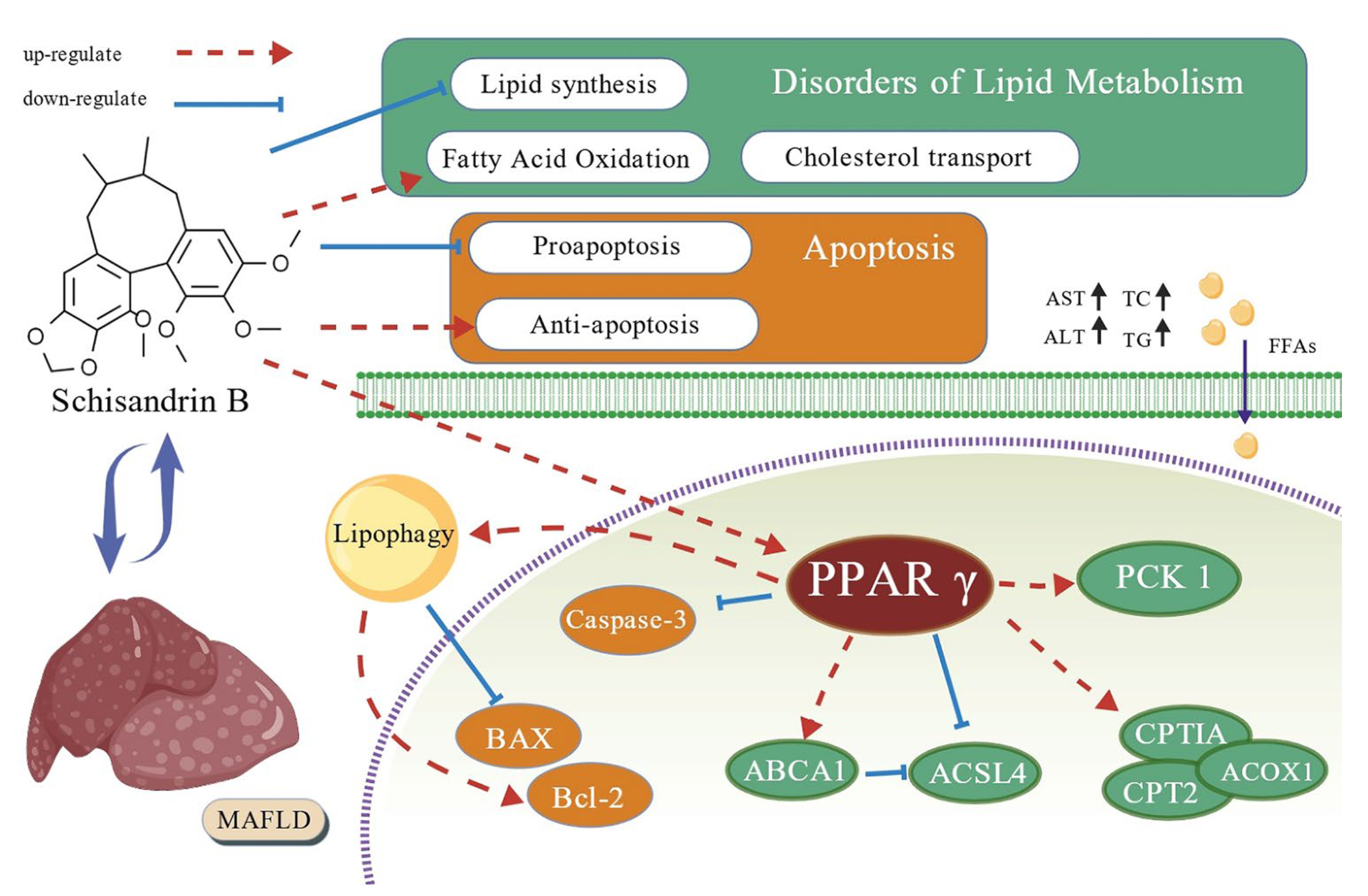
Schisandrin B Alleviates Lipid Metabolism Disorders and Apoptosis of MAFLD via Modulation of PPARγ-PCK1 and Caspase-3 Signaling Pathways
Abstract
1. Introduction
2. Results
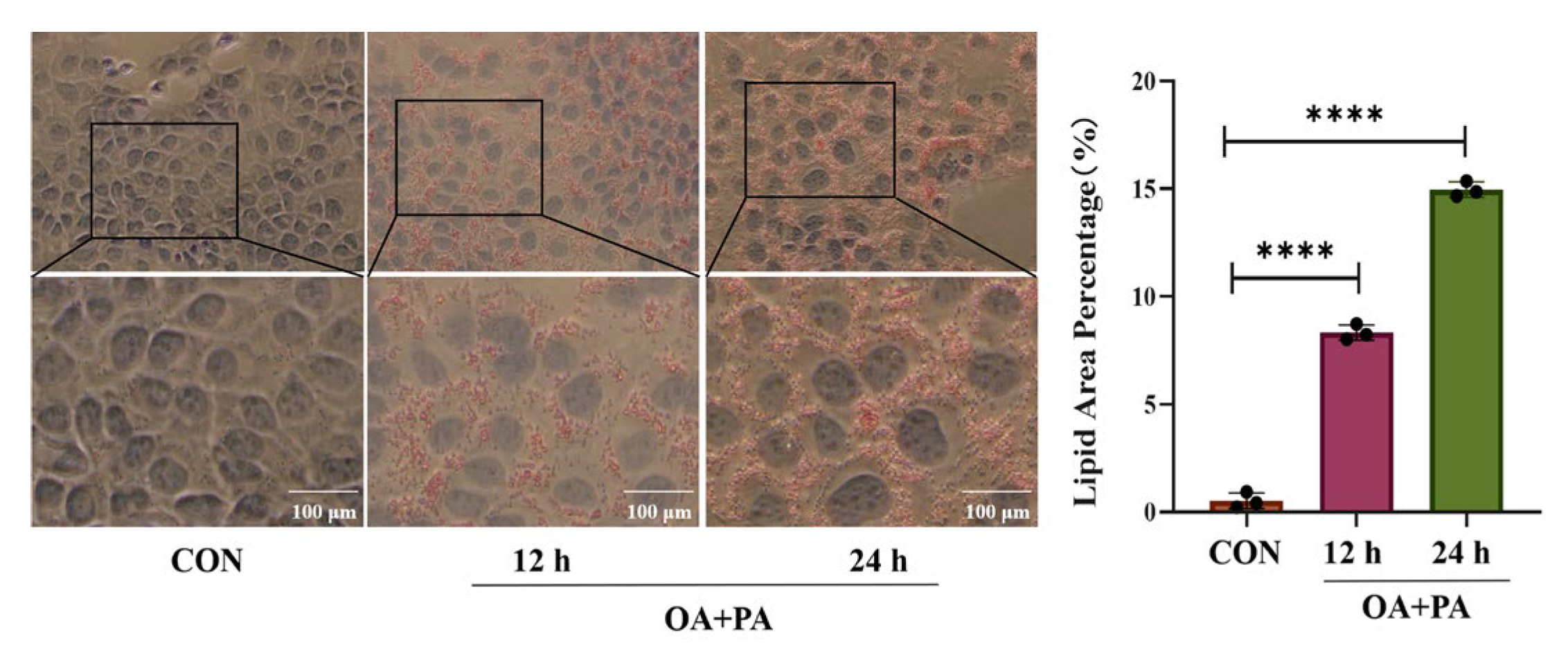
2.1. Effects of Different Induction Times on MAFLD Cell Model
2.2. Effects of Sch B on AML-12 Cell Activities
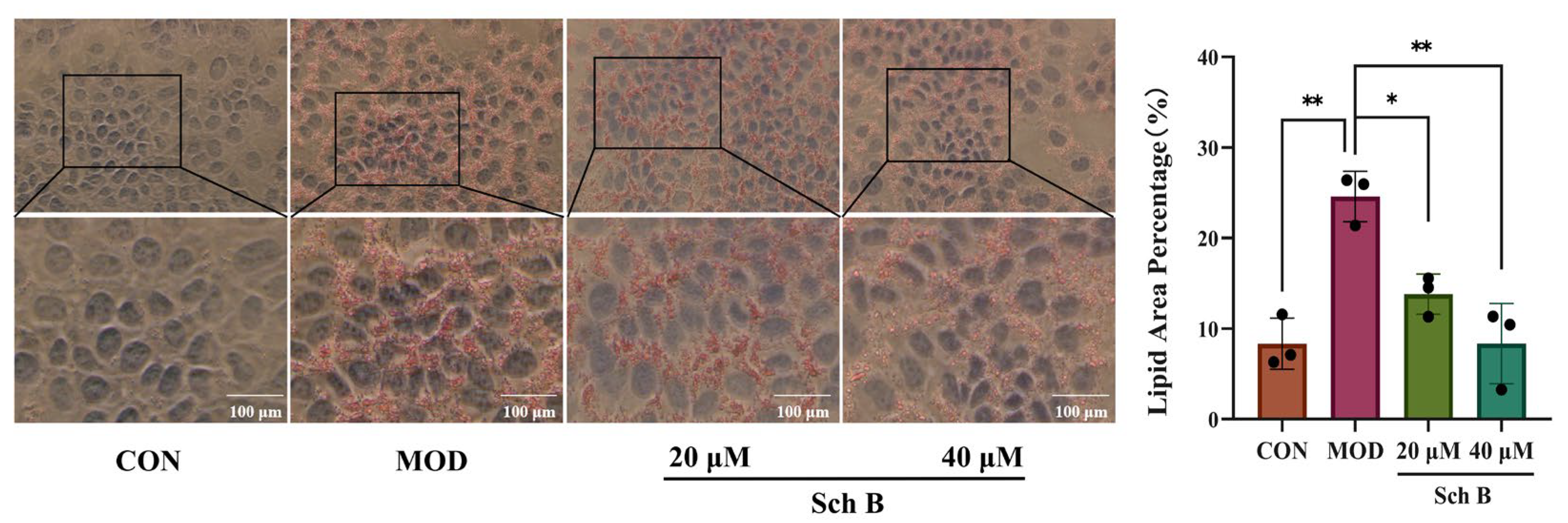
2.3. Histomorphological Results of AML-12 Cells by Oil Red Staining
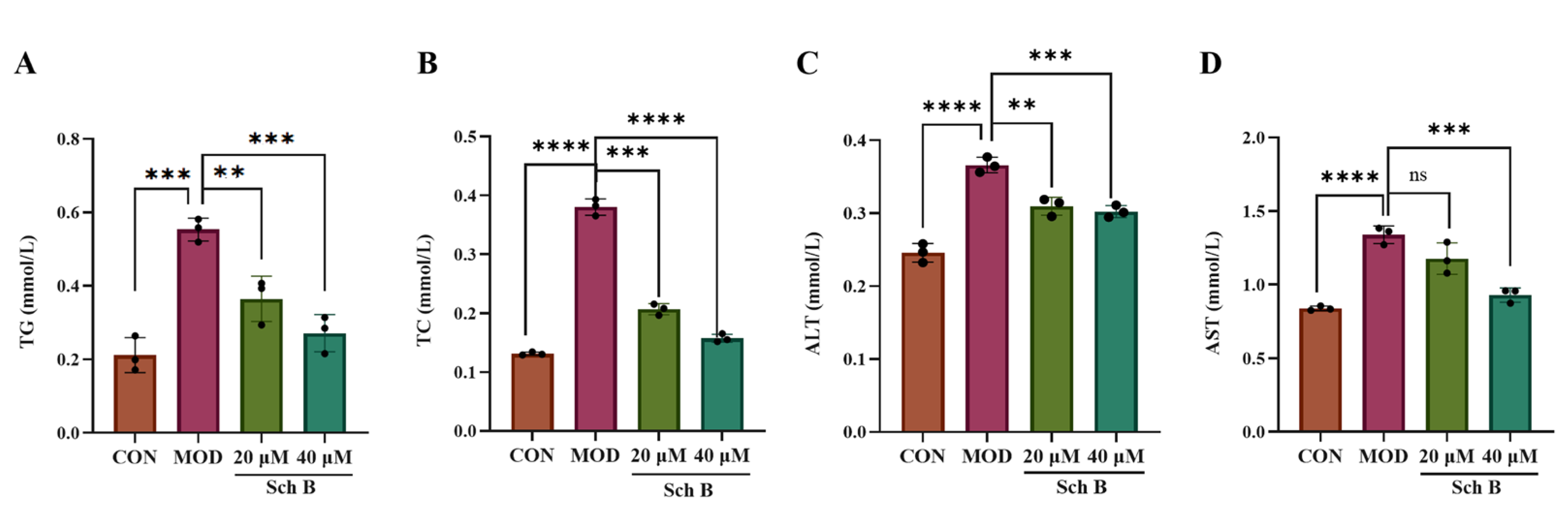
2.4. Effects of Sch B on TG, TC, ALT, and AST Levels in AML Cells
2.5. Network Pharmacology Prediction
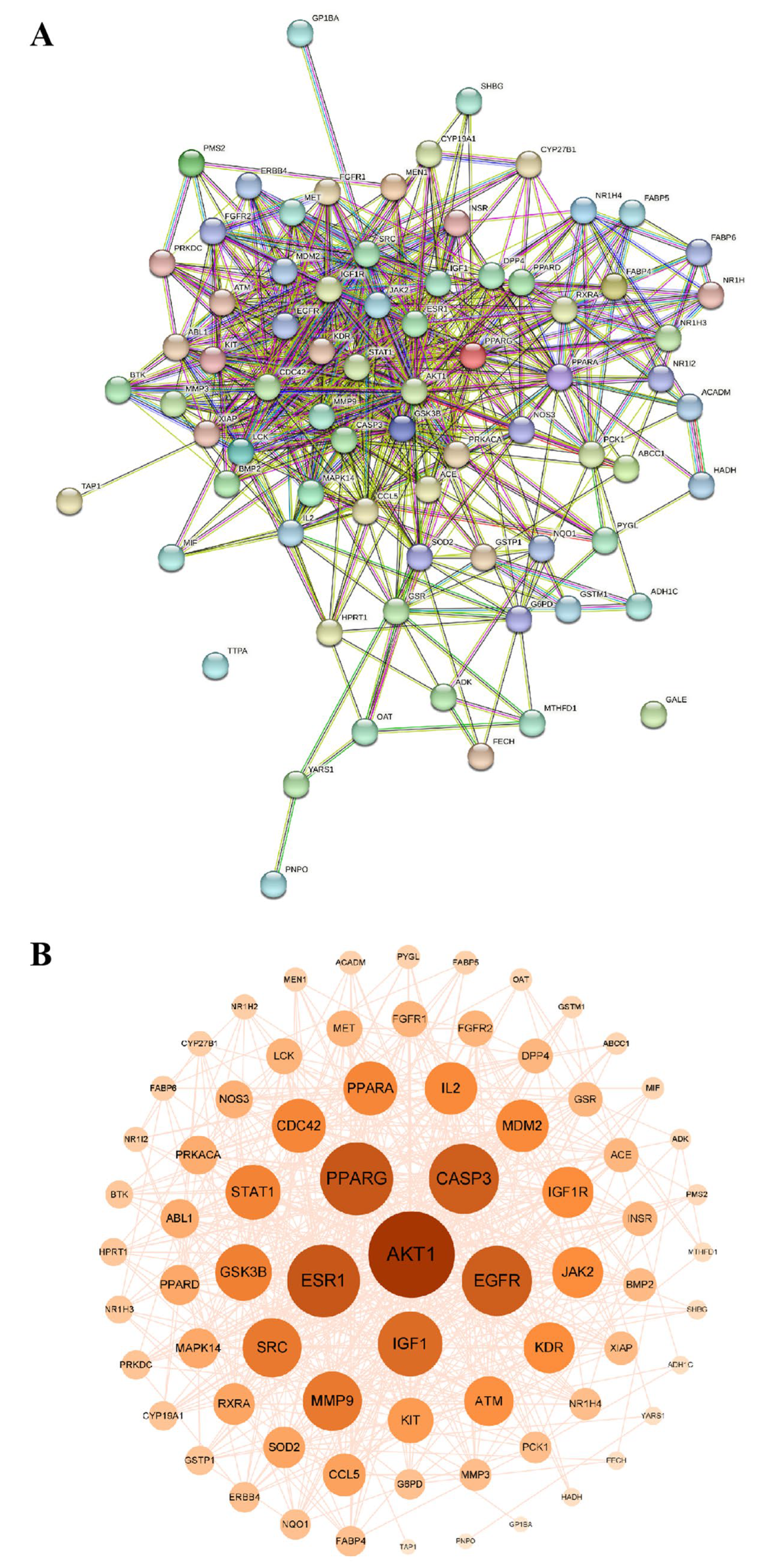
2.5.1. Acquisition of Intersecting Targets
2.5.2. Construction and Analysis of Protein–Protein Interaction Network (PPI)
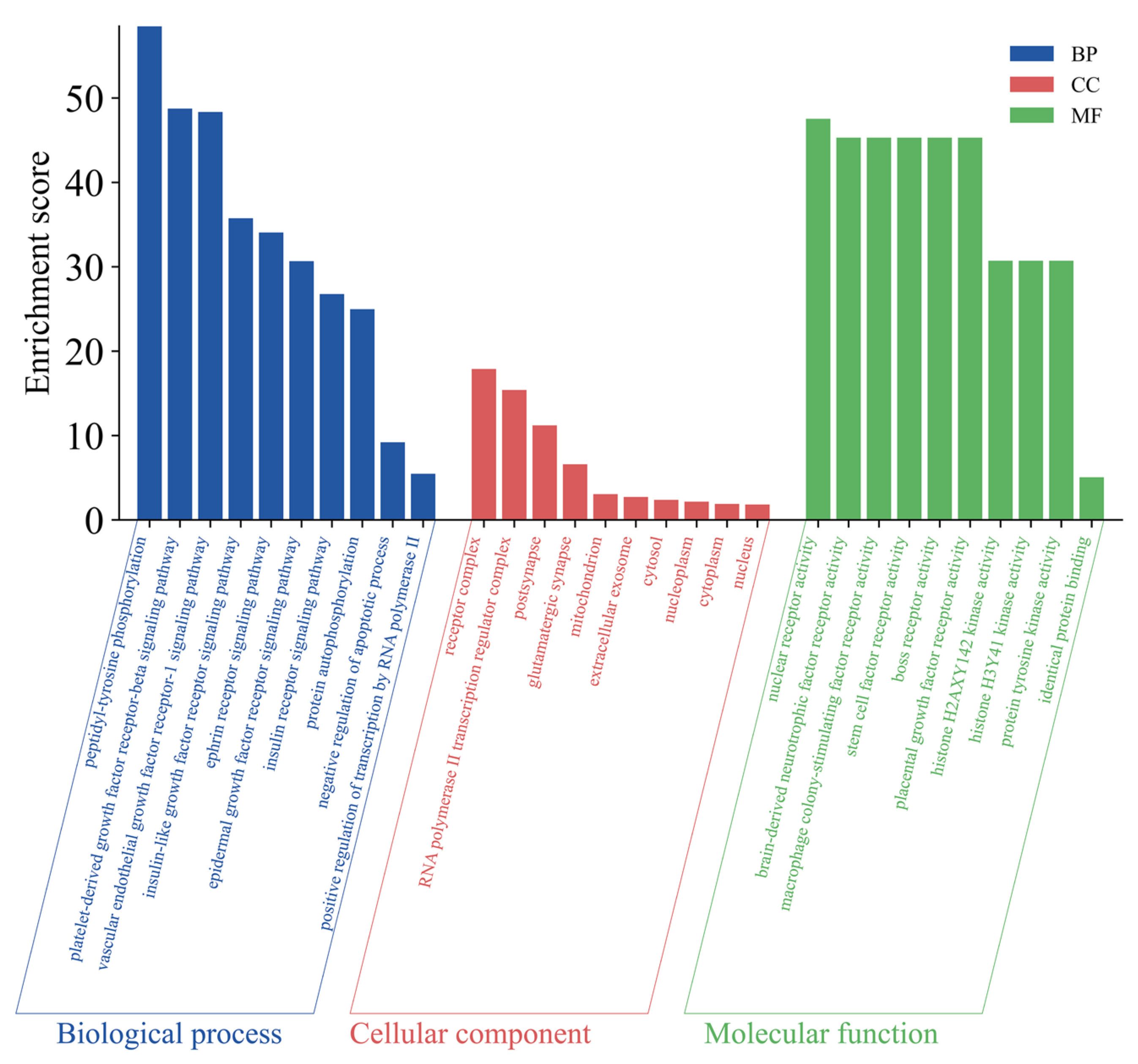
2.5.3. Gene Ontology (GO) Enrichment Analysis
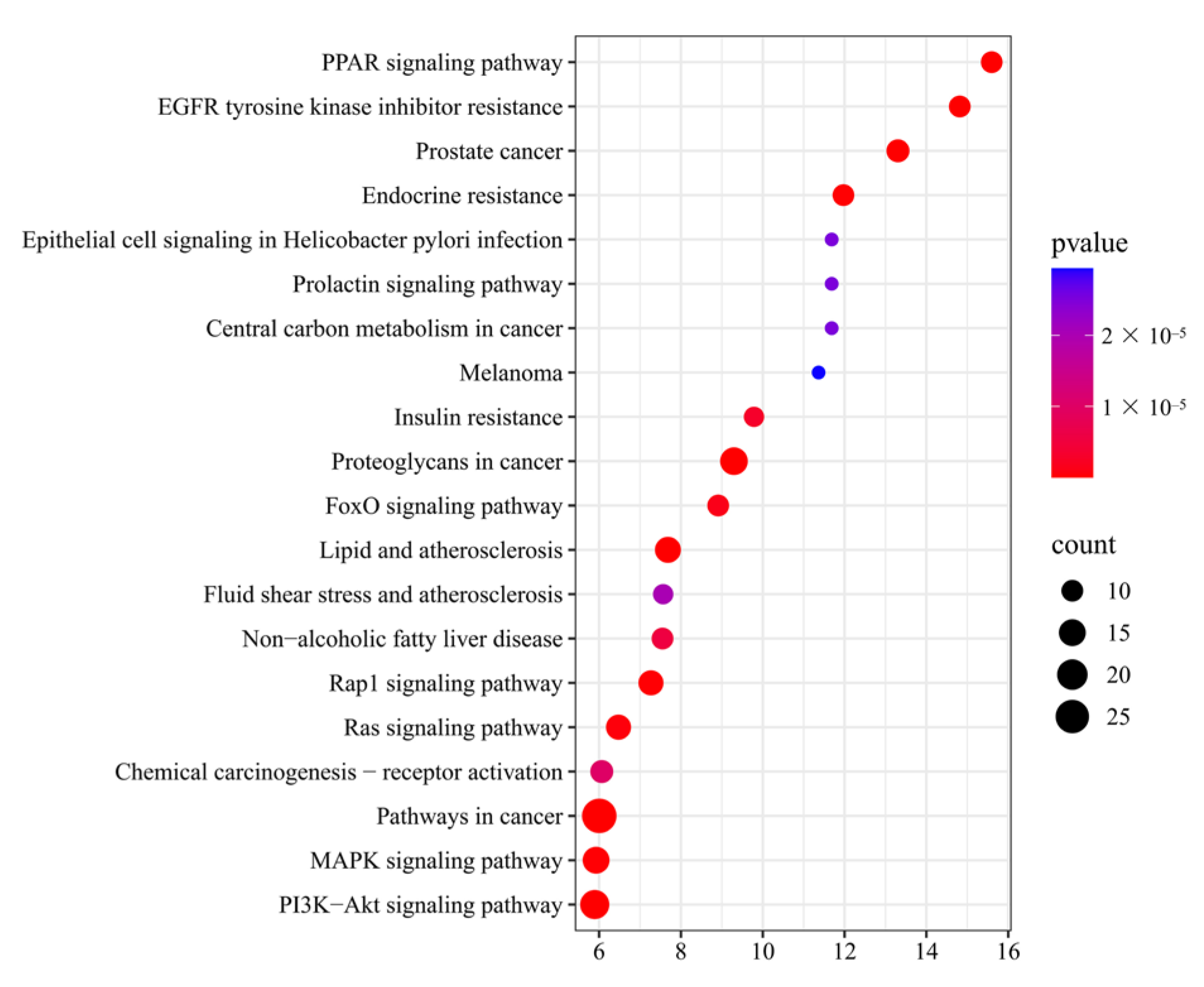
2.5.4. Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Enrichment Analysis
2.6. Molecular Docking Simulation Results
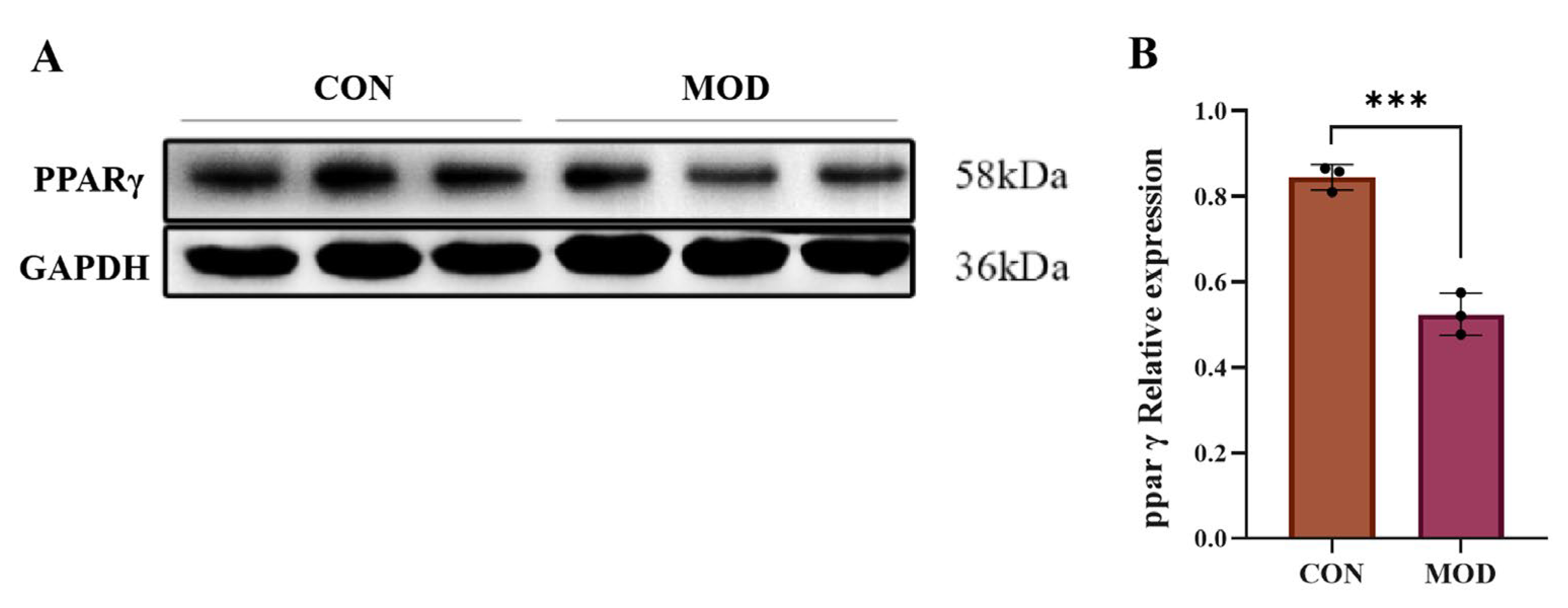
2.7. Expression of PPARγ Protein in MAFLD Cell Model
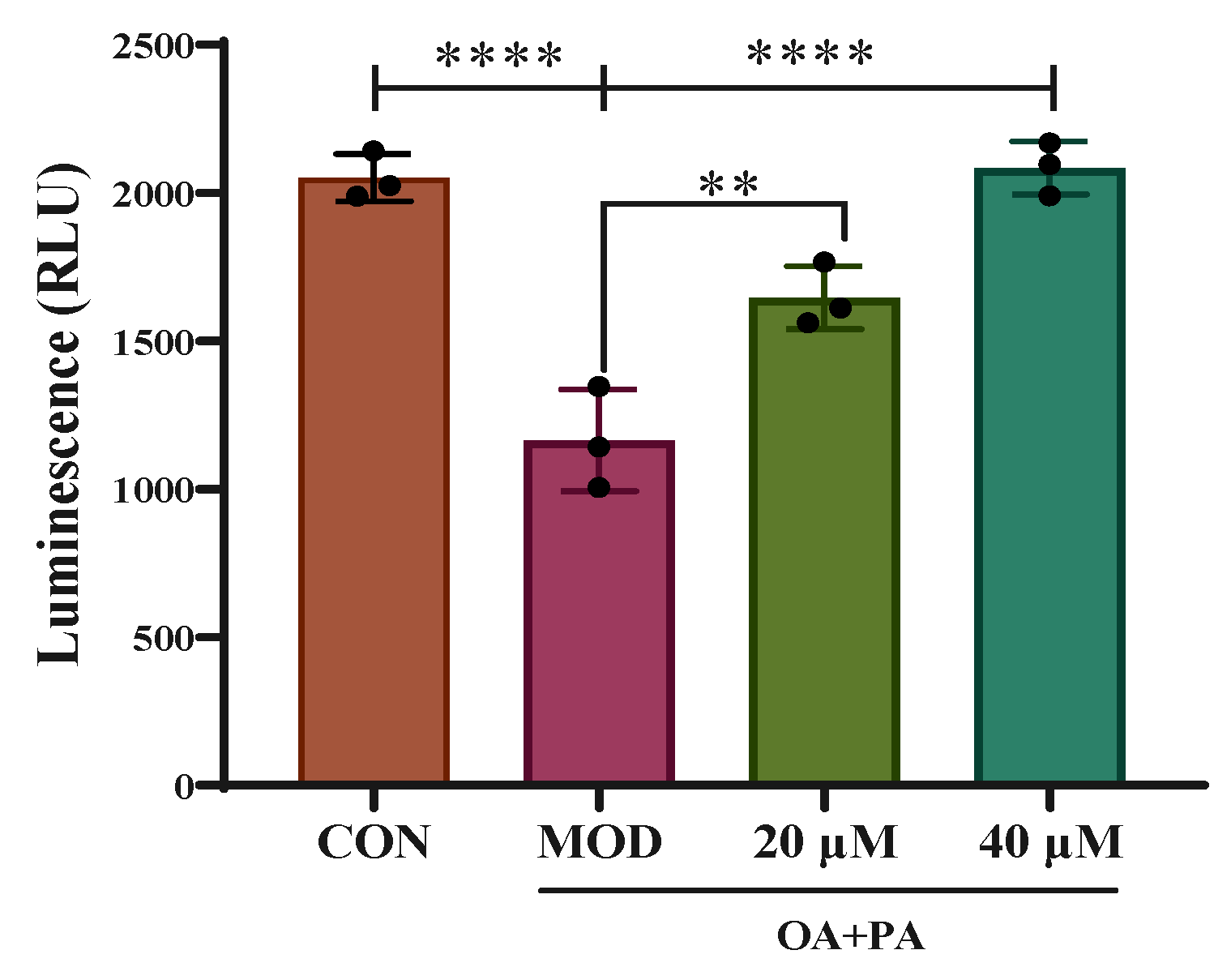
2.8. Detection of PPAR γ Luciferase Reporter Gene
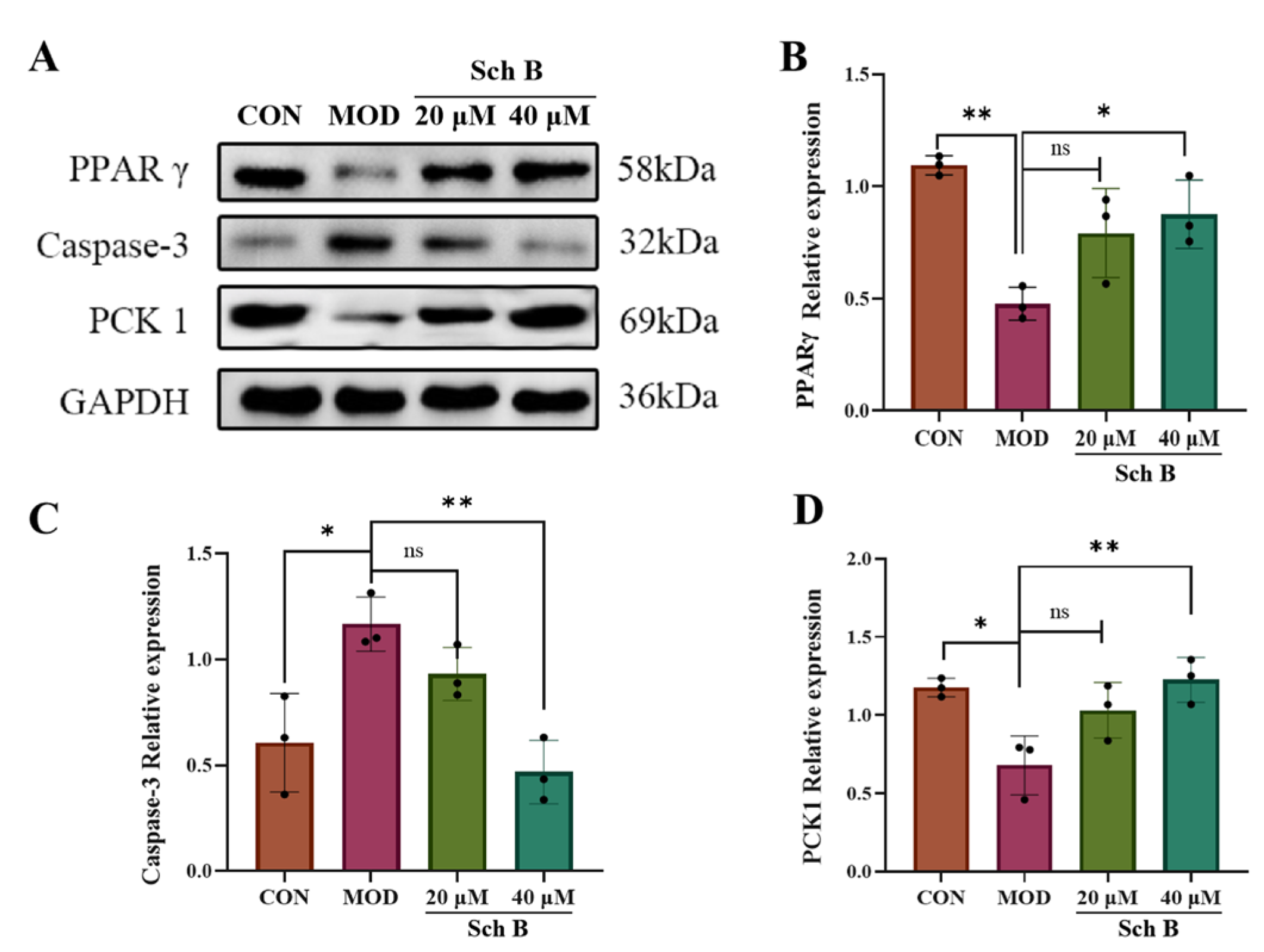
2.9. Effects of Sch B on Expression of PPARγ Pathway-Related Proteins
2.10. Effects of Sch B on the Expression of Lipid Metabolism-Related Proteins
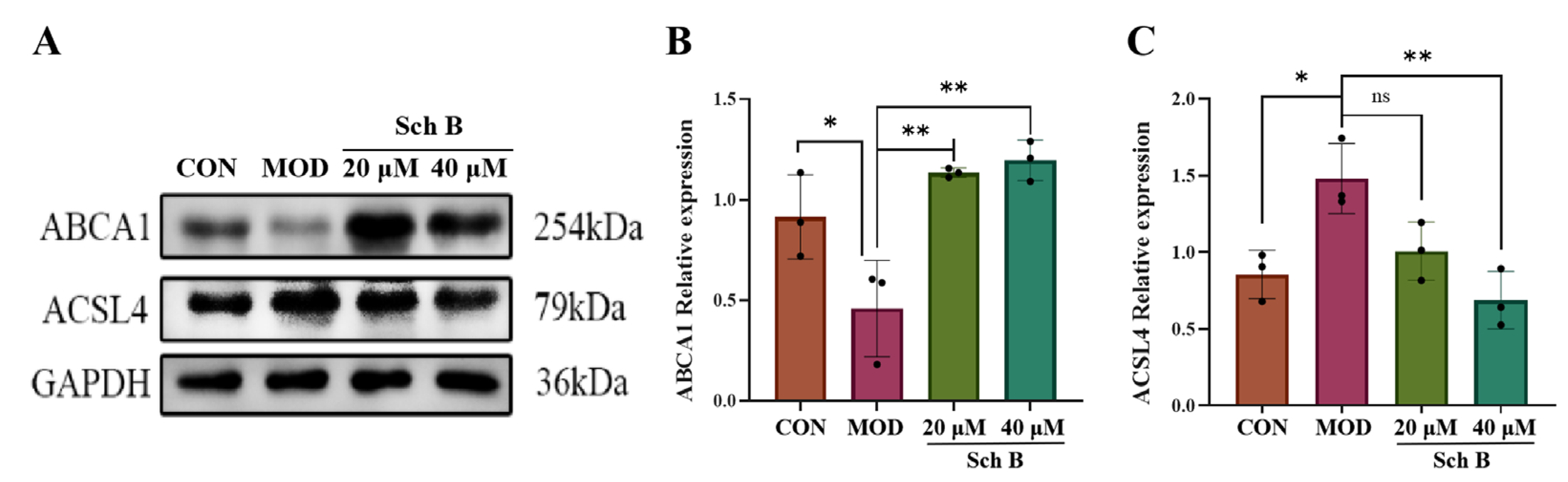
2.10.1. Effects of Sch B on the Expression of Lipid Synthesis and Cholesterol Metabolism-Related Proteins
2.10.2. Fatty Acid Oxidation
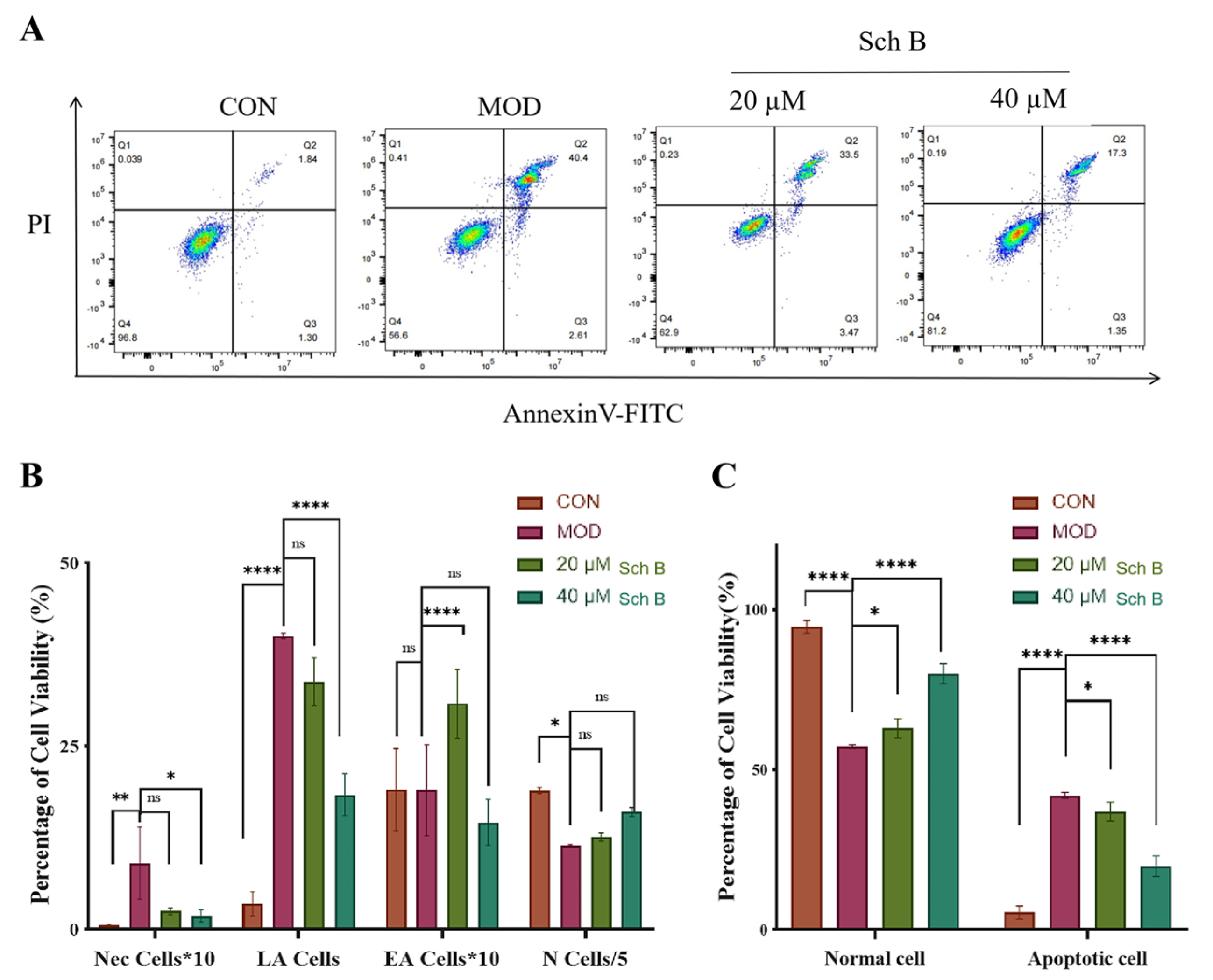
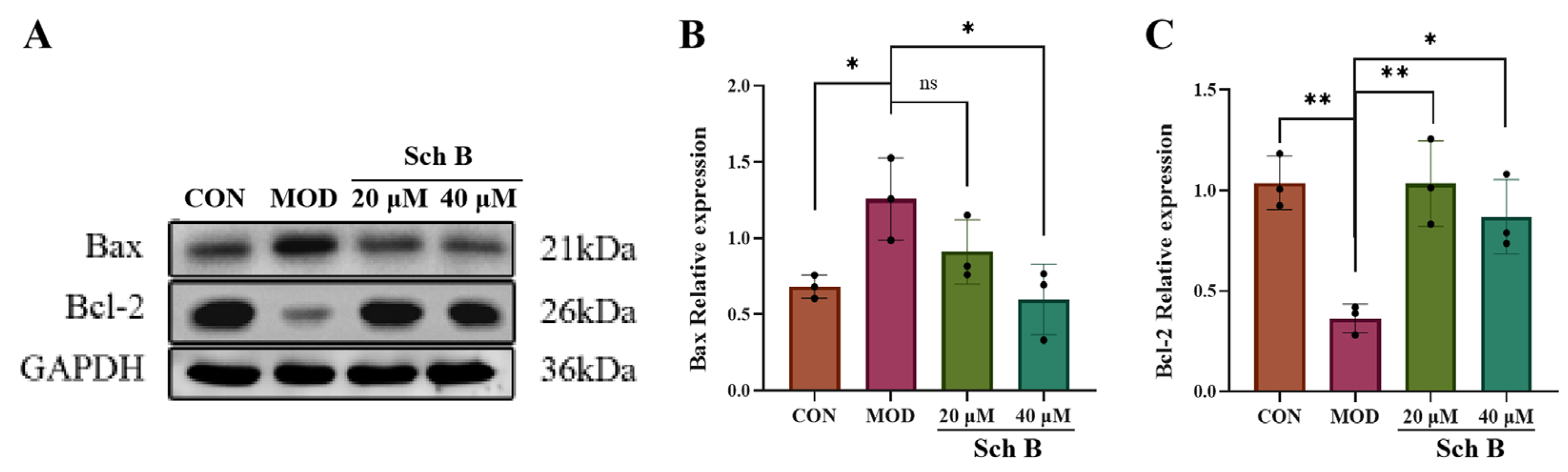
2.11. Effects of Sch B on Cell Apoptosis
2.11.1. Annexin V-FITC/PI Detection of Cell Apoptosis
2.11.2. Effects of Sch B on Expression of Apoptosis-Related Proteins
3. Discussion
4. Materials and Methods
4.1. Experimental Drugs and Reagents
4.2. Cytology Experiments
4.2.1. AML-12 Cell Culture
4.2.2. Establishment of MAFLD Cell Model
4.3. CCK-8 Detection of Sch B Activity
4.4. Detection of TG and TC
4.5. Determination of ALT and AST
4.6. Cell Oil Red O Staining Detection
4.7. Prediction of Targets by Network Pharmacology Methods
4.7.1. Acquisition of Intersection Targets of Sch B and MAFLD
4.7.2. Construction of Sch B Target Network
4.7.3. Construction of Target PPI
4.7.4. GO Biological Process Enrichment and KEGG Pathway Analysis
4.8. Molecular Docking and Visualization Analysis
4.9. Dual Luciferase Reporter Gene Detection
4.10. Cell Apoptosis Detection by Annexin V-FITC/PI Dual Staining
4.11. Protein Immunoblot Results
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| Sch B | Schisandrin B |
| MAFLD | Metabolic-associated fatty liver disease |
| PPAγ | Peroxisome proliferator-activated receptor gamma |
| OA | Oleic acid |
| PA | Palmitic acid |
| TC | Total cholesterol |
| TG | Triglycerides |
| ALT | Alanine aminotransferase |
| AST | Aspartate aminotransferase |
| PPI | Protein–protein interaction network |
| BC | Betweenness centrality |
| CC | Compact centrality |
| DC | Degree centrality |
| GO | Gene Ontology |
| BP | Biological processes |
| MF | Molecular function |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
References
- Li, Y.X.; Zhou, Y.Y.; Wang, L.Y.; Lin, X.Q.; Mao, M.H.; Yin, S.Q.; Zhu, L.; Jiao, Y.F.; Yu, W.F.; Gao, P.; et al. Emerging trends and hotspots in the links between the gut microbiota and MAFLD from 2002 to 2021: A bibliometric analysis. Front. Endocrinol. 2022, 13, 990953. [Google Scholar] [CrossRef]
- Gamboa, C.M.; Wang, Y.J.; Xu, H.T.; Kalemba, K.; Wondisford, F.E.; Sabaawy, H.E. Optimized 3D Culture of Hepatic Cells for Liver Organoid Metabolic Assays. Cells 2021, 10, 3280. [Google Scholar] [CrossRef] [PubMed]
- Doustmohammadian, A.; Nezhadisalami, A.; Tameshke, F.S.; Motamed, N.; Maadi, M.; Farahmand, M.; Sohrabi, M.; Clark, C.C.T.; Ajdarkosh, H.; Faraji, A.H.; et al. A randomized triple-blind controlled clinical trial evaluation of sitagliptin in the treatment of patients with non-alcoholic fatty liver diseases without diabetes. Front. Med. 2022, 9, 937554. [Google Scholar] [CrossRef] [PubMed]
- Mun, J.; Kim, S.; Yoon, H.G.; You, Y.; Kim, O.K.; Choi, K.C.; Lee, Y.H.; Lee, J.; Park, J.; Jun, W. Water Extract of Curcuma longa L. Ameliorates Non-Alcoholic Fatty Liver Disease. Nutrients 2019, 11, 2536. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.W.; Wang, W.S.; Shen, C.Z.; Wang, X.H.; Pu, Z.C.; Yin, Q. Network pharmacology for systematic understanding of Schisandrin B reduces the epithelial cells injury of colitis through regulating pyroptosis by AMPK/Nrf2/NLRP3 inflammasome. Aging 2021, 13, 23193–23209. [Google Scholar] [CrossRef]
- Zhao, F.; Pan, H.H.; Chen, X.Y.; Miao, S. Study on the Role of Schisandrin B in Ameliorating Hepatic Ischemia-Reperfusion Injury by Modulating Hepatocyte Autophagy. Iran. J. Pharm. Res. 2025, 24, 157033. [Google Scholar] [CrossRef]
- Li, C.; Ji, K.B.; Kim, E.Y.; Choi, H.Y.; Yoon, J.H.; Zhang, X.; Kim, R.E.; Park, K.S.; Liu, H.; Kim, M.K. Schisandrin B mitigates oxidative damage in early porcine embryos exposed to Ochratoxin A via NRF2 pathway activation. Anim. Reprod. Sci. 2025, 279, 107889. [Google Scholar] [CrossRef]
- Yan, L.S.; Zhang, S.F.; Luo, G.; Cheng, B.C.; Zhang, C.; Wang, Y.W.; Qiu, X.Y.; Zhou, X.H.; Wang, Q.G.; Song, X.L.; et al. Schisandrin B mitigates hepatic steatosis and promotes fatty acid oxidation by inducing autophagy through AMPK/mTOR signaling pathway. Metabolism 2022, 131, 155200. [Google Scholar] [CrossRef]
- Ma, R.J.; Zhan, Y.K.; Zhang, Y.M.; Wu, L.G.; Wang, X.; Guo, M. Schisandrin B ameliorates non-alcoholic liver disease through anti-inflammation activation in diabetic mice. Drug Dev. Res. 2022, 83, 735–744. [Google Scholar] [CrossRef]
- Kwan, H.Y.; Niu, X.Y.; Dai, W.L.; Tong, T.J.; Chao, X.J.; Su, T.; Chan, C.L.; Lee, K.C.; Fu, X.Q.; Yi, H.; et al. Lipidomic-based investigation into the regulatory effect of Schisandrin B on palmitic acid level in non-alcoholic steatotic livers. Sci. Rep. 2015, 5, 9114. [Google Scholar] [CrossRef]
- Wagner, N.; Wagner, K.D. The Role of PPARs in Disease. Cells 2020, 9, 2367. [Google Scholar] [CrossRef]
- Chen, H.; Tan, H.B.; Wan, J.; Zeng, Y.; Wang, J.C.; Wang, H.C.; Lu, X.J. PPAR-γ signaling in nonalcoholic fatty liver disease: Pathogenesis and therapeutic targets. Pharmacol. Ther. 2023, 245, 108391. [Google Scholar] [CrossRef]
- Gross, B.; Pawlak, M.; Lefebvre, P.; Staels, B. PPARs in obesity-induced T2DM, dyslipidaemia and NAFLD. Nat. Rev. 2017, 13, 36–49. [Google Scholar] [CrossRef] [PubMed]
- Ji, J.; Wu, L.W.; Feng, J.; Mo, W.H.; Wu, J.Y.; Yu, Q.; Li, S.N.; Zhang, J.; Dai, W.Q.; Xu, X.F.; et al. Cafestol preconditioning attenuates apoptosis and autophagy during hepatic ischemia-reperfusion injury by inhibiting ERK/PPARγ pathway. Int. Immunopharmacol. 2020, 84, 106529. [Google Scholar] [CrossRef] [PubMed]
- Xiang, S.H.; Chen, K.; Xu, L.; Guo, C.Y. Bergenin exerts hepatoprotective effects by inhibiting the release of inflammatory factors, apoptosis and autophagy via the PPAR-γ pathway. Drug Des. Dev. Ther. 2020, 14, 129–143. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Dai, W.Q.; Mao, Y.Q.; Wu, L.W.; Li, J.J.; Chen, K.; Yu, Q.; Kong, R.; Li, S.N.; Zhang, J.; et al. Simvastatin re-sensitizes hepatocellular carcinoma cells to sorafenib by inhibiting HIF-1α/PPAR-γ/PKM2-mediated glycolysis. J. Exp. Clin. Cancer Res. 2020, 39, 24. [Google Scholar] [CrossRef]
- Zhang, H.; Chen, Q.S.; Dahan, A.; Xue, J.Y.; Wei, L.W.; Tan, W.F.; Zhang, G.Q. Transcriptomic analyses reveal the molecular mechanisms of schisandrin B alleviates CCl(4)-induced liver fibrosis in rats by RNA-sequencing. Chem. Biol. Interact. 2019, 309, 108675. [Google Scholar] [CrossRef]
- Liu, N.; Zheng, J.X.; Zhuang, Y.S.; Zhou, Z.K.; Zhao, J.H.; Yang, L. Anti-Inflammatory Effects of Schisandrin B on LPS-Stimulated BV2 Microglia via Activating PPAR-γ. Inflammation 2017, 40, 1006–1011. [Google Scholar] [CrossRef]
- Sun, J.H.; Lin, H.; Wang, K.; Zeng, Q.C.; Li, H.; Wang, C.M.; Chen, J.G. Study of Schisandra chinensis in Treating Metabolic Associated Fatty Liver Disease Based on Network Pharmacology and Molecular Docking Technology. J. Beihua Univ. (Nat. Sci.) 2022, 23, 768–774. [Google Scholar]
- Wan, J.C.; Lang, C.J.; Gao, M.; Liu, F.L.; Feng, X.Y.; Li, H.; Wang, C.M.; Sun, J.H. Schisandrin B alleviates metabolic associated fatty liver disease by regulating the PPARγ signaling pathway and gut microbiota in mice. Front. Pharmacol. 2025, 16, 1583307. [Google Scholar] [CrossRef]
- Zhang, M.; Bai, X.; Du, Q.; Xu, J.J.; Wang, D.Q.; Chen, L.; Dong, K.T.; Chen, Z.Y.; Yang, J.H. The Different Mechanisms of Lipid Accumulation in Hepatocytes Induced by Oleic Acid/Palmitic Acid and High-Fat Diet. Molecules 2023, 28, 6714. [Google Scholar] [CrossRef]
- Wang, Y.P.; Nakajima, T.; Gonzalez, F.J.; Tanaka, N. PPARs as Metabolic Regulators in the Liver: Lessons from Liver-Specific PPAR-Null Mice. Int. J. Mol. Sci. 2020, 17, 2061. [Google Scholar] [CrossRef]
- Beale, E.G.; Harvey, B.J.; Forest, C. PCK1 and PCK2 as candidate diabetes and obesity genes. Cell Biochem. Biophys. 2007, 48, 89–95. [Google Scholar] [CrossRef]
- Tuo, L.; Xiang, J.; Pan, X.M.; Hu, J.L.; Tang, H.; Liang, L.; Xia, J.; Hu, Y.; Zhang, W.; Huang, A.L.; et al. PCK1 negatively regulates cell cycle progression and hepatoma cell proliferation via the AMPK/p27Kip1 axis. J. Exp. Clin. Cancer Res. 2019, 38, 50. [Google Scholar] [CrossRef]
- Fan, T.J.; Han, L.H.; Cong, R.S.; Liang, J. Caspase family proteases and apoptosis. Acta Biochim. Biophys. Sin. 2005, 37, 719–727. [Google Scholar] [CrossRef] [PubMed]
- Alturaif, S.A.; Alhusaini, A.; Sarawi, W.; Hasan, I.; Alsaab, J.; Ali, R.; Mohammed, R.; Alotaibi, S.S.; Almutairi, F.; Alsaif, S.; et al. Indole-3-Acetic Acid: Promising Protective Agent Against Methotrexate-Induced Liver Injury via Modulation of TLR4/NF-κB/Caspase-3 Pathway. Pharmaceuticals 2025, 18, 828. [Google Scholar] [CrossRef] [PubMed]
- Lam, T.; Harmancey, R.; Vasquez, H.; Gilbert, B.; Patel, N.; Hariharan, V.; Lee, A.; Covey, M.; Taegtmeyer, H. Reversal of intramyocellular lipid accumulation by lipophagy and a p62-mediated pathway. Cell Death Discov. 2016, 2, 16061. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.W.; Zou, Y.; Chen, J.; Cui, C.; Song, J.; Yang, M.M.; Gao, J.; Hu, H.Q.; Xia, L.Q.; Wang, L.M.; et al. Didymin alleviates metabolic dysfunction-associated fatty liver disease (MAFLD) via the stimulation of Sirt1-mediated lipophagy and mitochondrial biogenesis. J. Transl. Med. 2023, 19, 921. [Google Scholar] [CrossRef]
- Kan, C.F.K.; Singh, A.B.; Dong, B.; Shende, V.R.; Liu, J.W. PPARδ activation induces hepatic long-chain acyl-CoA synthetase 4 expression in vivo and in vitro. Biochim. Biophys. Acta 2015, 1851, 577–587. [Google Scholar] [CrossRef]
- Ding, K.Y.; Liu, C.B.; Li, L.; Yang, M.; Jiang, N.; Luo, S.L.; Sun, L. Acyl-CoA synthase ACSL4: An essential target in ferroptosis and fatty acid metabolism. Chin. Med. J. 2023, 136, 2521–2537. [Google Scholar] [CrossRef]
- Gwon, M.H.; Im, Y.S.; Seo, A.R.; Kim, K.Y.; Moon, H.R.; Yun, J.M. Phenethyl Isothiocyanate Protects against High Fat/Cholesterol Diet-Induced Obesity and Atherosclerosis in C57BL/6 Mice. Nutrients 2020, 12, 3657. [Google Scholar] [CrossRef]
- Schlaepfer, I.R.; Joshi, M. CPT1A-mediated Fat Oxidation, Mechanisms, and Therapeutic Potential. Endocrinology 2020, 161, bqz046. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.X.; Tan, J.; Cao, Z.Y.; Cai, Y.W.; Huang, Z.Q.; Chen, Z.T.; He, W.B.; Liu, X.; Jiang, Y.; Gao, Q.Y.; et al. Sirt5 improves cardiomyocytes fatty acid metabolism and ameliorates cardiac lipotoxicity in diabetic cardiomyopathy via CPT2 de-succinylation. Redox Biol. 2024, 73, 103184. [Google Scholar] [CrossRef]
- Chen, X.F.; Tian, M.X.; Sun, R.Q.; Zhang, M.L.; Zhou, L.S.; Jin, L.; Chen, L.L.; Zhou, W.J.; Duan, K.L.; Chen, Y.J.; et al. SIRT5 inhibits peroxisomal ACOX1 to prevent oxidative damage and is downregulated in liver cancer. EMBO Rep. 2018, 19, e45124. [Google Scholar] [CrossRef]
- Lin, K.N.; Zhao, W.; Huang, S.Y.; Li, H. Grape seed proanthocyanidin extract induces apoptosis of HL-60/ADR cells via the Bax/Bcl-2 caspase-3/9 signaling pathway. Transl. Cancer Res. 2021, 10, 3939–3947. [Google Scholar] [CrossRef]
- Li, T.; Zhang, L.; Jin, C.; Xiong, Y.; Cheng, Y.Y.; Chen, K. Pomegranate flower extract bidirectionally regulates the proliferation, differentiation and apoptosis of 3T3-L1 cells through regulation of PPARγ expression mediated by PI3K-AKT signaling pathway. Biomed. Pharmacother. 2020, 131, 110769. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, M.; Liu, F.; Feng, X.; Wang, M.; Zhang, Z.; Li, H.; Wang, C.; Sun, J. Schisandrin B Alleviates Lipid Metabolism Disorders and Apoptosis of MAFLD via Modulation of PPARγ-PCK1 and Caspase-3 Signaling Pathways. Pharmaceuticals 2025, 18, 1441. https://doi.org/10.3390/ph18101441
Gao M, Liu F, Feng X, Wang M, Zhang Z, Li H, Wang C, Sun J. Schisandrin B Alleviates Lipid Metabolism Disorders and Apoptosis of MAFLD via Modulation of PPARγ-PCK1 and Caspase-3 Signaling Pathways. Pharmaceuticals. 2025; 18(10):1441. https://doi.org/10.3390/ph18101441
Chicago/Turabian StyleGao, Meng, Feilong Liu, Xiyuan Feng, Mengyang Wang, Zhihong Zhang, He Li, Chunmei Wang, and Jinghui Sun. 2025. "Schisandrin B Alleviates Lipid Metabolism Disorders and Apoptosis of MAFLD via Modulation of PPARγ-PCK1 and Caspase-3 Signaling Pathways" Pharmaceuticals 18, no. 10: 1441. https://doi.org/10.3390/ph18101441
APA StyleGao, M., Liu, F., Feng, X., Wang, M., Zhang, Z., Li, H., Wang, C., & Sun, J. (2025). Schisandrin B Alleviates Lipid Metabolism Disorders and Apoptosis of MAFLD via Modulation of PPARγ-PCK1 and Caspase-3 Signaling Pathways. Pharmaceuticals, 18(10), 1441. https://doi.org/10.3390/ph18101441





