In Silico Design and Computational Elucidation of Hypothetical Resveratrol–Curcumin Hybrids as Potential Cancer Pathway Modulators
Abstract
1. Introduction
2. Results
2.1. DFT-Based Electronic Properties and Spectroscopic Analyses
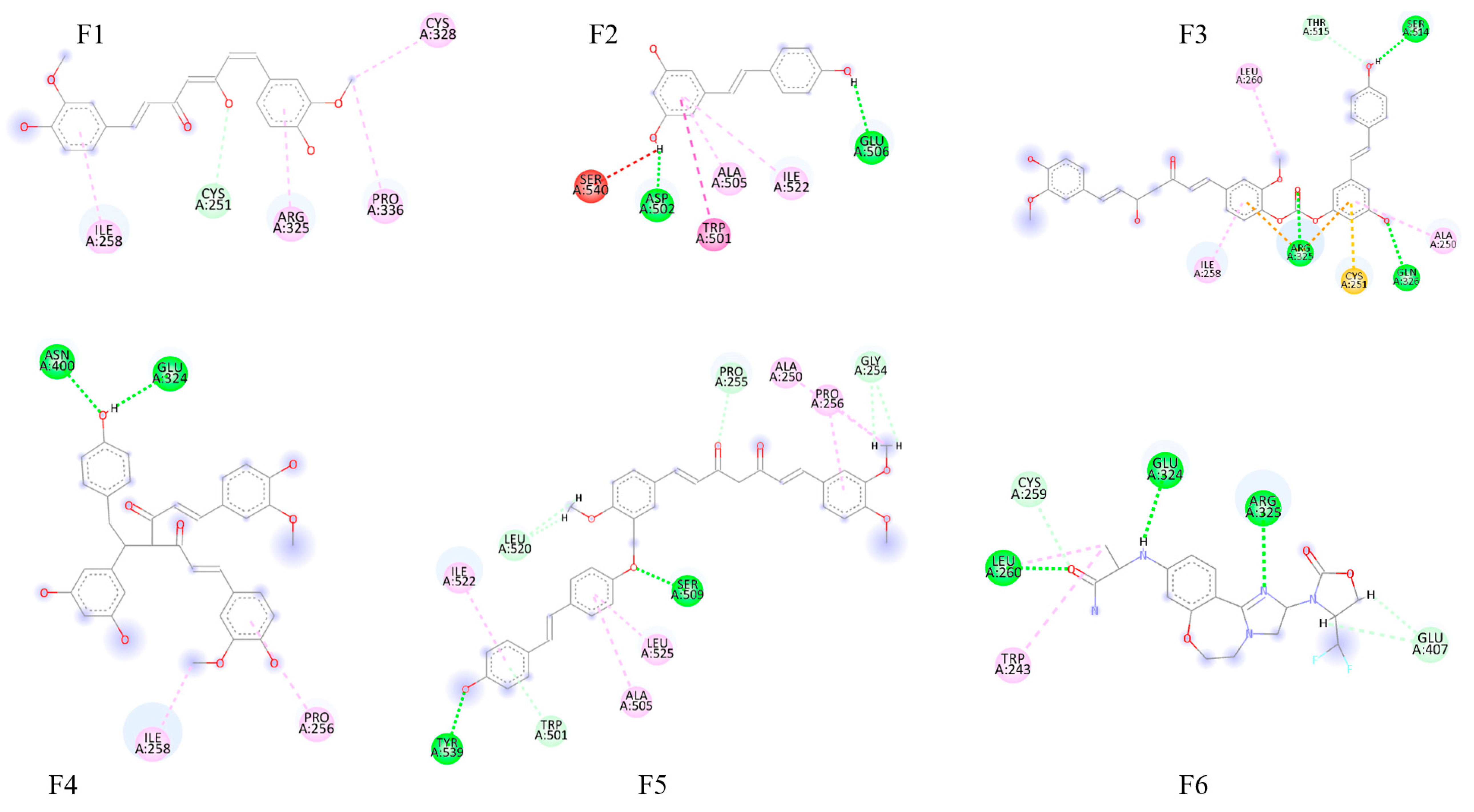
2.2. Molecular Docking Results
2.3. Molecular Dynamics (MD) Simulation
2.4. Binding Free Energy Analysis Using Molecular Mechanics/Poisson–Boltzmann Surface Area (MM/PBSA)
3. Discussion
3.1. Density Functional Theory (DFT) Analysis
3.1.1. Frontier Molecular Orbitals (FMO) Analysis
3.1.2. Molecular Electrostatic Potential (MEP) Analysis
3.1.3. FTIR (Fourier Transform Infrared) Spectroscopy Analysis
| Prominent Theoretical Peaks (cm−1) * | Experimental Peak (cm−1) | Assignment | ||
|---|---|---|---|---|
| ELRC-LC | ELRC-SC | EtLRC | ||
| 1650, 1654, 1722, 1828 | 1650, 1656, 1730, 1748 | 1631, 1724 | 1639 (Curcumin) [39]; 1752 (Resveratrol) [40] | C=O stretching (ester; conjugated carbonyl) |
| 1620, 1625, 1643, 1612, 1616, 1636 | 1616, 1619, 1627, 1637, 1635, 1648, 1653 | 1626, 1651, 1549, 1607, 1652 | 1639 (Curcumin) [39]; 1600–1640 (Resveratrol) [40] | C=C aromatic stretching (phenyl ring) |
| 1139, 1219 | 1023, 1271 | 1079 (Curcumin) [39]; 1215 (Resveratrol) [40] | C–O phenolic; C–O–C ester | |
| 3013, 3014 | 3005, 3013 | 3013, 3016, 3019 | 2960, 2922 (Curcumin) [39] | C–H aliphatic (CH2, CH3 asymmetric stretching) |
| 3740, 3798–3802 | 3742, 3794, 3793, 3797, 3800 | 3797 | 3265 (Curcumin) [39]; 3174–3248 (Resveratrol) [40] | O–H phenolic stretching (hydrogen bonded; broad) |
3.2. Molecular Docking and Interaction Analysis
| Receptors | ||||
|---|---|---|---|---|
| Ligands | AKT1 | MAPK | STAT3 | |
| Resveratrol analogues | Resveratrol | −8.3 | −7.6 | −6.0 |
| ZINC000000006787 | −8.3 | −7.5 | −5.7 | |
| ZINC000003978779 | −8.3 | −7.2 | −5.4 | |
| ZINC000004098633 | −8.2 | −8.2 | −6.9 | |
| ZINC000015112534 | −9.6 | −7.6 | −7.2 | |
| ZINC000015112536 | −9.7 | −8.3 | −6.7 | |
| ZINC000015112538 | −9.8 | −8.1 | −6.7 | |
| ZINC000015112540 | −9.8 | −7.1 | −6.3 | |
| ZINC000035653092 | −9.7 | −7.4 | −6.4 | |
| ZINC000040977346 | −9.0 | −7.4 | −5.5 | |
| ZINC000085612047 | −8.1 | −9.5 | −6.8 | |
| ZINC000095620822 | −10.6 | −7.3 | −6.7 | |
| ZINC000100823225 | −8.8 | −10.3 | −7.9 | |
| ZINC000100823227 | −11 | −9.1 | −6.8 | |
| ZINC000100823228 | −11.2 | −9.3 | −8.0 | |
| ZINC000100827960 | −11.3 | −8.8 | −6.9 | |
| ZINC000100827962 | −11.0 | −8.8 | −8.1 | |
| ZINC000100827965 | −11.0 | −9.0 | −8.2 | |
| ZINC000230079510 | −10.9 | −9.9 | −7.5 | |
| ZINC000230079516 | −11.8 | −10.3 | −8.3 | |
| ZINC000230079520 | −10.6 | −10.2 | −8.0 | |
| ZINC000230079525 | −10.8 | −10.2 | −7.5 | |
| ZINC000230097101 | −11.3 | −9.2 | −7.2 | |
| ZINC000230097106 | −10.8 | −8.8 | −7.7 | |
| ZINC000230097112 | −10.8 | −8.5 | −7.4 | |
| ZINC000230097119 | −11.0 | −9.0 | −7.4 | |
| Curcumin analogues | Curcumin | −9.8 | −6.6 | −5.6 |
| ZINC000000899824 | −11.0 | −7.4 | −5.6 | |
| ZINC000014948330 | −9.1 | −7.0 | −5.3 | |
| ZINC000016527488 | −9.3 | −6.8 | −5.4 | |
| ZINC000019816066 | −9.7 | −7.7 | −6.4 | |
| ZINC000085926636 | −9.0 | −6.4 | −5.2 | |
| ZINC000150366575 | −10.8 | −7.4 | −6.9 | |
| ZINC000150366578 | −10.3 | −7.7 | −6.6 | |
| ZINC000150366582 | −10.2 | −6.9 | −6.6 | |
| ZINC000150366588 | −10.8 | −7.1 | −6.8 | |
| ZINC000150368101 | −10.2 | −8.3 | −7.1 | |
| ZINC000150368109 | −11.0 | −9.0 | −7.4 | |
| ZINC000150368115 | −10.8 | −8.9 | −6.9 | |
| ZINC000150368122 | −9.8 | −9.3 | −8.1 | |
| ZINC000150368128 | −10.2 | −7.1 | −6.1 | |
| ZINC000150368132 | −9.7 | −7.6 | −6.1 | |
| ZINC000150368137 | −10.4 | −8.1 | −7.0 | |
| ZINC000150368142 | −8.8 | −7.7 | −7.4 | |
| Hybrid Molecules of Curcumin-Resveratrol | ELRC-LC | −11.4 | −9.6 | −5.7 |
| ELRC-SC | −9.5 | −7.8 | −7.2 | |
| EtLRC | −10.8 | −9.3 | −7.6 | |
| Inavolisib | −9.9 | −8.1 | −7.2 | |
3.3. MD Simulation Analysis
3.3.1. Conformational Flexibility Analysis
3.3.2. Radius of Rotation (Rg)
3.3.3. Binding Free Energy MM/PBSA Analysis
| Receptors | Ligands | VDWAALS | EEL | Total (ΔGbind) | |||
|---|---|---|---|---|---|---|---|
| Frame Types | Initial | Last | Initial | Last | Initial | Last | |
| AKT1 | Curcumin | −44.97 | −46.02 | −28.71 | −0.70 | −15.65 | −13.38 |
| Resveratrol | −28.99 | −27.56 | −11.45 | −16.70 | −4.54 | −12.46 | |
| ELRC-LC | −70.94 | −69.99 | −34.00 | −30.53 | 7.91 | −22.11 | |
| ELRC-SC | −56.97 | −44.73 | −28.13 | −71.10 | 1.63 | −22.95 | |
| EtLRC | −72.42 | −75.42 | −6.32 | −29.64 | 1.31 | −26.50 | |
| Inavolisib | −47.83 | −26.48 | −63.91 | −71.86 | 7.16 | −16.06 | |
| MAPK | Curcumin | −39.67 | −41.70 | −43.20 | −39.95 | −17.70 | −32.28 |
| Resveratrol | −26.20 | −25.51 | −31.47 | −37.46 | −12.66 | −17.20 | |
| ELRC-LC | −50.37 | −38.24 | −45.44 | −50.66 | −12.51 | −26.21 | |
| ELRC-SC | −66.23 | −47.65 | −29.87 | −52.83 | 1.77 | −36.29 | |
| EtLRC | −57.92 | −28.68 | −17.8 | −52.03 | −10.98 | −29.38 | |
| Inavolisib | −40.62 | −19.36 | 41.74 | −65.61 | 37.29 | −4.63 | |
| STAT3 | Curcumin | −36.94 | −32.55 | −7.35 | 2.84 | −15.80 | −17.60 |
| Resveratrol | −22 | −20.39 | −24.65 | −40.82 | −11.64 | −20.57 | |
| ELRC-LC | −43.67 | −37.52 | −18.09 | −26.89 | −7.53 | −14.94 | |
| ELRC-SC | −42.89 | −19.89 | −22.80 | −18.35 | −15.41 | −17.83 | |
| EtLRC | −51.74 | −61.97 | −17.75 | −21.62 | −22.25 | −35.40 | |
| Inavolisib | −29.94 | −26.60 | −55.96 | −114.74 | −6.68 | −29.14 | |
4. Materials and Methods
4.1. De Novo Hybrid Drug Design
4.2. Geometry Optimization Using Density Functional Theory (DFT)
4.3. Molecular Docking
4.4. Molecular Dynamics Simulation
4.5. MM/PBSA-Based Binding Free Energy Calculations
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| RMSD | Root Mean Square Deviation |
| AKT1 | AKT serine/threonine kinase 1 |
| MAPK | Mitogen-Activated Protein Kinases |
| STAT3 | Signal Transducer and Activator of Transcription 3 |
| RMSF | Root Mean Square Fluctuation |
| Rg | Radius of Gyration |
| MM/PBSA | Molecular Mechanics Poisson-Boltzmann Surface Area |
| ELRC-SC | Ester-Linked Resveratrol–Curcumin Short-Chain |
| ELRC-LC | Ester-Linked Resveratrol–Curcumin Long-Chain |
| EtRC | Ester-Linked Resveratrol–Curcumin |
| DFT | Density Functional Theory |
| MD | Molecular Dynamics |
| NF-κB | Nuclear Factor kappa B |
| EGFR | Epidermal Growth Factor Receptor |
| ADMET | Absorption, Distribution, Metabolism, Excretion, and Toxicity |
| TGFβ3 | Tumor Protein p53, Transforming Growth Factor Beta 3 |
References
- Kiri, S.; Ryba, T. Cancer, metastasis, and the epigenome. Mol. Cancer 2024, 23, 154. [Google Scholar] [CrossRef] [PubMed]
- Zafar, A.; Khatoon, S.; Khan, M.J.; Abu, J.; Naeem, A. Advancements and limitations in traditional anti-cancer therapies: A comprehensive review of surgery, chemotherapy, radiation therapy, and hormonal therapy. Discov. Oncol. 2025, 16, 607. [Google Scholar] [CrossRef]
- Panda, S.S.; Biswal, B.K. The phytochemical plumbagin: Mechanism behind its “pleiotropic” nature and potential as an anticancer treatment. Arch. Toxicol. 2024, 98, 3585–3601. [Google Scholar] [CrossRef]
- Wang, P.P.; He, C.X.; Shuai, Y.Y.; Zhang, H.J.; Zhang, L.; Ao, H.; Peng, W.; Zhang, H. The Combination of Natural Products: A Promising Therapeutical Way for Management of Breast Cancers. Phytother. Res. 2025, 39, 3886–3902. [Google Scholar] [CrossRef]
- Saeed, M.E.M.; Yucer, R.; Dawood, M.; Hegazy, M.F.; Drif, A.; Ooko, E.; Kadioglu, O.; Seo, E.J.; Kamounah, F.S.; Titinchi, S.J.; et al. In Silico and In Vitro Screening of 50 Curcumin Compounds as EGFR and NF-kappaB Inhibitors. Int. J. Mol. Sci. 2022, 23, 3966. [Google Scholar] [CrossRef]
- Ameer, S.F.; Mohamed, M.Y.; Elzubair, Q.A.; Sharif, E.A.M.; Ibrahim, W.N. Curcumin as a novel therapeutic candidate for cancer: Can this natural compound revolutionize cancer treatment? Front. Oncol. 2024, 14, 1438040. [Google Scholar] [CrossRef]
- Kaur, K.; Al-Khazaleh, A.K.; Bhuyan, D.J.; Li, F.; Li, C.G. A Review of Recent Curcumin Analogues and Their Antioxidant, Anti-Inflammatory, and Anticancer Activities. Antioxidants 2024, 13, 1092. [Google Scholar] [CrossRef]
- Feng, C.; Xia, Y.; Zou, P.; Shen, M.; Hu, J.; Ying, S.; Pan, J.; Liu, Z.; Dai, X.; Zhuge, W.; et al. Curcumin analog L48H37 induces apoptosis through ROS-mediated endoplasmic reticulum stress and STAT3 pathways in human lung cancer cells. Mol. Carcinog. 2017, 56, 1765–1777. [Google Scholar] [CrossRef]
- Ardiansah, B.; Hardhani, M.R.; Putera, D.D.S.R.; Wukirsari, T.; Cahyana, A.H.; Jia, J.; Khan, M.M. Design, synthesis, and antioxidant evaluation of monocarbonyl curcumin analogues tethered 1,2,3-triazole scaffold. Case Stud. Chem. Environ. Eng. 2023, 8, 100425. [Google Scholar] [CrossRef]
- Mandalapu, D.; Saini, K.S.; Gupta, S.; Sharma, V.; Yaseen Malik, M.; Chaturvedi, S.; Bala, V.; Hamidullah; Thakur, S.; Maikhuri, J.P.; et al. Synthesis and biological evaluation of some novel triazole hybrids of curcumin mimics and their selective anticancer activity against breast and prostate cancer cell lines. Bioorg. Med. Chem. Lett. 2016, 26, 4223–4232. [Google Scholar] [CrossRef] [PubMed]
- Flint, A.L.; Hansen, D.W.; Brown, L.D.; Stewart, L.E.; Ortiz, E.; Panda, S.S. Modified Curcumins as Potential Drug Candidates for Breast Cancer: An Overview. Molecules 2022, 27, 8891. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.R.; Yesmin, T.; Prapty, A.N.; Biswash, M.A.R.; Rana, M.S.; Rashid, M.H.O. Natural Environmental Sources of Resveratrol and Its Therapeutic Role in Cancer Prevention. Aust. Herbal. Insight 2024, 7, 1–11. [Google Scholar] [CrossRef]
- Hedayati, N.; Safari, M.H.; Milasi, Y.E.; Kahkesh, S.; Farahani, N.; Khoshnazar, S.M.; Dorostgou, Z.; Alaei, E.; Alimohammadi, M.; Rahimzadeh, P.; et al. Modulation of the PI3K/Akt signaling pathway by resveratrol in cancer: Molecular mechanisms and therapeutic opportunity. Discov. Oncol. 2025, 16, 669. [Google Scholar] [CrossRef]
- Jang, J.Y.; Im, E.; Kim, N.D. Mechanism of Resveratrol-Induced Programmed Cell Death and New Drug Discovery against Cancer: A Review. Int. J. Mol. Sci. 2022, 23, 3689. [Google Scholar] [CrossRef] [PubMed]
- Chang, W.S.; Tsai, C.W.; Yang, J.S.; Hsu, Y.M.; Shih, L.C.; Chiu, H.Y.; Bau, D.T.; Tsai, F.J. Resveratrol inhibited the metastatic behaviors of cisplatin-resistant human oral cancer cells via phosphorylation of ERK/p-38 and suppression of MMP-2/9. J. Food Biochem. 2021, 45, e13666. [Google Scholar] [CrossRef]
- Schmidt, B.; Ferreira, C.; Alves Passos, C.L.; Silva, J.L.; Fialho, E. Resveratrol, Curcumin and Piperine Alter Human Glyoxalase 1 in MCF-7 Breast Cancer Cells. Int. J. Mol. Sci. 2020, 21, 5244. [Google Scholar] [CrossRef]
- Ferraz da Costa, D.C.; Campos, N.P.C.; Santos, R.A.; Guedes-da-Silva, F.H.; Martins-Dinis, M.; Zanphorlin, L.; Ramos, C.; Rangel, L.P.; Silva, J.L. Resveratrol prevents p53 aggregation in vitro and in breast cancer cells. Oncotarget 2018, 9, 29112–29122. [Google Scholar] [CrossRef]
- Horgan, X.J.; Tatum, H.; Brannan, E.; Paull, D.H.; Rhodes, L.V. Resveratrol analogues surprisingly effective against triple-negative breast cancer, independent of ERalpha. Oncol. Rep. 2019, 41, 3517–3526. [Google Scholar] [CrossRef] [PubMed]
- Fragopoulou, E.; Gkotsi, K.; Petsini, F.; Gioti, K.; Kalampaliki, A.D.; Lambrinidis, G.; Kostakis, I.K.; Tenta, R. Synthesis and Biological Evaluation of Resveratrol Methoxy Derivatives. Molecules 2023, 28, 5547. [Google Scholar] [CrossRef]
- Liu, X.; Pei, J.; Li, J.; Zhu, H.; Zheng, X.; Zhang, X.; Ruan, B.; Chen, L. Recent Advances in Resveratrol Derivatives: Structural Modifications and Biological Activities. Molecules 2025, 30, 958. [Google Scholar] [CrossRef]
- Simon, A.; Allais, D.P.; Duroux, J.L.; Basly, J.P.; Durand-Fontanier, S.; Delage, C. Inhibitory effect of curcuminoids on MCF-7 cell proliferation and structure-activity relationships. Cancer Lett. 1998, 129, 111–116. [Google Scholar] [CrossRef]
- Du, Q.; Hu, B.; An, H.M.; Shen, K.P.; Xu, L.; Deng, S.; Wei, M.M. Synergistic anticancer effects of curcumin and resveratrol in Hepa1-6 hepatocellular carcinoma cells. Oncol. Rep. 2013, 29, 1851–1858. [Google Scholar] [CrossRef]
- de Freitas Silva, M.; Coelho, L.F.; Guirelli, I.M.; Pereira, R.M.; Ferreira-Silva, G.A.; Graravelli, G.Y.; Horvath, R.O.; Caixeta, E.S.; Ionta, M.; Viegas, C. Synthetic resveratrol-curcumin hybrid derivative inhibits mitosis progression in estrogen positive MCF-7 breast cancer cells. Toxicol. Vitr. 2018, 50, 75–85. [Google Scholar] [CrossRef]
- Moreno, Q.G.; Herrera, R.A.; Yepes, A.F.; Naranjo, T.W.; Cardona, G.W. Proapoptotic Effect and Molecular Docking Analysis of Curcumin-Resveratrol Hybrids in Colorectal Cancer Chemoprevention. Molecules 2022, 27, 3486. [Google Scholar] [CrossRef]
- Brockmueller, A.; Buhrmann, C.; Moravejolahkami, A.R.; Shakibaei, M. Resveratrol and p53: How are they involved in CRC plasticity and apoptosis? J. Adv. Res. 2024, 66, 181–195. [Google Scholar] [CrossRef] [PubMed]
- Boulmokh, Y.; Belguidoum, K.; Meddour, F.; Amira-Guebailia, H. Enhanced antioxidant properties of novel curcumin derivatives: A comprehensive DFT computational study. Struct. Chem. 2023, 35, 825–839. [Google Scholar] [CrossRef]
- Lima, I.T.; Gomes, R.F.C.; Paura, E.N.C.; Provasi, P.F.; Gester, R.; Rodrigues da Cunha, A. Exploring the molecular solvatochromism, stability, reactivity, and non-linear optical response of resveratrol. J. Mol. Model. 2024, 30, 314. [Google Scholar] [CrossRef] [PubMed]
- Rudrapal, M.; Eltayeb, W.A.; Rakshit, G.; El-Arabey, A.A.; Khan, J.; Aldosari, S.M.; Alshehri, B.; Abdalla, M.J.S.r. Dual synergistic inhibition of COX and LOX by potential chemicals from Indian daily spices investigated through detailed computational studies. Sci. Rep. 2023, 13, 8656. [Google Scholar] [CrossRef]
- Ahsan, M.J.; Choudhary, K.; Ali, A.; Ali, A.; Azam, F.; Almalki, A.H.; Santali, E.Y.; Bakht, M.A.; Tahir, A.; Salahuddin. Synthesis, DFT Analyses, Antiproliferative Activity, and Molecular Docking Studies of Curcumin Analogues. Plants 2022, 11, 2835. [Google Scholar] [CrossRef]
- Adindu, E.A.; Godfrey, O.C.; Agwupuye, E.I.; Ekpong, B.O.; Agurokpon, D.C.; Ogbodo, S.E.; Benjamin, I.; Louis, H. Structural analysis, reactivity descriptors (HOMO-LUMO, ELF, NBO), effect of polar (DMSO, EtOH, H2O) solvation, and libido-enhancing potential of resveratrol by molecular docking. Chem. Phys. Impact 2023, 7, 100296. [Google Scholar] [CrossRef]
- Vennelakanti, V.; Qi, H.W.; Mehmood, R.; Kulik, H.J. When are two hydrogen bonds better than one? Accurate first-principles models explain the balance of hydrogen bond donors and acceptors found in proteins. Chem. Sci. 2021, 12, 1147–1162. [Google Scholar] [CrossRef]
- Rasouli, A.; Jamali, Y.; Tajkhorshid, E.; Bavi, O.; Pishkenari, H.N. Mechanical properties of ester- and ether-DPhPC bilayers: A molecular dynamics study. J. Mech. Behav. Biomed. Mater. 2021, 117, 104386. [Google Scholar] [CrossRef] [PubMed]
- Elangovan, N.; Arumugam, N.; Santhamoorthy, M.; Thomas, R. Intermolecular Forces in Bioactive Resveratrol Complexes with Alcohols: A Study of Stability and Electronic Structure. J. Phys. Chem. B 2025, 129, 1966–1975. [Google Scholar] [CrossRef] [PubMed]
- Hazarika, R.; Kalita, B. Elucidating the therapeutic activity of selective curcumin analogues: DFT-based reactivity analysis. Struct. Chem. 2021, 32, 1701–1715. [Google Scholar] [CrossRef]
- Nandhini, M.; Pitchumani Violet Mary, C.; Gopinath, S.; Vijayakumar, S. Structure based interaction and molecular dynamics studies of cysteine protease Cathepsin B against curcumin and resveratrol. J. Biomol. Struct. Dyn. 2024, 42, 1–11. [Google Scholar] [CrossRef]
- Hernandez, C.; Moreno, G.; Herrera, R.A.; Cardona, G.W. New Hybrids Based on Curcumin and Resveratrol: Synthesis, Cytotoxicity and Antiproliferative Activity against Colorectal Cancer Cells. Molecules 2021, 26, 2661. [Google Scholar] [CrossRef]
- de Souza, F.N.; de Lima, H.B.; de Souza, L.R.; Oliveira, G.S.; de Paula da Silva, C.H.; Pereira, A.C.s.M.; da Silva Hage-Melim, L.I. Design of Multitarget Natural Products Analogs with Potential Anti-Alzheimer’s Activity. Curr. Comput.-Aided Drug Des. 2022, 18, 120–149. [Google Scholar]
- Wang, L.; Sun, C. Regulating the antioxidant activity of resveratrol through photoisomerization in different solvents: Insights from experiment and DFT calculation. J. Mol. Struct. 2025, 1340, 142603. [Google Scholar] [CrossRef]
- Prabaningdyah, N.K.; Riyanto, S.; Rohman, A. Application of FTIR spectroscopy and multivariate calibration for analysis of curcuminoid in syrup formulation. J. Appl. Pharm. Sci. 2018, 8, 172–179. [Google Scholar] [CrossRef][Green Version]
- Vázquez, B.E.R.; Rodríguez-Beas, C.; Iñiguez-Palomares, R.A.; Santacruz-Ortega, H.; Mendoza-Cruz, R.; Bazán-Díaz, L.S.; Soberanes, Y.; Rodríguez-León, E.; Navarro, R.E. Spectroscopic analysis and nuclear magnetic resonance for silver nanoparticles synthesized with trans-resveratrol and cis-resveratrol. Colloid. Polym. Sci. 2022, 300, 465–475. [Google Scholar] [CrossRef]
- de Deus, W.F.; de Franca, B.M.; Forero, J.S.B.; Granato, A.E.C.; Ulrich, H.; Doria, A.; Amaral, M.M.; Slabon, A.; Rodrigues, B.V.M. Curcuminoid-Tailored Interfacial Free Energy of Hydrophobic Fibers for Enhanced Biological Properties. ACS Appl. Mater. Interfaces 2021, 13, 24493–24504. [Google Scholar] [CrossRef]
- Liu, J.; Liu, X.; Cui, X.; Qin, J.; Wu, M.; Fu, L.; Shi, M.; Wang, D.; Liang, L. Investigation on the properties and structures of resveratrol-derived epoxy thermosets cured with an active ester. Polym. Chem. 2023, 14, 1665–1679. [Google Scholar] [CrossRef]
- Pistone, A.; de Gaetano, A.; Piperopoulos, E.; Abate, C. Effect of Sodium Hydroxide and Tripolyphosphate on Curcumin Release from Chitosan-Based Macroparticles. Materials 2023, 16, 5850. [Google Scholar] [CrossRef]
- Dai, F.; Zhuang, Q.; Huang, G.; Deng, H.; Zhang, X. Infrared Spectrum Characteristics and Quantification of OH Groups in Coal. ACS Omega 2023, 8, 17064–17076. [Google Scholar] [CrossRef]
- Szelag, M.; Czerwoniec, A.; Wesoly, J.; Bluyssen, H.A. Identification of STAT1 and STAT3 specific inhibitors using comparative virtual screening and docking validation. PLoS ONE 2015, 10, e0116688. [Google Scholar] [CrossRef]
- Micale, N.; Molonia, M.S.; Citarella, A.; Cimino, F.; Saija, A.; Cristani, M.; Speciale, A. Natural Product-Based Hybrids as Potential Candidates for the Treatment of Cancer: Focus on Curcumin and Resveratrol. Molecules 2021, 26, 4665. [Google Scholar] [CrossRef]
- Moreno-Quintero, G.; Castrillon-Lopez, W.; Herrera-Ramirez, A.; Yepes-Perez, A.F.; Quintero-Saumeth, J.; Cardona-Galeano, W. Synthesis and Chemopreventive Potential of 5-FU/Genistein Hybrids on Colorectal Cancer Cells. Pharmaceuticals 2022, 15, 1299. [Google Scholar] [CrossRef]
- Alkhathami, A.G.; Alshahrani, M.Y.; Alshehri, S.A.; Nasir, N.; Wahab, S. Curcumin as a potential inhibitor of TGFbeta3 computational insights for breast cancer therapy. Sci. Rep. 2025, 15, 2871. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; You, L.; Nepovimova, E.; Psotka, M.; Malinak, D.; Valko, M.; Sivak, L.; Korabecny, J.; Heger, Z.; Adam, V.; et al. Inhibitors of phosphoinositide 3-kinase (PI3K) and phosphoinositide 3-kinase-related protein kinase family (PIKK). J. Enzym. Inhib. Med. Chem. 2023, 38, 2237209. [Google Scholar] [CrossRef] [PubMed]
- Baby, K.; Maity, S.; Mehta, C.H.; Nayak, U.Y.; Shenoy, G.G.; Pai, K.S.R.; Harikumar, K.B.; Nayak, Y. Computational drug repurposing of Akt-1 allosteric inhibitors for non-small cell lung cancer. Sci. Rep. 2023, 13, 7947. [Google Scholar] [CrossRef] [PubMed]
- Santhoshkumar, R.; Yusuf, A. In silico structural modeling and analysis of physicochemical properties of curcumin synthase (CURS1, CURS2, and CURS3) proteins of Curcuma longa. J. Genet. Eng. Biotechnol. 2020, 18, 24. [Google Scholar] [CrossRef]
- Sahu, R.K.; Verma, V.V.; Kumar, A.; Tandon, S.; Chandra Das, B.; Hedau, S.T. In silico prediction and interaction of resveratrol on methyl-CpG binding proteins by molecular docking and MD simulations study. RSC Adv. 2022, 12, 11493–11504. [Google Scholar] [CrossRef]
- Raschka, S.; Wolf, A.J.; Bemister-Buffington, J.; Kuhn, L.A. Protein–ligand interfaces are polarized: Discovery of a strong trend for intermolecular hydrogen bonds to favor donors on the protein side with implications for predicting and designing ligand complexes. J. Comput.-Aided Mol. Des. 2018, 32, 511–528. [Google Scholar] [CrossRef]
- Barratt, E.; Bingham, R.J.; Warner, D.J.; Laughton, C.A.; Phillips, S.E.; Homans, S.W. Van der Waals interactions dominate ligand-protein association in a protein binding site occluded from solvent water. J. Am. Chem. Soc. 2005, 127, 11827–11834. [Google Scholar] [CrossRef]
- Desantis, F.; Miotto, M.; Di Rienzo, L.; Milanetti, E.; Ruocco, G. Spatial organization of hydrophobic and charged residues affects protein thermal stability and binding affinity. Sci. Rep. 2022, 12, 12087. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Hong, W.; Wang, X.; Xu, C.; Jiang, Y.; Du, J.; Miao, D.; Xiao, G. Preparation and structure–property relationship of flexible aramid films with enhanced strength by introducing asymmetric and symmetric aromatic ether bond structures. Polym. Chem. 2023, 14, 3906–3915. [Google Scholar] [CrossRef]
- Maurer, M.; Oostenbrink, C. Water in protein hydration and ligand recognition. J. Mol. Recognit. 2019, 32, e2810. [Google Scholar] [CrossRef] [PubMed]
- Marchenko, E.I.; Fateev, S.A.; Ordinartsev, A.A.; Ivlev, P.A.; Goodilin, E.A.; Tarasov, A.B. Relative distance from the center of mass—A new structural descriptor linking the structure of organic cations with inorganic framework distortions in layered hybrid halide perovskites. Mendeleev Commun. 2022, 32, 315–316. [Google Scholar] [CrossRef]
- Qin, H.; Lim, L.; Song, J. Protein dynamics at Eph receptor-ligand interfaces as revealed by crystallography, NMR and MD simulations. BMC Biophys. 2012, 5, 2. [Google Scholar] [CrossRef]
- Liu, K.; Kokubo, H. Exploring the Stability of Ligand Binding Modes to Proteins by Molecular Dynamics Simulations: A Cross-docking Study. J. Chem. Inf. Model. 2017, 57, 2514–2522. [Google Scholar] [CrossRef]
- Kalin, S.; Comert Onder, F. Discovery of potential RSK1 inhibitors for cancer therapy using virtual screening, molecular docking, molecular dynamics simulation, and MM/GBSA calculations. J. Biomol. Struct. Dyn. 2025, 43, 1424–1444. [Google Scholar] [CrossRef]
- Noorbakhsh Varnosfaderani, S.M.; Sadat Haeri, M.; Arian, A.S.; Yousefi Rad, A.; Yazdanpour, M.; Mojahedian, F.; Yaghoubzad-Maleki, M.; Zalpoor, H.; Baziyar, P.; Nabi-Afjadi, M. Fighting against amyotrophic lateral sclerosis (ALS) with flavonoids: A computational approach to inhibit superoxide dismutase (SOD1) mutant aggregation. J. Biomol. Struct. Dyn. 2025, 43, 419–436. [Google Scholar] [CrossRef]
- Matricon, P.; Suresh, R.R.; Gao, Z.G.; Panel, N.; Jacobson, K.A.; Carlsson, J. Ligand design by targeting a binding site water. Chem. Sci. 2020, 12, 960–968. [Google Scholar] [CrossRef]
- Grabarczyk, M.; Justynska, W.; Czpakowska, J.; Smolinska, E.; Bielenin, A.; Glabinski, A.; Szpakowski, P. Role of Plant Phytochemicals: Resveratrol, Curcumin, Luteolin and Quercetin in Demyelination, Neurodegeneration, and Epilepsy. Antioxidant 2024, 13, 1363. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Jiao, Y.; Yang, M.; Wu, L.; Long, G.; Hu, W. Network Pharmacology, Molecular Docking and Molecular Dynamics to Explore the Potential Immunomodulatory Mechanisms of Deer Antler. Int. J. Mol. Sci. 2023, 24, 10370. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Warthaka, M.; Yan, C.; Kaoud, T.S.; Piserchio, A.; Ghose, R.; Ren, P.; Dalby, K.N. A model of a MAPK*substrate complex in an active conformation: A computational and experimental approach. PLoS ONE 2011, 6, e18594. [Google Scholar] [CrossRef] [PubMed]
- Weiss, R.G.; Chudoba, R.; Setny, P.; Dzubiella, J. Affinity, kinetics, and pathways of anisotropic ligands binding to hydrophobic model pockets. J. Chem. Phys. 2018, 149, 094902. [Google Scholar] [CrossRef]
- Bhandari, S.V.; Kuthe, P.V.; Patil, S.M.; Nagras, O.G.; Sarkate, A.P.; Chaudhari, S.Y.; Surve, S.V. Molecular Docking, Pharmacokinetic and Molecular Simulation Analysis of Novel Mono-Carbonyl Curcumin Analogs as L858R/T790M/C797S Mutant EGFR Inhibitors. Chem. Biodivers. 2023, 20, e202301081. [Google Scholar] [CrossRef]
- Asl, F.D.; Altememy, D.; Khosravian, P.; Rezaee, M.; Saffari-Chaleshtori, J. Evaluation Of Curcumin Effects On Bad, Bak, And Bim: A Molecular Dynamics Simulation Study. J. Pharm. Negat. Results 2022, 13, 8–14. [Google Scholar] [CrossRef]
- Novak, J.; Tseilikman, V.E.; Tseilikman, O.B.; Lazuko, S.S.; Belyeva, L.E.; Rahmani, A.; Fedotova, J. Can Resveratrol Influence the Activity of 11beta-Hydroxysteroid Dehydrogenase Type 1? A Combined In Silico and In Vivo Study. Pharmaceuticals 2023, 16, 251. [Google Scholar] [CrossRef]
- Silalahi, T.; Alwi, I.; Suyatna, F.; Sartika, K.D.; Suwita, C.S. Curcumin’s Antioxidant Properties in Stable Coronary Artery Disease Patients Undergoing Percutaneous Coronary Intervention: A Randomized Controlled Trial. Indones. Biomed. J. 2022, 14, 66–73. [Google Scholar] [CrossRef]
- Shahidi, F.; Dissanayaka, C.S. Phenolic-protein interactions: Insight from in-silico analyses—A review. Food Prod. Process. Nutr. 2023, 5, 2. [Google Scholar] [CrossRef]
- Sowa-Kasprzak, K.; Toton, E.; Kujawski, J.; Olender, D.; Lisiak, N.; Zaprutko, L.; Rubis, B.; Kaczmarek, M.; Pawelczyk, A. Synthesis, Cytotoxicity and Molecular Docking of New Hybrid Compounds by Combination of Curcumin with Oleanolic Acid. Biomedicines 2023, 11, 1506. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Xu, Y.; Lan, J.; Fan, S.; Huang, J.; Xu, F. Structural Achievability of an NH-pi Interaction between Gln and Phe in a Crystal Structure of a Collagen-like Peptide. Biomolecules 2022, 12, 1433. [Google Scholar] [CrossRef]
- Rout, M.; Mishra, S.; Dey, S.; Singh, M.K.; Dehury, B.; Pati, S. Exploiting the potential of natural polyphenols as antivirals against monkeypox envelope protein F13 using machine learning and all-atoms MD simulations. Comput. Biol. Med. 2023, 162, 107116. [Google Scholar] [CrossRef]
- Akash, S.; Islam, M.R.; Bhuiyan, A.A.; Islam, M.N.; Bayil, I.; Saleem, R.M.; Albadrani, G.M.; Al-Ghadi, M.Q.; Abdel-Daim, M.M. In silico evaluation of anti-colorectal cancer inhibitors by Resveratrol derivatives targeting Armadillo repeats domain of APC: Molecular docking and molecular dynamics simulation. Front. Oncol. 2024, 14, 1360745. [Google Scholar] [CrossRef] [PubMed]
- Akkus, E.; Tayfuroglu, O.; Yildiz, M.; Kocak, A. Accurate Binding Free Energy Method from End-State MD Simulations. J. Chem. Inf. Model. 2022, 62, 4095–4106. [Google Scholar] [CrossRef]
- Wankowicz, S.A.; de Oliveira, S.H.; Hogan, D.W.; van den Bedem, H.; Fraser, J.S. Ligand binding remodels protein side-chain conformational heterogeneity. Elife 2022, 11, e74114. [Google Scholar] [CrossRef]
- Amezian, D.; Nauen, R.; Van Leeuwen, T. The role of ATP-binding cassette transporters in arthropod pesticide toxicity and resistance. Curr. Opin. Insect Sci. 2024, 63, 101200. [Google Scholar] [CrossRef]
- Ma, L.; Kuang, Z.; Zhang, H.; Wan, Y.; Guo, Y.; Xia, A.; Li, Y. Modulating the Charge Transfer Coupling in Boron-Dipyrromethene Homodimers by pi-Bridge Units. J. Phys. Chem. B 2025, 129, 3428–3435. [Google Scholar] [CrossRef]
- Yasir, M.; Park, J.; Han, E.T.; Han, J.H.; Park, W.S.; Chun, W. Investigating the Inhibitory Potential of Flavonoids against Aldose Reductase: Insights from Molecular Docking, Dynamics Simulations, and gmx_MMPBSA Analysis. Curr. Issues Mol. Biol. 2024, 46, 11503–11518. [Google Scholar] [CrossRef]
- Roy, L.D.; Kumar, J.; Krishnamurthy, G.; Gour, P.; Arland, S.E.; Rahman, N. Phytogenic One-pot Synthesis and Spectroscopic Characterization of Novel Mono Benzylated Resveratrol Hybrid Molecule Using Extracted Resveratrol from Green Grape Peels: In Silico ADMET Study and In Vitro Antitumor Activities Against Breast Cancer Cells. Curr. Bioact. Compd. 2023, 19, 82–92. [Google Scholar] [CrossRef]
- Diamanti, A.; Ganase, Z.; Grant, E.; Armstrong, A.; Piccione, P.M.; Rea, A.M.; Richardson, J.; Galindo, A.; Adjiman, C.S. Mechanism, kinetics and selectivity of a Williamson ether synthesis: Elucidation under different reaction conditions. React. Chem. Eng. 2021, 6, 1195–1211. [Google Scholar] [CrossRef]
- Boulaamane, Y.; Touati, I.; Qamar, I.; Ahmad, I.; Patel, H.; Chandra, A.; Britel, M.R.; Maurady, A. In silico Discovery of Dual Ligands Targeting MAO-B and AA2AR from African Natural Products Using Pharmacophore Modelling, Molecular Docking, and Molecular Dynamics Simulations. Chem. Afr. 2024, 7, 4337–4359. [Google Scholar] [CrossRef]
- Sumathi, S.; Karthik, N.; Jeyavijayan, S. Computational and Spectroscopic Insights Into 4-Methoxychalcone as a Potential Acetylcholinesterase Inhibitor: A DFT and Molecular Docking Approach. Int. J. Quantum Chem. 2025, 125, e70092. [Google Scholar] [CrossRef]
- Turner, N.C.; Im, S.A.; Saura, C.; Juric, D.; Loibl, S.; Kalinsky, K.; Schmid, P.; Loi, S.; Sunpaweravong, P.; Musolino, A.; et al. Inavolisib-Based Therapy in PIK3CA-Mutated Advanced Breast Cancer. N. Eng. J. Med. 2024, 391, 1584–1596. [Google Scholar] [CrossRef]
- Li, W.; Luo, R.; Liu, Z.; Li, X.; Zhang, C.; Huang, J.; Wang, Z.; Chen, J.; Ding, H.; Zhou, X.; et al. Anti-inflammatory effects of resveratrol in treating interstitial cystitis/bladder pain syndrome: A multi-faceted approach integrating network pharmacology, molecular docking, and experimental validation. Mol. Divers. 2025, 29, 2489–2497. [Google Scholar] [CrossRef]
- Zhang, Y.; Forli, S.; Omelchenko, A.; Sanner, M.F. AutoGridFR: Improvements on AutoDock Affinity Maps and Associated Software Tools. J. Comput. Chem. 2019, 40, 2882–2886. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Bahadar, N.; Bahadar, S.; Sajid, A.; Wahid, M.; Ali, G.; Alghamdi, A.; Zada, H.; Khan, T.; Ullah, S.; Sun, Q. Epigallocatechin gallate and curcumin inhibit Bcl-2: A pharmacophore and docking based approach against cancer. Breast Cancer Res. 2024, 26, 114. [Google Scholar] [CrossRef]
- Baba, H.; Bouqdayr, M.; Jouimyi, M.R.; Elmessaoudi-Idrissi, M.; Kettani, A. A Simple Overview for Proteins Molecular Dynamics Simulations Using GROMACS. In International Conference on Advanced Intelligent Systems for Sustainable Development; Springer Nature: Cham, Switzerland, 2023; pp. 355–363. [Google Scholar]









| Hybrid Molecules | HOMO | LUMO | GAP | χ | η | σ | μ | ω |
|---|---|---|---|---|---|---|---|---|
| ELRC-LC | −5.5786 | −2.3468 | 3.2318 | 3.9627 | 1.6159 | 0.6189 | −3.9627 | 4.8589 |
| ELRC-SC | −5.6607 | −2.0453 | 3.6154 | 3.8530 | 1.8077 | 0.5532 | −3.8530 | 4.1062 |
| EtLRC | −5.1430 | −2.3158 | 2.8272 | 3.7294 | 1.4136 | 0.7074 | −3.7294 | 4.9195 |
| Receptors | PDB ID | Grid Center Coordinates (Å) | Grid Box Dimensions (Å) * | Key Binding Site Residues |
|---|---|---|---|---|
| AKT1 | 8UW9 | X: 10.950, Y: 10.531, Z: −32.793 | 30 × 30 × 30 | Thr211 |
| MAPK1 | 8ZJV | X: −26.719, Y: 17.156, Z: 4.932 | 30 × 30 × 30 | Asp106, Met108, Asp111, Ser153, Asp167 |
| STAT3 | 6NUQ | X: 10.076, Y: 51.043, Z: 5.082 | 30 × 30 × 30 | Arg609, Ser611, Glu612, Ser613, Ser636, Glu638, Gln644, Tyr657 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sazlı, N.; Karataş, D. In Silico Design and Computational Elucidation of Hypothetical Resveratrol–Curcumin Hybrids as Potential Cancer Pathway Modulators. Pharmaceuticals 2025, 18, 1473. https://doi.org/10.3390/ph18101473
Sazlı N, Karataş D. In Silico Design and Computational Elucidation of Hypothetical Resveratrol–Curcumin Hybrids as Potential Cancer Pathway Modulators. Pharmaceuticals. 2025; 18(10):1473. https://doi.org/10.3390/ph18101473
Chicago/Turabian StyleSazlı, Nil, and Deniz Karataş. 2025. "In Silico Design and Computational Elucidation of Hypothetical Resveratrol–Curcumin Hybrids as Potential Cancer Pathway Modulators" Pharmaceuticals 18, no. 10: 1473. https://doi.org/10.3390/ph18101473
APA StyleSazlı, N., & Karataş, D. (2025). In Silico Design and Computational Elucidation of Hypothetical Resveratrol–Curcumin Hybrids as Potential Cancer Pathway Modulators. Pharmaceuticals, 18(10), 1473. https://doi.org/10.3390/ph18101473






