, P.; Rajak, H. Development of hydroxamic acid derivatives as anticancer agents with the application of 3D-QSAR, docking, and molecular dynamics simulation studies. Med. Chem. Res. 2018, 27, 2100–2115. [Google Scholar] [CrossRef]
- Rudrapal, M.; Eltayeb, W.A.; Rakshit, G.; El-Arabey, A.A.; Khan, J.; Aldosari, S.M.; Alshehri, B.; Abdalla, M. Dual synergistic inhibition of COX and LOX by potential chemicals from Indian daily spices investigated through detailed computational studies. Sci. Rep. 2023, 13, 8656. [Google Scholar] [CrossRef]
- Ivanova, L.; Tammiku-Taul, J.; Garcia-Sosa, A.T.; Sidorova, Y.; Saarma, M.; Karelson, M. Molecular dynamics simulations of the interactions between glial cell line-derived neurotrophic factor family receptor GFRα1 and small-molecule ligands. ACS Omega 2018, 3, 11407–11414. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.S.; Ranjan, S.; Sharma, C.S.; Singh, H.P.; Kalra, S.; Kumar, N. Computational investigation of potential inhibitors of novel coronavirus 2019 through structure-based virtual screening, molecular dynamics, and density functional theory studies. J. Biomol. Struct. Dyn. 2021, 39, 4449–4461. [Google Scholar] [CrossRef]
- Yuan, Y.; Deng, J.; Cui, Q. Molecular dynamics simulations establish the molecular basis for the broad allostery hotspot distributions in the tetracycline repressor. J. Am. Chem. Soc. 2022, 144, 10870–10887. [Google Scholar] [CrossRef]
- AbdElmoniem, N.; Abdallah, M.H.; Mukhtar, R.M.; Moutasim, F.; Ahmed, A.R.; Edris, A.; Ibraheem, W.; Makki, A.A.; Elshamly, E.M.; Elhag, R.; et al. Identification of novel natural dual HDAC and Hsp90 inhibitors for metastatic TNBC using e-pharmacophore modeling, molecular docking, and molecular dynamics studies. Molecules 2023, 28, 1771. [Google Scholar] [CrossRef]
- Singh, S.; Kumar, S.; Dhanasingh, I. In-silico identification of flavonoids based inhibitors against Sortase-A from Enterococcus faecalis (Ef). J. Microbiol. Biotechnol. Food Sci. 2024, 14, e11256. [Google Scholar] [CrossRef]
- Jiao, J.; Fang, H.; Wang, X.; Guan, P.; Yuan, Y.; Xu, W. Design, synthesis and preliminary biological evaluation of N-hydroxy-4-(3-phenylpropanamido)benzamide (HPPB) derivatives as novel histone deacetylase inhibitors. Eur. J. Med. Chem. 2009, 44, 4470–4476. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Wang, L.; Hou, X.; Fu, H.; Song, W.; Tang, W.; Fang, H. Design, synthesis and preliminary bioactivity studies of 1,2-dihydrobenzo[d]isothiazol-3-one-1,1-dioxide hydroxamic acid derivatives as novel histone deacetylase inhibitors. Bioorganic Med. Chem. 2014, 22, 1529–1538. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Sun, Y.; Liu, J.; Wu, X.; Chen, L.; Ma, L.; Wu, P. Histone deacetylase inhibitor sodium butyrate suppresses DNA double strand break repair induced by etoposide more effectively in MCF-7 cells than in HEK293 cells. BMC Biochem. 2015, 16, 2. [Google Scholar] [CrossRef]
- Zhao, N.; Yang, F.; Han, L.; Qu, Y.; Ge, D.; Zhang, H. Development of Coumarin-Based Hydroxamates as Histone Deacetylase Inhibitors with Antitumor Activities. Molecules 2020, 25, 717. [Google Scholar] [CrossRef]
- Wu, Q.; Cheng, Z.; Zhu, J.; Xu, W.; Peng, X.; Chen, C.; Li, W.; Wang, F.; Cao, L.; Yi, X.; et al. Suberoylanilide hydroxamic acid treatment reveals crosstalks among proteome, ubiquitylome and acetylome in non-small cell lung cancer A549 cell line. Sci. Rep. 2015, 5, 9520. [Google Scholar] [CrossRef]
- Kholiya, F.; Chatterjee, S.; Bhojani, G.; Sen, S.; Barkume, M.; Kasinathan, N.K.; Kode, J.; Meena, R. Seaweed polysaccharide derived bioaldehyde nanocomposite: Potential application in anticancer therapeutics. Carbohydr. Polym. 2020, 240, 116282. [Google Scholar] [CrossRef] [PubMed]




 | ||||
|---|---|---|---|---|
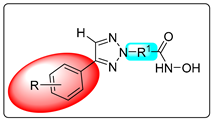
| Compd | R | R1 (Linker) | GI50 (μg/mL) a | |
| A-549 (Human Lung Cancer Cell Line) | MCF-7 (Human Breast Cancer Cell Line) | |||
| VI(a) | p-OCH3 phenyl | 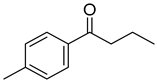 | 19.35 | 15.13 |
| VI(b) | p-NO2 phenyl | <10 | <10 | |
| VI(c) | p-Cl phenyl | <10 | <10 | |
| VI(d) | p-CH3 phenyl | 14.69 | 12.91 | |
| VI(e) | p-OH phenyl | <10 | <10 | |
| VI(f) | p-F phenyl | 26.92 | <10 | |
| VI(g) | 3,4-diOCH3 phenyl | 53.00 | 15.00 | |
| VI(h) | p-Cl phenyl |  | >80 | 50.00 |
| VI(i) | p-OCH3 phenyl | <10 | <10 | |
| VI(j) | p-OH phenyl | 50.00 | 17.00 | |
| VI(k) | p-CH3 phenyl | >80 | 14.86 | |
| VI(l) | p-Cl phenyl | 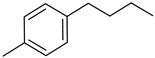 | >80 | >80 |
| VI(m) | p-CH3 phenyl | >80 | >80 | |
| VI(n) | p-OH phenyl | >80 | >80 | |
| VI(o) | p-OCH3 phenyl | 62.00 | >80 | |
| ADR | - | - | <10 | <10 |
| Comp. No. | IC50 Values a (μg/mL) | |
|---|---|---|
| HDAC1 | HDAC6 | |
| VI(b) | 8.04 | 9.21 |
| VI(c) | 7.87 | 11.76 |
| VI(e) | 9.60 | 6.57 |
| VI(i) | 3.06 | 4.08 |
| SAHA b | 0.08 | 0.09 |
| Sr. No. | Compd. Code | Dock Score (kcal/mol) | |
|---|---|---|---|
| HDAC1: 1C3S | HDAC6: 5EEI | ||
| 1 | VI(a) | −9.002 | −8.449 |
| 2 | VI(b) | −8.301 | −7.096 |
| 3 | VI(c) | −9.446 | −7.065 |
| 4 | VI(d) | −8.562 | −7.646 |
| 5 | VI(e) | −8.343 | −6.718 |
| 6 | VI(f) | −9.308 | −7.937 |
| 7 | VI(g) | −8.398 | −7.841 |
| 8 | VI(h) | −9.883 | −9.718 |
| 9 | VI(i) | −10.691 | −11.373 |
| 10 | VI(j) | −10.482 | −10.358 |
| 11 | VI(k) | −9.951 | −9.581 |
| 12 | VI(l) | −9.770 | −9.509 |
| 13 | VI(m) | −10.047 | −10.201 |
| 14 | VI(n) | −8.629 | −10.073 |
| 15 | VI(o) | −9.395 | −10.349 |
| 16 | SAHA | −9.724 | −11.261 |
| MM/GBSA Energy Descriptors (kcal mol−1) | 1C3S-VI(i) | 5EEI-VI(i) | ||
|---|---|---|---|---|
| 0 ns | 100 ns | 0 ns | 100 ns | |
| MMGBSA_dG_Bind | −76.1329 | −91.8075 | −55.2446 | −59.4738 |
| MMGBSA_dG_Bind_Coulomb | −36.8718 | −37.8267 | 4.5912 | 38.9528 |
| MMGBSA_dG_Bind_Hbond | −1.4332 | −0.9737 | −1.5613 | −0.2904 |
| MMGBSA_dG_Bind_Lipo | −22.7017 | −32.4736 | −25.5294 | −19.6282 |
| MMGBSA_dG_Bind_Packing | −10.228 | −1.876 | −0.000019 | 0 |
| MMGBSA_dG_Bind_Solv_GB | 28.0751 | 23.7755 | −0.0757 | −35.1282 |
| MMGBSA_dG_Bind_vdW | −35.9366 | −47.8496 | −37.0841 | −20.7722 |
| Complex_Energy | −11,968.88 | −11,986.90 | −23,399.1 | −23,378.5 |
| Complex_Coulomb | −11,349.10 | −11,349.80 | −23,184.5 | −23,147 |
| Complex_Hbond | −162.939 | −162.48 | −351.914 | −350.643 |
| Complex_Lipo | −2045.13 | −2053.54 | −4366.95 | −4361.11 |
| Complex_Packing | −21.9893 | −13.449 | −25.4552 | −25.4552 |
| Complex_Solv_GB | −2737.34 | −3745.27 | −3372.43 | −3409.2 |
| Complex_vdW | −1254.67 | −1263.47 | −2654.44 | −2629.52 |
| Parameters | VI(j) (Best Docked Compound Among the Series) | VI(i) (Compound with Good Anticancer Activity) | SAHA (Marketed HDAC Inhibitor) |
|---|---|---|---|
| Total energy (eV) | −26,909.72 | −27,978.74 | −23,962.34 |
| HOMO energy (eV) | −6.49 | −6.40 | −6.39 |
| LUMO energy (eV) | −1.62 | −1.59 | −1.43 |
| Band gap (eV) | 4.87 | 4.81 | 4.96 |
| Dipole moment | 5.19 | 6.49 | 2.93 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shirbhate, E.; Koch, B.; Singh, V.; Dubey, A.; Yasin, H.K.A.; Rajak, H. Heteroaryl-Capped Hydroxamic Acid Derivatives with Varied Linkers: Synthesis and Anticancer Evaluation with Various Apoptosis Analyses in Breast Cancer Cells, Including Docking, Simulation, DFT, and ADMET Studies. Pharmaceuticals 2025, 18, 1148. https://doi.org/10.3390/ph18081148
Shirbhate E, Koch B, Singh V, Dubey A, Yasin HKA, Rajak H. Heteroaryl-Capped Hydroxamic Acid Derivatives with Varied Linkers: Synthesis and Anticancer Evaluation with Various Apoptosis Analyses in Breast Cancer Cells, Including Docking, Simulation, DFT, and ADMET Studies. Pharmaceuticals. 2025; 18(8):1148. https://doi.org/10.3390/ph18081148
Chicago/Turabian StyleShirbhate, Ekta, Biplob Koch, Vaibhav Singh, Akanksha Dubey, Haya Khader Ahmad Yasin, and Harish Rajak. 2025. "Heteroaryl-Capped Hydroxamic Acid Derivatives with Varied Linkers: Synthesis and Anticancer Evaluation with Various Apoptosis Analyses in Breast Cancer Cells, Including Docking, Simulation, DFT, and ADMET Studies" Pharmaceuticals 18, no. 8: 1148. https://doi.org/10.3390/ph18081148
APA StyleShirbhate, E., Koch, B., Singh, V., Dubey, A., Yasin, H. K. A., & Rajak, H. (2025). Heteroaryl-Capped Hydroxamic Acid Derivatives with Varied Linkers: Synthesis and Anticancer Evaluation with Various Apoptosis Analyses in Breast Cancer Cells, Including Docking, Simulation, DFT, and ADMET Studies. Pharmaceuticals, 18(8), 1148. https://doi.org/10.3390/ph18081148







