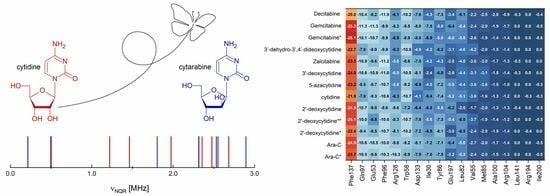
Butterfly Effect in Cytarabine: Combined NMR-NQR Experiment, Solid-State Computational Modeling, Quantitative Structure-Property Relationships and Molecular Docking Study
Abstract
1. Introduction
2. Results and Discussion
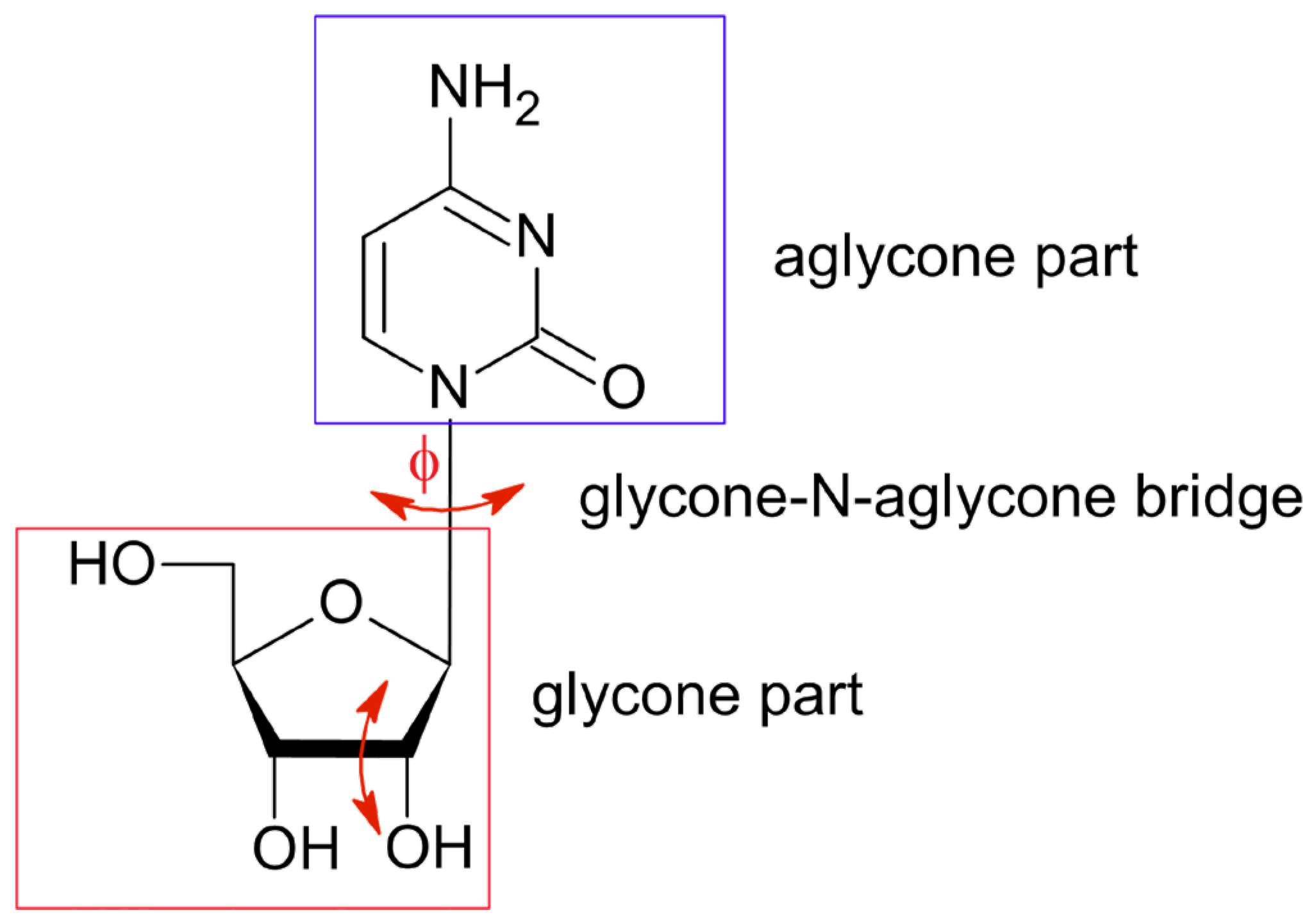
2.1. 1H-14N NQDR Spectrum
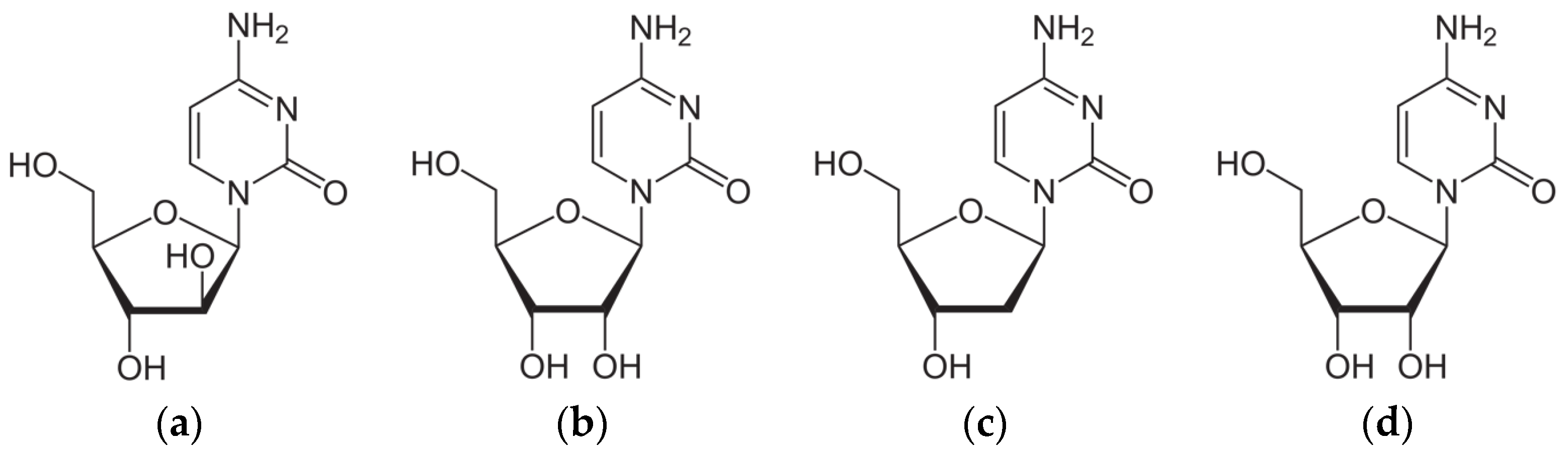
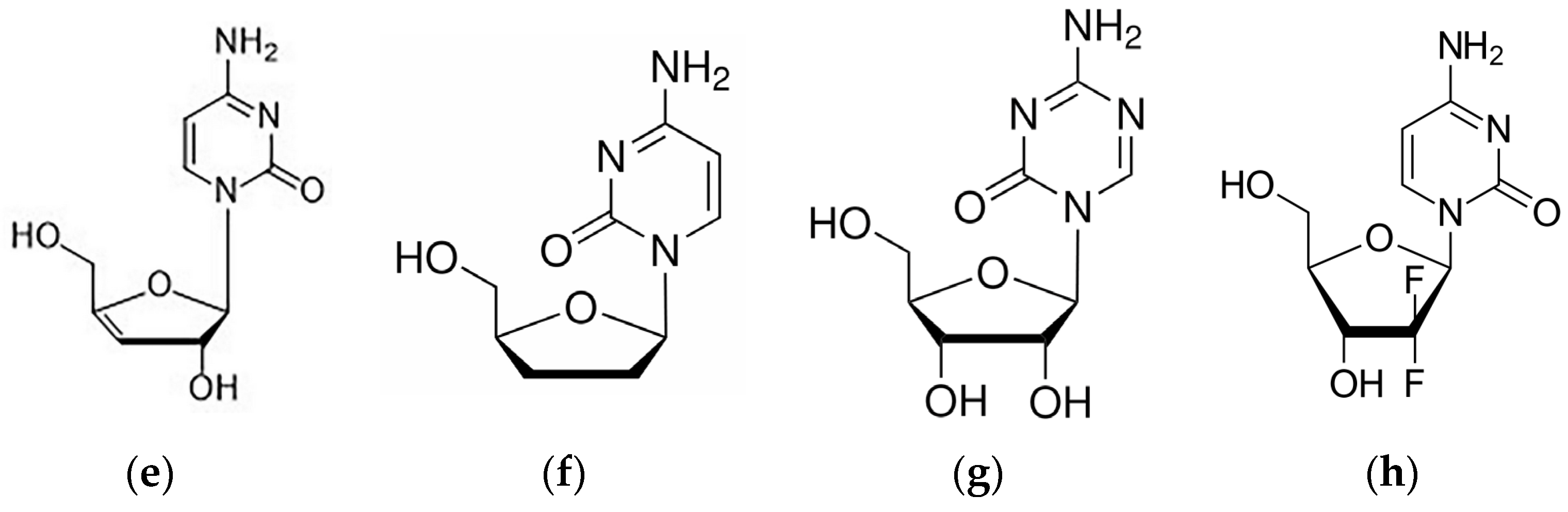
2.2. Intermolecular Interactions Pattern in Crystal
2.2.1. Binding Motifs
2.2.2. Strength of the Interactions
2.2.3. Differences in Binding Modes in the Solid State
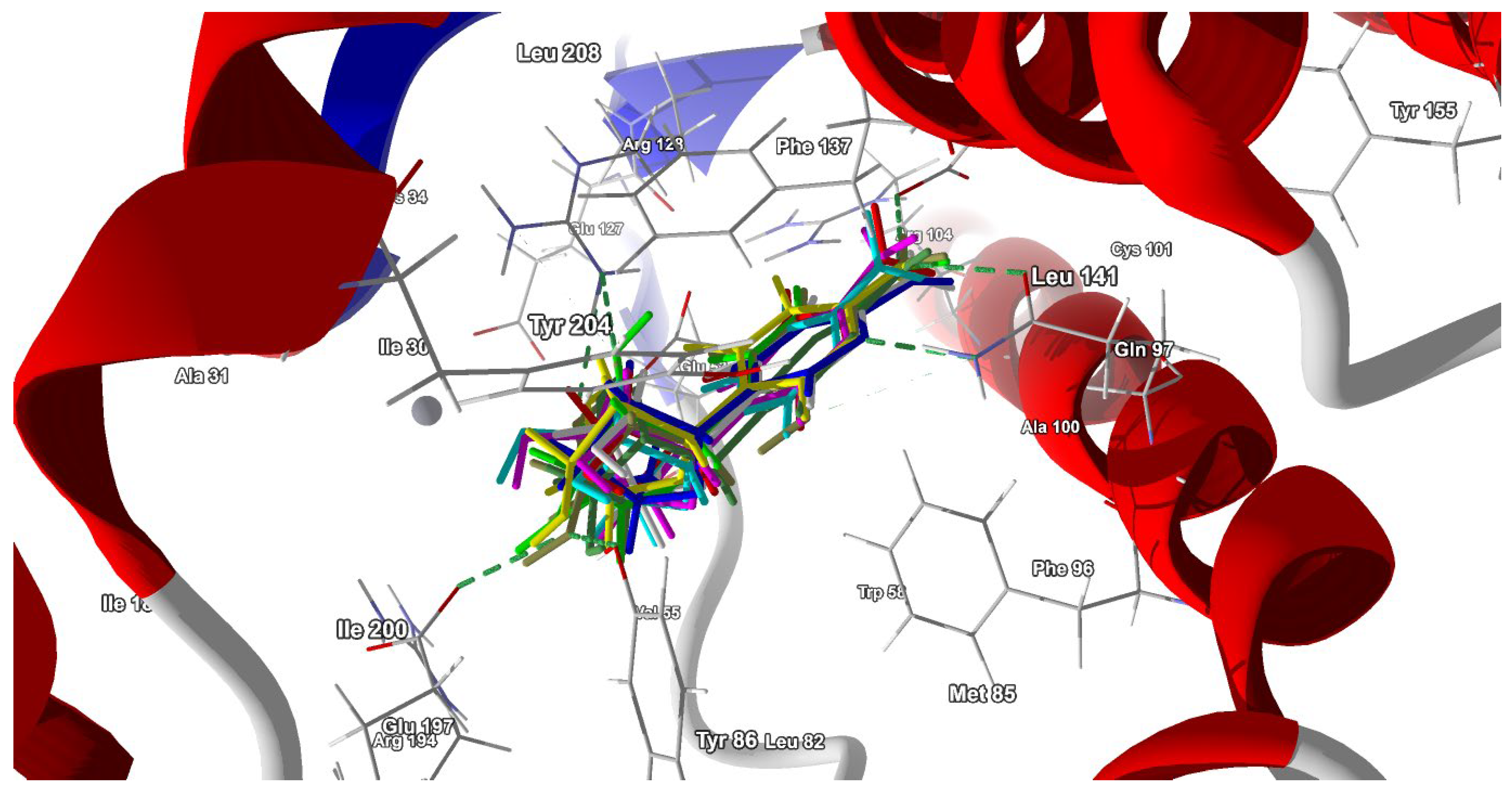
2.3. Binding to Deoxycytidine Kinase (dCK)
2.3.1. Molecular Docking Results
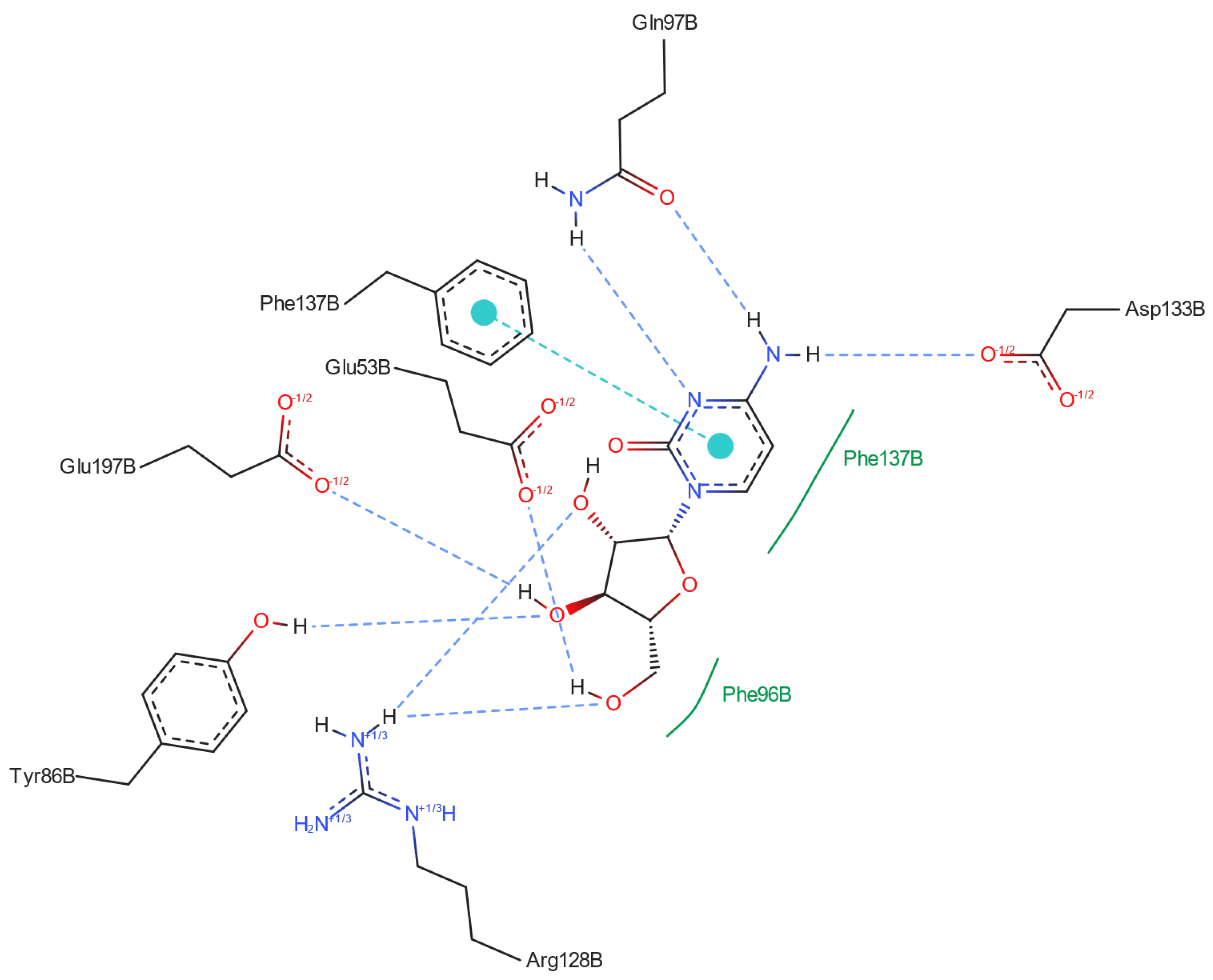
2.3.2. dCK-ligand Binding Modes
2.4. Relative Reactivity of the Ligands
>2′-deoxycytidine > Gemcitabine
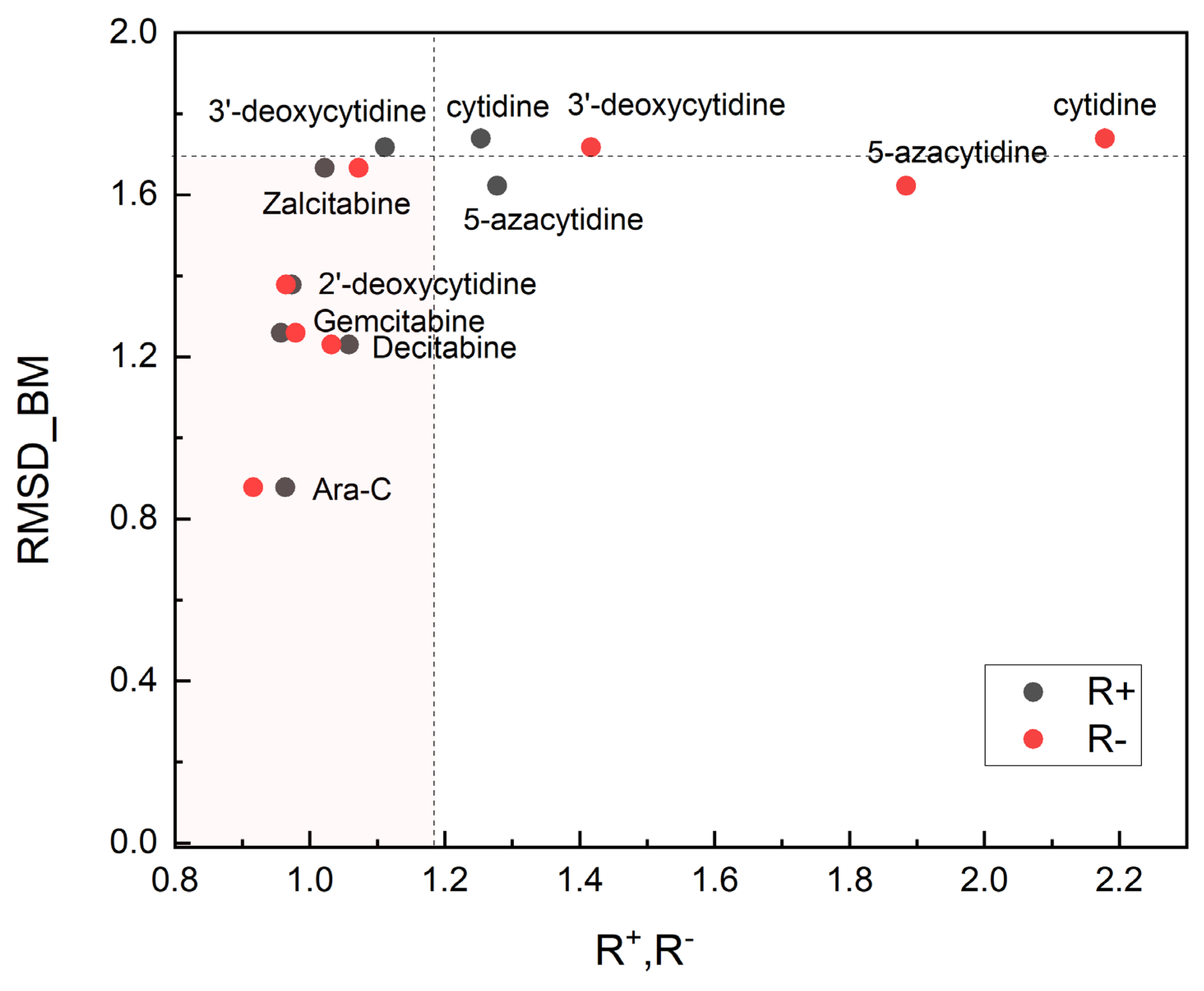
2.5. Quantitative Structure–Property Relationships (Binding Affinity, Relative Reactivity and Biological Activity)
3. Materials and Method
3.1. Material
3.2. Methods
3.2.1. Nuclear Quadrupole Resonance (NQR)
- (i)
- Polarization field for time , where is the proton spin-lattice relaxation time in the field (an equilibrium proton magnetization condition).
- (ii)
- Relaxation (also called “mixing”) field for time , where the proton magnetization relaxes towards the equilibrium value with a time constant .
- (iii)
- Acquisition field , at which the proton NMR signal amplitude, which is proportional to the proton magnetization at the end of interval, is measured.
3.2.2. Computational Modeling-Density Functional Theory
Single Molecules and Clusters
Solid-State
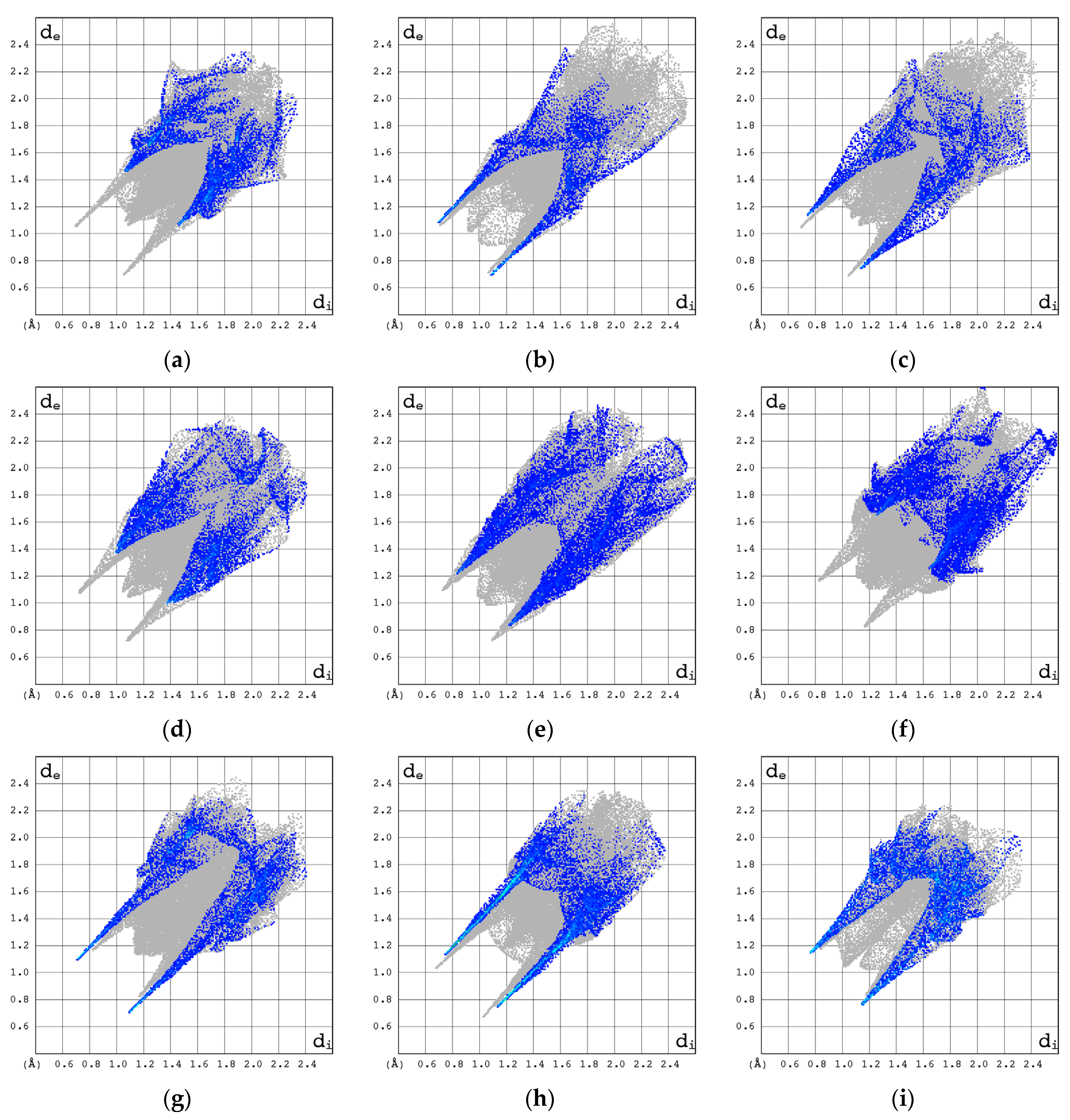
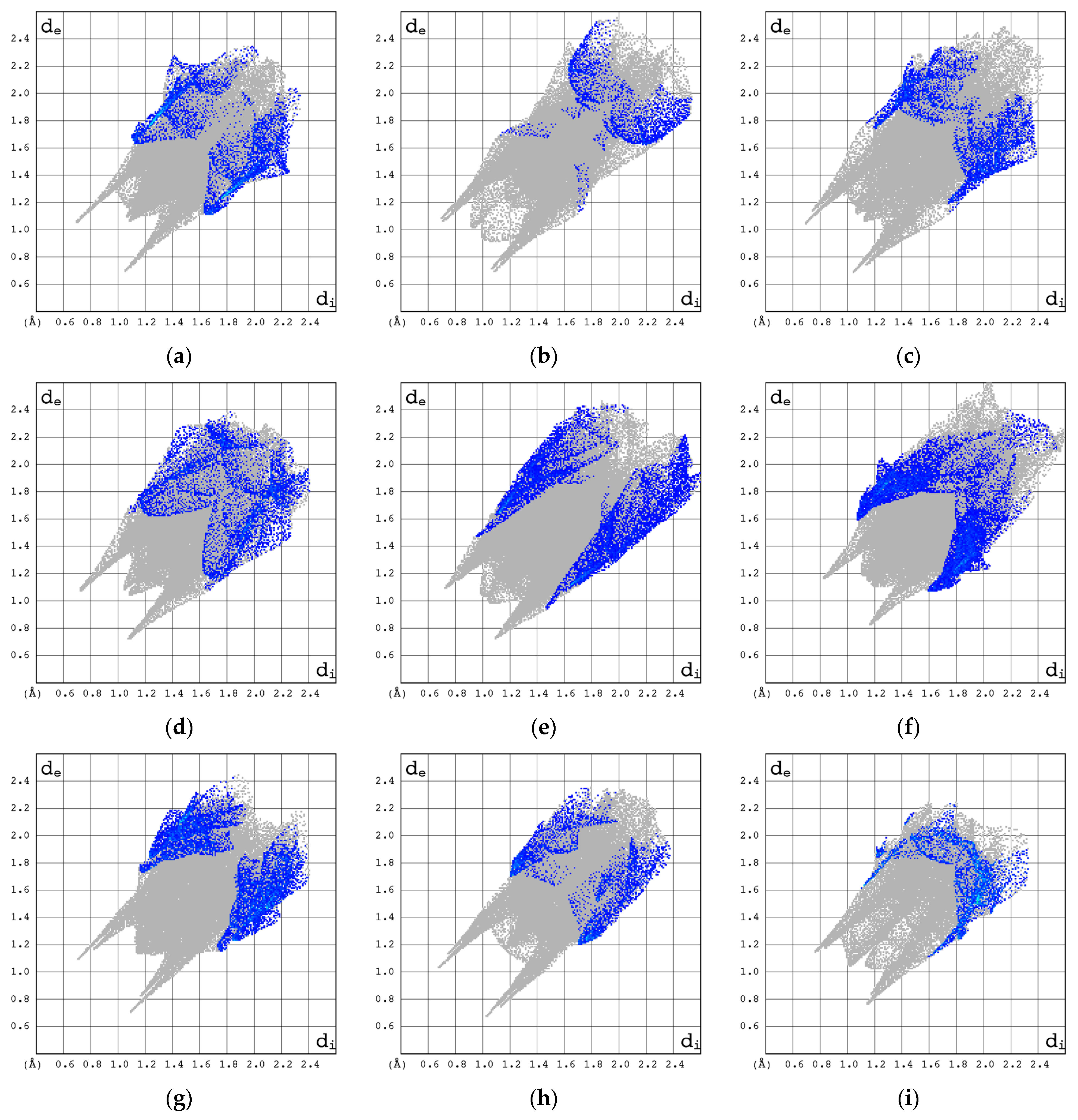
3.2.3. Hirshfeld Surfaces (3D HS)
3.2.4. Molecular Docking
3.2.5. Comparison of the Differences in the Binding Modes
Root-Mean-Square Deviation of the Binding Mode
Grid Heatmaps
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bergmann, W.; Feeney, R.J. The Isolation of a New Thymine Pentoside from Sponges. J. Am. Chem. Soc. 1950, 72, 2809–2810. [Google Scholar] [CrossRef]
- Walwick, E.; Roberts, W.; Dekker, C. Cyclisation during the Phosphorylation of Uridine and Cytidine by Polyphosphoric Acid-A New Route to the O-2, 2′-Cyclonucleosides. Proc. Chem. Soc. Lond. 1959, 1959, 84. [Google Scholar]
- Eriksson, S.; Munch-Petersen, B.; Johansson, K.; Ecklund, H. Structure and Function of Cellular Deoxyribonucleoside Kinases. Cell. Mol. Life Sci. CMLS 2002, 59, 1327–1346. [Google Scholar] [CrossRef] [PubMed]
- Graham, F.L.; Whitmore, G.F. Studies in Mouse L-Cells on the Incorporation of 1-Beta-D-Arabinofuranosylcytosine into DNA and on Inhibition of DNA Polymerase by 1-Beta-D-Arabinofuranosylcytosine 5′-Triphosphate. Cancer Res. 1970, 30, 2636–2644. [Google Scholar] [PubMed]
- Gruffaz, M.; Zhou, S.; Vasan, K.; Rushing, T.; Michael, Q.L.; Lu, C.; Jung, J.U.; Gao, S.-J. Repurposing Cytarabine for Treating Primary Effusion Lymphoma by Targeting Kaposi’s Sarcoma-Associated Herpesvirus Latent and Lytic Replications. mBio 2018, 9, e00756-18. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Dai, P.; Ding, W.; Granstein, R.D.; Shuman, S. Vaccinia Virus Infection Attenuates Innate Immune Responses and Antigen Presentation by Epidermal Dendritic Cells. J. Virol. 2006, 80, 9977–9987. [Google Scholar] [CrossRef] [PubMed]
- Hamada, A.; Kawaguchi, T.; Nakano, M. Clinical Pharmacokinetics of Cytarabine Formulations. Clin. Pharmacokinet. 2002, 41, 705–718. [Google Scholar] [CrossRef] [PubMed]
- Lichtman, M.A. A Historical Perspective on the Development of the Cytarabine (7 days) and Daunorubicin (3days) Treatment Regimen for Acute Myelogenous Leukemia: 2013 the 40th Anniversary of 7+3. Blood Cells Mol. Dis. 2013, 50, 119–130. [Google Scholar] [CrossRef] [PubMed]
- Hoshi, A.; Kanzawa, F.; Kuretani, K. Antitumor Activity of Cyclocytidine in a Variety of Tumors. Jpn. J. Cancer Res. 1972, 63, 353–360. [Google Scholar] [CrossRef]
- Nakahara, W.; Tokuzen, R. Effect of 2,2′-o-Cyclocytidine on Transplanted Lymphocytic Sarcoma and Reticulum Cell Sarcoma in Mice. Jpn. J. Cancer Res. 1972, 63, 379–381. [Google Scholar] [CrossRef]
- Stoehr, M.; Friedel, G.; Engel, P.; Goerttler, K.; Futterman, G. Cytostatic and Cytotoxic Response of Ehrlich Ascites Tumor Cells In Vivo on Chronic Treatment with Cytarabine, Bleomycin, and Peplomycin. Arzneim. Forsch. 1984, 34, 451–454. [Google Scholar]
- Raut, L.S. Novel Formulation of Cytarabine and Daunorubicin: A New Hope in AML Treatment. South Asian J. Cancer 2015, 4, 38–40. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Duan, C.; Chen, L.; Chen, S. Efficacy and Safety of Different Doses of Cytarabine in Consolidation Therapy for Adult Acute Myeloid Leukemia Patients: A Network Meta-Analysis. Sci. Rep. 2017, 7, 9509. [Google Scholar] [CrossRef] [PubMed]
- Puty, T.C.; Sarraf, J.S.; Do Carmo Almeida, T.C.; Filho, V.C.B.; de Carvalho, L.E.W.; Fonseca, F.L.A.; Adami, F. Evaluation of the impact of single-nucleotide polymorphisms on treatment response, survival and toxicity with cytarabine and anthracyclines in patients with acute myeloid leukaemia: A systematic review protocol. Syst. Rev. 2019, 8, 109. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.Y.; Ge, S.S.; Huang, Y.H. Efficacy and safety of cladribine, low-dose cytarabine and venetoclax in relapsed/refractory acute myeloid leukemia: Results of a pilot study. Blood Cancer J. 2024, 14, 12. [Google Scholar] [CrossRef] [PubMed]
- Hong, M.; Zhu, H.; Sun, Q.; Zhu, Y.; Miao, Y.; Yang, H.; Qiu, H.R.; Li, J.Y.; Qian, S.X. Decitabine in combination with low-dose cytarabine, aclarubicin and G-CSF tends to improve prognosis in elderly patients with high-risk AML. Aging 2020, 12, 5792–5811. [Google Scholar] [CrossRef] [PubMed]
- Bader, R.F.W. Atoms in Molecules: A Quantum Theory; The International Series of Monographs on Chemistry; Clarendon Press: Oxford, NY, USA, 1990; ISBN 978-0-19-855168-3. [Google Scholar]
- Bader, R.F.W. A Quantum Theory of Molecular Structure and Its Applications. Chem. Rev. 1991, 91, 893–928. [Google Scholar] [CrossRef]
- Zheng, X.; Yang, S.; Chen, F.; Wu, S.; Li, W. The Efficacy and Safety of Cytarabine on Newly Diagnosed Primary Central Nervous System Lymphoma: A Systematic Review and Meta-Analysis. Front. Oncol. 2020, 31, 1213. [Google Scholar] [CrossRef] [PubMed]
- Estey, E.H. Acute myeloid leukemia: 2021 update on risk-stratification and management. Am. J. Hematol. 2020, 95, 1368–1398. [Google Scholar] [CrossRef]
- Wang, X.; Liu, D.; Chen, E.; Wang, L.; Zhao, N.; Zhou, L.; Tong, J.; Xue, L.; Zhang, L.; Geng, L.; et al. High-dose cytarabine monotherapy is superior to standard-dose cytarabine-based multiagent sequential treatment cycle for consolidation treatment in adult (14–59 years) AML patients according to European Leukemia Net 2022 risk stratification. Front. Oncol. 2023, 12, 1070588. [Google Scholar] [CrossRef]
- Johnson, E.R.; Keinan, S.; Mori-Sánchez, P.; Contreras-García, J.; Cohen, A.J.; Yang, W. Revealing Noncovalent Interactions. J. Am. Chem. Soc. 2010, 132, 6498–6506. [Google Scholar] [CrossRef] [PubMed]
- Spackman, M.A.; Jayatilaka, D. Hirshfeld Surface Analysis. CrystEngComm 2009, 11, 19–32. [Google Scholar] [CrossRef]
- Parr, R.G.; Szentpály, L.V.; Liu, S. Electrophilicity Index. J. Am. Chem. Soc. 1999, 121, 1922–1924. [Google Scholar] [CrossRef]
- Seliger, J.; Zagar, V.; Blinc, R. A New Highly Sensitive 1H-14N Nuclear-Quadrupole Double-Resonance Technique. J. Magn. Reson. Ser. A 1994, 106, 214–222. [Google Scholar] [CrossRef]
- Seliger, J.; Žagar, V.; Blinc, R. 1H-14N Nuclear Quadrupole Double Resonance with Multiple Frequency Sweeps. Z. Nat. A 1994, 49, 31–34. [Google Scholar] [CrossRef]
- Edmonds, D.T.; Speight, P.A. Nuclear Quadrupole Resonance of 14N in Pyrimidines, Purines and Their Nucleosides. J. Magn. Reson. 1972, 6, 265–273. [Google Scholar] [CrossRef]
- Hiyama, Y.; Watanabe, N.; Maruizumi, T.; Niki, E. Laboratory Nuclear Quadrupole Resonance Spectrometer Using Adiabatic Demagnetization. Nippon Kagaku Kaishi 1979, 8, 961–968. [Google Scholar] [CrossRef]
- Hiyama, Y.; Roy, S.; Cohen, J.S.; Torchia, D.A. Solid-State Deuterium NMR Study of Thymidine. Base Rigidity and Ribose Ring Flexibility in Deoxynucleosides. J. Am. Chem. Soc. 1989, 111, 8609–8613. [Google Scholar] [CrossRef]
- Tougard, P.; Lefebvre-Soubeyran, O. Stéréochimie de La 1-β-D-Arabinofuranosyl-Cytosine. Acta Cryst. 1974, 30, 86–89. [Google Scholar] [CrossRef]
- Sato, S.; Yoshida, T.; Hori, T. Determination of Absolute Configurations of Light-Atom Molecules by Means of Direct Detection of Bijvoet Differences. Enantiomer 2002, 7, 271–281. [Google Scholar] [CrossRef]
- Young, D.W.; Wilson, H.R. The Crystal and Molecular Structure of 2′-Deoxycytidine. Acta Cryst. 1975, 31, 961–965. [Google Scholar] [CrossRef]
- Karthe, P.; Gautham, N.; Kumar, A.; Katti, S.B. β-D-3′-Deoxyadenosine (Cordycepin). Acta Cryst. C 1997, 53, 1694–1696. [Google Scholar] [CrossRef]
- Silverton, J.V.; Quinn, F.R.; Haugwitz, R.D.; Todaro, L.J. Structures of Two Dideoxynucleosides: 2′,3′-Dideoxyadenosine and 2′,3′-Dideoxycytidine. Acta Crystallogr. C Cryst. Struct. Commun. 1988, 44, 321–324. [Google Scholar] [CrossRef] [PubMed]
- Ito, S.; White, F.J.; Okunishi, E.; Aoyama, Y.; Yamano, A.; Sato, H.; Ferrara, J.D.; Jasnowski, M.; Meyer, M. Structure Determination of Small Molecule Compounds by an Electron Diffractometer for 3D ED/MicroED. CrystEngComm 2021, 23, 8622–8630. [Google Scholar] [CrossRef]
- Etter, M.C. Encoding and decoding hydrogen-bond patterns of organic compounds. Acc. Chem. Res. 2002, 23, 120–126. [Google Scholar] [CrossRef]
- Etter, M.C. Hydrogen bonds as design elements in organic chemistry. J. Phys. Chem. 2002, 95, 4601–4610. [Google Scholar] [CrossRef]
- Post, M.L.; Birnbaum, G.I.; Huber, C.P.; Shugar, D. α-Nucleosides in Biological Systems. Biochim. Biophys. Acta 1977, 479, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Ward, D.L. β-Cytidine, C9H13N3O5. Acta Crystallogr. C 1993, 49, 1789–1792. [Google Scholar] [CrossRef]
- Coome, J.A.; Goeta, A.E.; Howard, J.A.K.; Probert, M.R. Masquerade: Removing Non-Sample Scattering from Integrated Reflection Intensities. J. Appl. Crystallogr. 2012, 45, 292–298. [Google Scholar] [CrossRef]
- Furberg, S.; Peterson, C.S.; Rømming, C. A Refinement of the Crystal Structure of Cytidine. Acta Cryst. 1965, 18, 313–320. [Google Scholar] [CrossRef]
- Budow-Busse, S.; Chai, Y.; Müller, S.L.; Daniliuc, C.; Seela, F. The α-D-Anomer of 2′-Deoxycytidine: Crystal Structure, Nucleoside Conformation and Hirshfeld Surface Analysis. Acta Cryst. C 2021, 77, 202–208. [Google Scholar] [CrossRef] [PubMed]
- Birnbaum, G.I.; Lin, T.-S.; Prusoff, W.H. Unusual Structural Features of 2′,3′-Dideoxycytidine, an Inhibitor of the HIV (AIDS) Virus. Biochem. Biophys. Res. Commun. 1988, 151, 608–614. [Google Scholar] [CrossRef] [PubMed]
- Brus, J.; Czernek, J.; Kobera, K.; Urbanova, M.; Abbrent, S.; Husak, M. Predicting the Crystal Structure of Decitabine by Powder NMR Crystallography: Influence of Long-Range Molecular Packing Symmetry on NMR Parameters. Cryst. Growth Des. 2016, 16, 7102–7111. [Google Scholar] [CrossRef]
- Maris, T. Crystal Structure Database. CCDC 2001551: Experimental Crystal Structure Determination. 2020. Available online: https://www.ccdc.cam.ac.uk/structures/search?id=doi:10.5517/ccdc.csd.cc255s5b&sid=DataCite (accessed on 17 December 2023).
- Barker, D.L.; Marsh, R.E. The Crystal Structure of Cytosine. Acta Cryst. 1964, 17, 1581–1587. [Google Scholar] [CrossRef]
- McClure, R.J.; Craven, B.M. New Investigations of Cytosine and Its Monohydrate. Acta Cryst. B 1973, 29, 1234–1238. [Google Scholar] [CrossRef]
- Sridhar, B.; Nanubolu, J.B.; Ravikumar, K. The First Polymorph in the Family of Nucleobases: A Second Form of Cytosine. Acta Cryst. C 2015, 71, 128–135. [Google Scholar] [CrossRef] [PubMed]
- Sabini, E.; Ort, S.; Monnerjahn, C.; Konrad, M.; Lavie, A. Structure of Human dCK Suggests Strategies to Improve Anticancer and Antiviral Therapy. Nat. Struct. Mol. Biol. 2003, 10, 513–519. [Google Scholar] [CrossRef] [PubMed]
- Donigan, K.A.; McLenigan, M.P.; Yang, W.; Goodman, M.F.; Woodgate, R. The Steric Gate of DNA Polymerase ι Regulates Ribonucleotide Incorporation and Deoxyribonucleotide Fidelity. J. Biol. Chem. 2014, 289, 9136–9145. [Google Scholar] [CrossRef] [PubMed]
- Latosińska, M.; Latosińska, J.N. Favipiravir Analogues as Inhibitors of SARS-CoV-2 RNA-Dependent RNA Polymerase, Combined Quantum Chemical Modeling, Quantitative Structure–Property Relationship, and Molecular Docking Study. Molecules 2024, 29, 441. [Google Scholar] [CrossRef]
- Drenberg, C.D.; Shelat, A.; Dang, J.; Cotton, A.; Orwick, S.J.; Li, M.; Jeon, J.Y.; Fu, Q.; Buelow, D.R.; Pioso, M.; et al. A high-throughput screen indicates gemcitabine and JAK inhibitors may be useful for treating pediatric AML. Nat. Commun. 2019, 10, 2189. [Google Scholar] [CrossRef]
- Momparler, R.L. A Perspective on the Comparative Antileukemic Activity of 5-Aza-2′-deoxycytidine (Decitabine) and 5-Azacytidine (Vidaza). Pharmaceuticals 2012, 5, 875–881. [Google Scholar] [CrossRef]
- Dalvand, S.; Namdari, A.; Sepahvand, F.; Meshkibaf, M.H.; Ahmadpour, G. Investigation of Decitabine Effects on HDAC3 and HDAC7 mRNA Expression in NALM-6 and HL-60 Cancer Cell Lines. Rep. Biochem. Mol. Biol. 2021, 10, 420–428. [Google Scholar] [CrossRef] [PubMed]
- Leonard, S.M.; Perry, T.; Woodman, C.B.; Kearns, P. Sequential treatment with cytarabine and decitabine has an increased anti-leukemia effect compared to cytarabine alone in xenograft models of childhood acute myeloid leukemia. PLoS ONE 2014, 28, e87475. [Google Scholar] [CrossRef]
- Bergman, A.M.; Pinedo, H.M.; Jongsma, A.P.; Brouwer, M.; Ruiz van Haperen, V.W.; Veerman, G.; Leyva, A.; Eriksson, S.; Peters, G.J. Decreased resistance to gemcitabine (2′,2′-difluorodeoxycitidine) of cytosine arabinoside-resistant myeloblastic murine and rat leukemia cell lines: Role of altered activity and substrate specificity of deoxycytidine kinase. Biochem. Pharmacol. 1999, 57, 397–406. [Google Scholar] [CrossRef]
- Owens, J.K.; Shewach, D.S.; Ullman, B. Resistance to 1-β-d-arabinofuranosylcytosine in human T-lymphoblasts mediated by mutations within the deoxycytidine kinase gene. Cancer Res. 1992, 52, 2389–2393. [Google Scholar] [PubMed]
- Sebastiani, V.; Ricci, F.; Rubio-Viqueira, B.; Kulesza, P.; Yeo, C.J.; Hidalgo, M.; Klein, A.; Laheru, D.; Iacobuzio-Donahue, C.A. Immunohistochemical and genetic evaluation of deoxycytidine kinase in pancreatic cancer: Relationship to molecular mechanisms of gemcitabine resistance and survival. Clin. Cancer Res. 2006, 12, 2492–2497. [Google Scholar] [CrossRef]
- Guantay, L.; Garro, C.; Siri, S.; Pansa, M.F.; Ghidelli-Disse, S.; Paviolo, N.; Racca, A.; Nicotra, V.; Radu, C.; Bocco, J.L.; et al. Deoxycytidine kinase (dCK) inhibition is synthetic lethal with BRCA2 deficiency. Drug Resist. Updates 2023, 67, 100932. [Google Scholar] [CrossRef]
- Chen, B.Y.; Salas, J.R.; Trias, A.O.; Perez Rodriguez, A.; Tsang, J.E.; Guemes, M.; Le, T.M.; Galic, Z.; Shepard, H.M.; Steinman, L.; et al. Targeting deoxycytidine kinase improves symptoms in mouse models of multiple sclerosis. Immunology 2023, 168, 152–169. [Google Scholar] [CrossRef] [PubMed]
- Dash, S.; Ueda, T.; Komuro, A.; Honda, M.; Sugisawa, R.; Okada, H. Deoxycytidine kinase inactivation enhances gemcitabine resistance and sensitizes mitochondrial metabolism interference in pancreatic cancer. Cell Death Dis. 2024, 12, 131. [Google Scholar] [CrossRef]
- Stephenson, D.; Smith, J.A.S. Nitrogen-14 quadrupole cross-relaxation spectroscopy. Proc. R. Soc. Lond. Ser. A Math. Phys. Sci. 1988, 416, 149–178. [Google Scholar] [CrossRef]
- Latosińska, J.N.; Latosińska, M.; Seliger, J.; Žagar, V.; Apih, T.; Grieb, P. Elucidating the Role of Noncovalent Interactions in Favipiravir, a Drug Active against Various Human RNA Viruses; a 1H-14N NQDR/Periodic DFT/QTAIM/RDS/3D Hirshfeld Surfaces Combined Study. Molecules 2023, 28, 3308. [Google Scholar] [CrossRef] [PubMed]
- Gregorovič, A.G.; Apih, T.; Seliger, J. 1H–14N cross-relaxation spectrum analysis in sildenafil and sildenafil citrate. Solid State Nucl. Magn. Reson. 2016, 78, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 16, Revision C.01; Gaussian, Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Kohn, W.; Sham, L.J. Self-Consistent Equations Including Exchange and Correlation Effects. Phys. Rev. 1965, 140, A1133–A1138. [Google Scholar] [CrossRef]
- Levy, M. Universal Variational Functionals of Electron Densities, First-Order Density Matrices, and Natural Spin-Orbitals and Solution of the v-Representability Problem. Proc. Natl. Acad. Sci. USA 1979, 76, 6062–6065. [Google Scholar] [CrossRef]
- Zhao, Y.; Truhlar, D.G. The M06 Suite of Density Functionals for Main Group Thermochemistry, Thermochemical Kinetics, Noncovalent Interactions, Excited States, and Transition Elements: Two New Functionals and Systematic Testing of Four M06-Class Functionals and 12 Other Functionals. Theor. Chem. Acc. 2008, 120, 215–241. [Google Scholar] [CrossRef]
- Latosińska, J.N.; Latosińska, M.; Seliger, J.; Žagar, V. Exploring Partial Structural Disorder in Anhydrous Paraxanthine through Combined Experiment, Solid-State Computational Modelling, and Molecular Docking. Processes 2023, 11, 2740. [Google Scholar] [CrossRef]
- Latosińska, J.N.; Latosińska, M.; Koput, J. The Tautomeric Equilibria of Cytosine Studied by NQR Spectroscopy and HF, MP2 and DFT Calculations. J. Mol. Struct. 2003, 648, 9–18. [Google Scholar] [CrossRef]
- Koopmans, T. Über die Zuordnung von Wellenfunktionen und Eigenwerten zu den Einzelnen Elektronen Eines Atoms. Physica 1934, 1, 104–113. [Google Scholar] [CrossRef]
- Parr, R.G.; Pearson, R.G. Absolute Hardness: Companion Parameter to Absolute Electronegativity. J. Am. Chem. Soc. 1983, 105, 7512–7516. [Google Scholar] [CrossRef]
- Gázquez, J.L.; Cedillo, A.; Vela, A. Electrodonating and Electroaccepting Powers. J. Phys. Chem. A 2007, 111, 1966–1970. [Google Scholar] [CrossRef]
- Chattaraj, P.K.; Chakraborty, A.; Giri, S. Net Electrophilicity. J. Phys. Chem. A 2009, 113, 10068–10074. [Google Scholar] [CrossRef] [PubMed]
- Clark, S.J.; Segall, M.D.; Pickard, C.J.; Hasnip, P.J.; Probert, M.I.J.; Refson, K.; Payne, M.C. First Principles Methods Using CASTEP. Z. Kristallogr. Cryst. Mater. 2005, 220, 567–570. [Google Scholar] [CrossRef]
- Perdew, J.P.; Burke, K.; Ernzerhof, M. Generalized Gradient Approximation Made Simple. Phys. Rev. Lett. 1996, 77, 3865–3868. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, W. Comment on “Generalized Gradient Approximation Made Simple”. Phys. Rev. Lett. 1998, 80, 890. [Google Scholar] [CrossRef]
- Tkatchenko, A.; Scheffler, M. Accurate Molecular Van Der Waals Interactions from Ground-State Electron Density and Free-Atom Reference Data. Phys. Rev. Lett. 2009, 102, 073005. [Google Scholar] [CrossRef] [PubMed]
- Pickard, C.J.; Mauri, F. All-electron magnetic response with pseudopotentials: NMR chemical shifts. Phys. Rev. B 2001, 63, 245101. [Google Scholar] [CrossRef]
- Monkhorst, H.J.; Pack, J.D. Special Points for Brillouin-Zone Integrations. Phys. Rev. B 1976, 13, 5188–5192. [Google Scholar] [CrossRef]
- Pyykkö, P. Year-2008 Nuclear Quadrupole Moments. Mol. Phys. 2008, 106, 1965–1974. [Google Scholar] [CrossRef]
- Spackman, M.A.; McKinnon, J.J. Fingerprinting Intermolecular Interactions in Molecular Crystals. CrystEngComm 2002, 4, 378–392. [Google Scholar] [CrossRef]
- Jelsch, C.; Ejsmont, K.; Huder, L. The Enrichment Ratio of Atomic Contacts in Crystals, an Indicator Derived from the Hirshfeld Surface Analysis. IUCrJ 2014, 1, 119–128. [Google Scholar] [CrossRef]
- McConkey, B.J.; Sobolev, V.; Edelman, M. The performance of current methods in ligand-protein docking. Curr. Sci. 2002, 83, 845–855. [Google Scholar]
- Meng, X.-Y.; Zhang, H.X.; Mezei, M.; Cui, M. Molecular Docking: A Powerful Approach for Structure-Based Drug Discovery. Curr. Comput. Aided Drug Des. 2011, 7, 146–157. [Google Scholar] [CrossRef]
- Pinzi, L.; Rastelli, G. Molecular docking: Shifting paradigms in drug discovery. Int. J. Mol. Sci. 2019, 20, 4331. [Google Scholar] [CrossRef]
- Latosińska, J.N.; Latosińska, M.; Maurin, J.K.; Orzeszko, A.; Kazimierczuk, Z. Quantum-Chemical Insight into Structure–Reactivity Relationship in 4,5,6,7-Tetrahalogeno-1H-benzimidazoles: A Combined X-ray, DSC, DFT/QTAIM, Hirshfeld Surface-Based, and Molecular Docking Approach. J. Phys. Chem. A 2014, 118, 2089–2106. [Google Scholar] [CrossRef]
- Rahim, F.; Ullah, H.; Hussain, R.; Taha, M.; Khan, S.; Nawaz, M.; Nawaz, F.; Gilani, S.J.; Bin Jumah, M.N. Thiadiazole based triazole/hydrazone derivatives: Synthesis, in vitro α-glucosidase inhibitory activity and in silico molecular docking study. J. Mol. Struct. 2023, 1287, 135619. [Google Scholar] [CrossRef]
- Khan, S.; Ullah, H.; Taha, M.; Rahim, F.; Sarfraz, M.; Iqbal, R.; Iqbal, N.; Hussain, R.; Ali Shah, S.A.; Ayub, K.; et al. Synthesis, DFT Studies, Molecular Docking and Biological Activity Evaluation of Thiazole-Sulfonamide Derivatives as Potent Alzheimer’s Inhibitors. Molecules 2023, 28, 559. [Google Scholar] [CrossRef]
- Hayat, S.; Ullah, H.; Rahim, F.; Ullah, I.; Taha, M.; Iqbal, N.; Khan, F.; Khan, M.S.; Ali Shah, S.A.; Wadood, A.; et al. Synthesis, biological evaluation and molecular docking study of benzimidazole derivatives as α-glucosidase inhibitors and anti-diabetes candidates. J. Mol. Struct. 2023, 1276, 134774. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef]
- Yang, J.; Chen, C. GEMDOCK: A Generic Evolutionary Method for Molecular Docking. Proteins 2004, 55, 288–304. [Google Scholar] [CrossRef]
- Kurkcuoglu, Z.; Koukos, P.I.; Citro, N.; Trellet, M.E.; Rodrigues, J.P.G.L.M.; Moreira, I.S.; Roel-Touris, J.; Melquiond, A.S.J.; Geng, C.; Schaarschmidt, J.; et al. Performance of HADDOCK and a simple contact-based protein-ligand binding affinity predictor in the D3R Grand Challenge 2. J. Comp. Aid. Mol. Des. 2018, 32, 175–185. [Google Scholar] [CrossRef]
- Vangone, A.; Schaarschmidt, J.; Koukos, P.; Geng, C.; Citro, N.; Trellet, M.E.; Xue, L.; Bonvin, A.M.J.J. Large-scale prediction of binding affinity in protein-small ligand complexes: The PRODIGY-LIG web server. Bioinformatics 2019, 35, 1585–1587. [Google Scholar] [CrossRef]
- Gehlhaar, D.K.; Bouzida, D.; Rejto, P.A. Fully Automated and Rapid Flexible Docking of Inhibitors Covalently Bound to Serine Proteases. In Evolutionary Programming VII; Porto, V.W., Saravanan, N., Waagen, D., Eiben, A.E., Eds.; LNCS: Marikina, Philippines, 1998; Volume 1447, pp. 449–461. [Google Scholar]
- Diedrich, K.; Krause, B.; Berg, O.; Rarey, M. PoseEdit: Enhanced ligand binding mode communication by interactive 2D diagrams. J. Comput. Aided Mol. Des. 2023, 37, 481–503. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
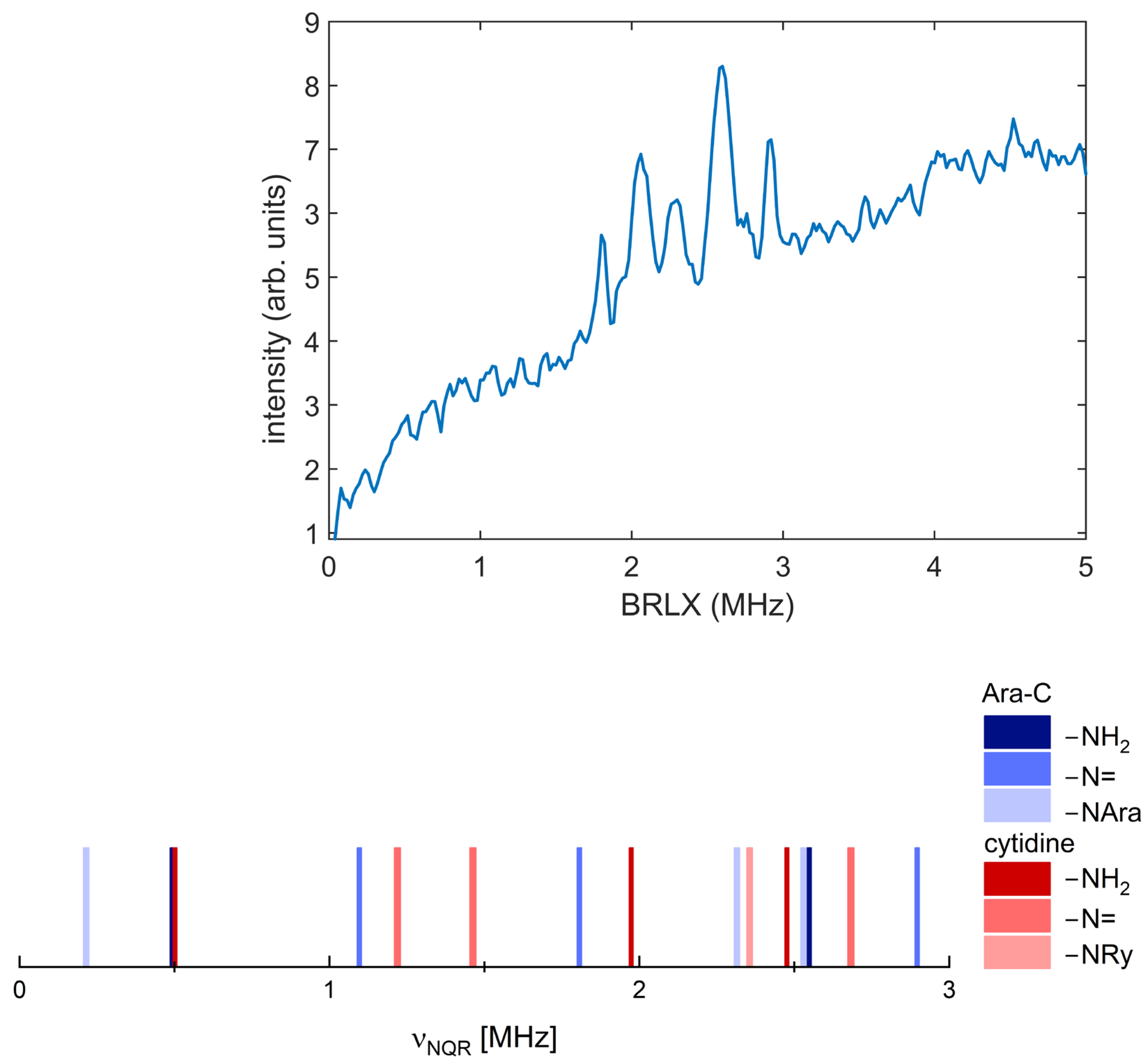









| Compound | Nitrogen Site | ν+ [MHz] | ν− [MHz] | ν0 [MHz] | |e2qQ/h| [MHz] | η | Assignment |
|---|---|---|---|---|---|---|---|
| Ara-C | N(1) | 2.555 | 2.055 | 0.500 | 3.073 | 0.325 | –NH2 |
| N(2) | 2.890 | 1.800 | 1.090 * | 3.147 | 0.712 | –N= | |
| N(3) | 2.530 | 2.315 | 0.215 | 3.230 | 0.133 | >N-sugar | |
| cytidine 1 | N(1) | 2.476 | 1.974 | 0.502 | 2.967 | 0.338 | –NH2 |
| N(2) | 2.683 | 1.463 | 1.220 | 2.769 | 0.883 | –N= | |
| N(3) | 2.356 | - | - | - | - | >N-sugar | |
| 5-azacytidine 1 | N(1) | 2.140 | 1.420 | 0.715 | 2.373 | 0.603 | –NH2 |
| N(2) | 2.740 | 2.120 * | 0.620 | 3.240 | 0.383 | –N(3)= 1 | |
| N(3) | 2.720 | 2.100 * | 0.620 | 3.243 | 0.386 | –N(5)= 1 | |
| N(4) | 2.200 | - | - | - | - | >N-sugar 1 | |
| cytosine 1,2 | N(1) | 2.492 | 1.922 | 0.570 | 2.491 | 0.388 | –NH2 |
| N(2) | 2.723 | 1.570 | 1.153 | 2.862 | 0.806 | –N= | |
| N(3) | 2.028 | 1.219 | 0.801 | 2.165 | 0.740 | >NH |
| Structure | Site | ν+ [MHz] | ν− [MHz] | ν0 [MHz] | e2qQ/h [MHz] | η | s, r2 |
|---|---|---|---|---|---|---|---|
| Ara-C 1 hydrogens optimized | –NH2 | 2.658 | 2.175 | 0.483 | −3.222 | 0.30 | 0.065, 0.997 |
| –N= | 2.835 | 1.970 | 0.865 | −3.203 | 0.54 | ||
| >N-sugar | 2.565 | 2.289 | 0.275 | −3.236 | 0.17 | ||
| Ara-C 1 X-ray hydrogens | –NH2 | 2.568 | 1.677 | 0.891 | −2.83 | 0.63 | |
| –N= | 2.844 | 2.031 | 0.813 | −3.25 | 0.50 | 0.235, 0.940 | |
| >N-sugar | 2.519 | 2.218 | 0.300 | −3.158 | 0.19 | ||
| β-cytidine 2 hydrogens optimized | –NH2 | 2.566 | 2.276 | 0.291 | −3.228 | 0.18 | 0.158, 0.974 |
| –N= | 2.699 | 1.619 | 1.080 | −2.879 | 0.75 | ||
| >N-sugar | 2.406 | 2.221 | 0.185 | −3.085 | 0.12 | ||
| β-cytidine 2 X-ray hydrogens | –NH2 | 2.506 | 1.718 | 0.788 | −2.816 | 0.56 | |
| –N= | 2.709 | 1.744 | 0.965 | −2.969 | 0.65 | 0.229, 0.921 | |
| >N-sugar | 2.335 | 2.113 | 0.222 | −2.965 | 0.15 | ||
| 2′-deoxycytidine form β 3 hydrogens optimized | –NH2 | 2.432 | 1.702 | 0.730 | −2.756 | 0.53 | - |
| –N= | 2.744 | 1.868 | 0.876 | −3075 | 0.57 | ||
| >N-sugar | 2.361 | 2.079 | 0.281 | −2.960 | 0.19 | ||
| 3′-deoxycytidine 4 hydrogens optimized | –NH2 | 3.267 | 2.481 | 0.786 | −3.832 | 0.41 | unreliable result due to poor quality structure |
| –NH2 | 4.473 | 4.020 | 0.453 | −5.662 | 0.16 | ||
| –N= | 2.741 | 1.728 | 1.013 | −2.979 | 0.68 | ||
| –N= | 2.945 | 2.298 | 0.647 | −3.495 | 0.37 | ||
| >N-sugar | 2.582 | 2.275 | 0.308 | −3.238 | 0.19 | ||
| >N-sugar | 2.201 | 1.965 | 0.236 | −2.777 | 0.17 | ||
| 3′-deoxycytidine single molecule optimized geometry | –NH2 | 3.178 | 2.876 | 0.303 | −4.036 | 0.15 | - |
| –N= | 2.958 | 2.216 | 0.742 | −3.449 | 0.43 | ||
| >N-sugar | 2.373 | 2.076 | 0.297 | −2.966 | 0.2 | ||
| 2′,3′-dideoxycytidine (Zalcitabine) 5 hydrogens optimized | –NH2 | 2.525 | 2.151 | 0.374 | −3.117 | 0.24 | - |
| –N= | 2.673 | 1.627 | 1.046 | −2.867 | 0.73 | ||
| >N-sugar | 2.317 | 2.153 | 0.164 | −2.980 | 0.11 | ||
| 5-azacytidine (Vidaza) structure predicted | –NH2 | 2.621 | 2.188 | 0.433 | −3.206 | 0.27 | 0.439, 0.832 |
| –N(3)= | 2.609 | 1.415 | 1.194 | −2.683 | 0.89 | ||
| –N(5)= | 2.842 | 2.453 | 0.388 | −3.530 | 0.22 | ||
| >N-sugar | 2.080 | 1.985 | 0.095 | −2.710 | 0.07 | ||
| 5-azacytidine (Vidaza) geometry optimized single molecule | –NH2 | 3.072 | 2.779 | 0.293 | −3.901 | 0.15 | 0.548, 0.763 |
| –N(3)= | 2.932 | 2.094 | 0.838 | −3.351 | 0.50 | ||
| –N(5)= | 2.763 | 2.338 | 0.425 | −3.401 | 0.25 | ||
| >N-sugar | 2.316 | 1.882 | 0.434 | −2.799 | 0.31 | ||
| cytosine 6 hydrogens optimized | –NH2 | 2.732 | 2.005 | 0.726 | −3.158 | 0.46 | 0.175, 0.955 |
| –N= | 2.643 | 1.643 | 1.000 | −2.857 | 0.70 | ||
| >NH | 2.243 | 1.537 | 0.706 | −2.520 | 0.56 |
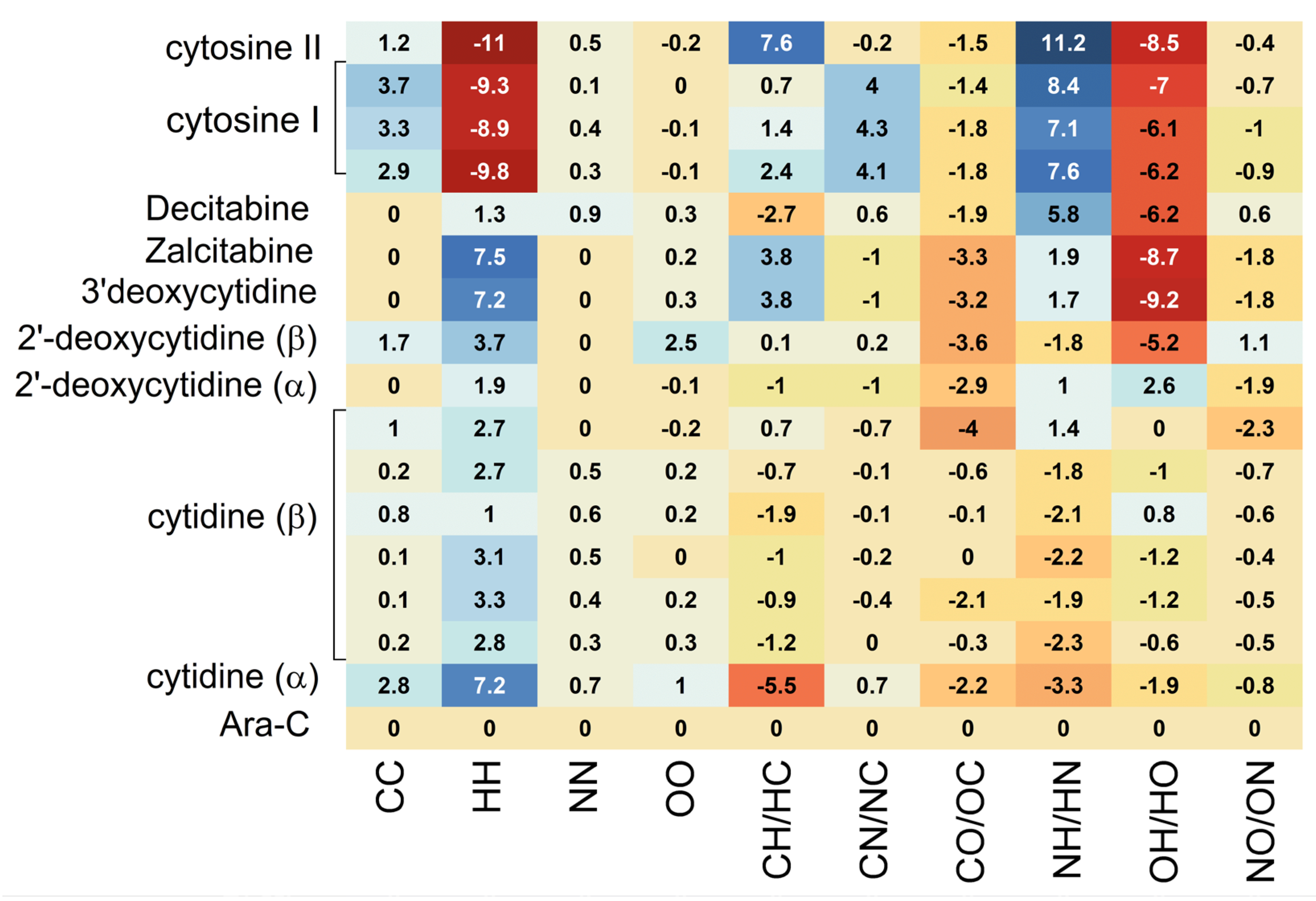
| Compound | Structure | Homonuclear | Heteronuclear | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CC | HH | NN | OO | CH/HC | CN/NC | CO/OC | NH/HN | OH/HO | NO/ON | ||
| Ara-C | 6.4%, 295 K [30] | 0 | 41.5 | 0 | 0.2 | 8.8 | 1.0 | 4.1 | 12 | 31.2 | 2.6 |
| α-cytidine | 3.3%, 295 K [38] | 2.8 | 48.7 | 0.7 | 1.2 | 3.3 | 1.7 | 1.9 | 8.7 | 29.3 | 1.8 |
| β-cytidine | 2.87%, 295 K [39] | 0.2 | 44.3 | 0.3 | 0.5 | 7.6 | 1.0 | 3.8 | 9.7 | 30.6 | 2.1 |
| 2.32%, 297.2 K [31] | 0.1 | 44.8 | 0.4 | 0.4 | 7.9 | 0.6 | 2.0 | 10.1 | 30.0 | 2.1 | |
| 4.57%, 2 K [40] | 0.1 | 44.6 | 0.5 | 0.2 | 7.8 | 0.8 | 4.1 | 9.8 | 30.0 | 2.2 | |
| 9.88%, 293 K [35] | 0.8 | 42.5 | 0.6 | 0.4 | 6.9 | 0.9 | 4.0 | 9.9 | 32.0 | 2.0 | |
| 5.6%, 295 K [41] | 0.2 | 44.2 | 0.5 | 0.4 | 8.1 | 0.9 | 3.5 | 10.2 | 30.2 | 1.9 | |
| 2′-deoxycytidine, α | 3.65%, 100 K [42] | 1.0 | 44.2 | 0 | 0 | 9.5 | 0.3 | 0.1 | 13.4 | 31.2 | 0.3 |
| 2′-deoxycytidine, β | 5.4%, 295 K [32] | 0 | 43.4 | 0 | 0.1 | 7.8 | 0.0 | 1.2 | 13 | 33.8 | 0.7 |
| 3′-deoxycytidine | 7.3%, 295 K [33] | 1.7 | 45.2 | 0 | 2.7 | 8.9 | 1.2 | 0.5 | 10.2 | 26.0 | 3.7 |
| 2′,3′-dideoxycytidine (Zalcitabine) | 5%, 295 K [43] | 0 | 48.7 | 0 | 0.5 | 12.6 | 0 | 0.9 | 13.7 | 22.0 | 0.8 |
| 3.7%, 295 K [34] | 0 | 49 | 0 | 0.4 | 12.6 | 0.0 | 0.8 | 13.9 | 22.5 | 0.8 | |
| 5-aza-2′-deoxycytidine (Decitabine) | 2.96% 150 K [44] | 0 | 42.8 | 0.9 | 0.5 | 6.1 | 1.6 | 2.2 | 17.8 | 25.0 | 3.2 |
| cytosine, form I | 7.0%, 295 K [45] | 2.9 | 31.7 | 0.3 | 0.1 | 11.2 | 5.1 | 2.3 | 19.6 | 25.0 | 1.7 |
| 3.1%, 295 K [46] | 3.3 | 32.6 | 0.4 | 0.1 | 10.2 | 5.3 | 2.3 | 19.1 | 25.1 | 1.6 | |
| 0.647%, 293 K [47] | 3.7 | 32.2 | 0.1 | 0.2 | 9.5 | 5 | 2.7 | 20.4 | 24.2 | 1.9 | |
| cytosine, form II | 5.15%, 294 K [48] | 1.2 | 30.5 | 0.5 | 0 | 16.4 | 0.8 | 2.6 | 23.2 | 22.7 | 2.2 |
| Structure | ED | RMSD |
|---|---|---|
| cytidine α | 10.58 | 3.34 |
| cytidine, β | 3.95 * | 1.25 * |
| 2′-deoxycytidine, α | 5.71 | 1.80 |
| 2′-deoxycytidine, β | 5.04 * | 1.59 * |
| 3′-deoxycytidine | 8.21 | 2.59 |
| 2′,3′-dideoxycytidine (Zalcitabine) | 12.91 * | 4.08 * |
| 5-aza-2′-deoxycytidine (Decitabine) | 9.29 | 2.94 |
| cytosine | 14.92 * | 4.27 * |
| Structure | Atom | C | H | N | O |
|---|---|---|---|---|---|
| Ara-C | Surface % | 6.95 | 67.5 | 7.05 | 18.4 |
| C | 0 | - | - | - | |
| H | 0.94 | 0.91 | - | - | |
| N | 1.02 | 1.26 | 0 | - | |
| O | 1.60 | 1.26 | 0.42 | 0.06 | |
| cytidine α-form | Surface % | 6.25 | 69.3 | 6.8 | 17.65 |
| C | 7.17 | - | - | - | |
| H | 0.38 | 1.01 | - | - | |
| N | 2.00 | 0.92 | 1.51 | - | |
| O | 0.86 | 1.19 | 0.75 | 0.39 | |
| cytidine β-form | Surface % | 6.20 | 68.25 | 5.80 | 17.75 |
| C | 0.26 | - | - | - | |
| H | 0.93 | 0.96 | - | - | |
| N | 0.71 | 1.09 | 0.87 | - | |
| O | 1.68 | 1.19 | 0.87 | 0.13 | |
| 2′-deoxycytidine α-form | Surface % | 5.9 | 71.27 | 7 | 15.77 |
| C | 2.87 | - | - | - | |
| H | 1.13 | 0.87 | - | - | |
| N | 0.36 | 1.34 | 0.00 | - | |
| O | 0.00 | 1.39 | 0.14 | 0.00 | |
| 2′-deoxycytidine β-form | Surface % | 4.5 | 70.7 | 6.8 | 17.9 |
| C | 0.00 | - | - | - | |
| H | 1.23 | 0.87 | - | - | |
| N | 0.00 | 1.35 | 0.00 | - | |
| O | 0.74 | 1.34 | 0.25 | 0.03 | |
| 3′-deoxycytidine | Surface % | 7 | 67.8 | 7.6 | 17.8 |
| C | 3.47 | - | - | - | |
| H | 0.94 | 0.98 | - | - | |
| N | 1.14 | 1.00 | 0.00 | - | |
| O | 0.20 | 1.08 | 1.38 | 0.85 | |
| Surface % | 6.7 | 73.5 | 7.3 | 12.5 | |
| Zalcitabine | C | 0.00 | - | - | - |
| H | 1.28 | 0.91 | - | - | |
| N | 0.00 | 1.29 | 0.44 | - | |
| O | 0.48 | 1.23 | 0.26 | 0.00 | |
| Surface % | 4.95 | 67.25 | 12.2 | 15.7 | |
| C | 0.00 | - | - | - | |
| Decitabine | H | 0.92 | 0.95 | - | - |
| N | 1.32 | 1.08 | 0.60 | - | |
| O | 1.42 | 1.18 | 0.84 | 0.20 | |
| cytosine | Surface % | 12.3 | 59.25 | 13.8 | 14.65 |
| C | 2.45 | - | - | - | |
| H | 0.65 | 0.92 | - | - | |
| N | 1.47 | 1.25 | 0.05 | - | |
| O | 0.75 | 1.39 | 0.49 | 0.09 |
| Structure | ED | RMSD |
|---|---|---|
| cytidine α | 7.33 (1.58 *) | 1.46 (0.26 *) |
| cytidine, β | 1.02 | 0.17 |
| 2′-deoxycytidine, α | 2.93 | 0.49 |
| 2′-deoxycytidine, β | 0.60 | 0.10 |
| 3′-deoxycytidine | 3.48 | 0.58 |
| 2′,3′-dideoxycytidine (Zalcitabine) | 0.74 | 0.12 |
| 5-aza-2′-deoxycytidine (Decitabine) | 0.72 | 0.12 |
| cytosine | 2.50 | 0.42 |
| Compound | Hydrogen Bond | RA∙∙∙B [Å] | <AHB [°] | Ee [kJ/mol] | Ep [kJ/mol] | Ed [kJ/mol] | Er [kJ/mol] | Etot [kJ/mol] | Moiety |
|---|---|---|---|---|---|---|---|---|---|
| Ara-C | OH∙∙∙O * | 2.650 | 155.45 | - | - | - | - | - | Ry |
| CH∙∙∙O * | 3.685 | 157.20 | - | - | - | - | - | Ry | |
| NH∙∙∙O | 2.983 | 163.70 | −32.7 | −8.9 | −21.1 | 28.1 | −42.3 | –NH2 | |
| NH∙∙∙O | 3.048 | 145.73 | −16.3 | −10.2 | −27 | 29 | −30.4 | –NH2 | |
| OH∙∙∙O | 3.003 | 159.64 | −33.4 | −9.3 | −23.4 | 30.5 | −43.7 | Ry | |
| OH∙∙∙O | 2.720 | 171.72 | −74.7 | −25.5 | −20.5 | 66.6 | −74.5 | Ry | |
| CH∙∙∙N | 3.652 | 159.76 | –N= | ||||||
| Cytidine α-form | CH∙∙∙O * | 2.662 | 101.87 | - | - | - | - | - | C(6)H/Ry |
| CH∙∙∙O * | 2.716 | 98.15 | - | - | - | - | - | =O/Ry | |
| NH∙∙∙O | 2.946 | 171.58 | −28.1 | −6.6 | −12.6 | 29.2 | −27.5 | –NH2 | |
| NH∙∙∙O | 3.183 | 144.97 | −30.4 | −8.7 | −15 | 18.7 | −40.0 | –NH2 | |
| CH∙∙∙O | 3.292 | 141.69 | =O | ||||||
| OH∙∙∙O | 2.674 | 151.65 | −45.3 | −9.3 | −13.1 | 49.6 | −35.6 | Ry | |
| OH∙∙∙N | 2.752 | 168.40 | −132.5 | −38.9 | −57 | 169.4 | −113.9 | –N= | |
| π···π stacking | 3.473 | - | rings | ||||||
| Cytidine β-form | CH∙∙∙O * | 3.233 | 151.50 | - | - | - | - | - | Ry |
| CH∙∙∙O * | 2.676 | 92.10 | - | - | - | - | - | =O/Ry | |
| CH∙∙∙O * | 2.347 | 97.60 | - | - | - | - | - | C(6)H/Ry | |
| NH∙∙∙O | 2.946 | 147.84 | −54.4 | −10.5 | −15.3 | 43.4 | −51.8 | –NH2 | |
| NH∙∙∙O | 2.923 | 145.81 | –NH2 | ||||||
| OH∙∙∙N | 2.860 | 174.31 | −114.8 | −35.9 | −24.6 | 115.0 | −98.3 | –N= | |
| OH∙∙∙O | 2.711 | 174.79 | Ry | ||||||
| CH∙∙∙O | 3.346 | 152.44 | 1.9 | −2.9 | −11.2 | 12.2 | −2.4 | C(5)/OH(Ry) | |
| 2′-deoxycytidine α form | CH∙∙∙O * | 2.689 | 85.46 | - | - | - | - | - | =O |
| OH∙∙∙O | 2.716 | 150.58 | −92.2 | −27 | −26.6 | 89 | −86.4 | =O | |
| NH∙∙∙O | 2.937 | 157.77 | –NH2 | ||||||
| CH∙∙∙O | 3.317 | 142.59 | Ry | ||||||
| NH∙∙∙O | 2.949 | 167.22 | −33.3 | −8.9 | −31.8 | 47.7 | −40.0 | –NH2 | |
| 2′-deoxycytidine β form | CH∙∙∙HO | 3.040 | 98.37 127.10 | - | - | - | - | - | - |
| CH∙∙∙O | 2.716 | 103.73 | - | - | - | - | - | =O/Ry | |
| CH∙∙∙O | 3.384 | 157.63 | - | - | - | - | - | - | |
| NH∙∙∙O | 2.967 | 141.59 | −83.2 | −20.3 | −17.2 | 64.3 | −78.3 | –NH2 | |
| NH∙∙∙N | 3.018 | 156.22 | –N= | ||||||
| NH∙∙∙O | 3.009 | 153.35 | −49.7 | −15.3 | −23.8 | 52.8 | −52.0 | –NH2 | |
| OH∙∙∙O | 3.153 | 137.88 | Ry | ||||||
| 3′-deoxycytidine | CH∙∙∙O * | 2.753 | 111.88 | - | - | - | - | - | C(6)H/Ry |
| CH∙∙∙O * | 2.687 | 95.40 | - | - | - | - | - | =O/Ry | |
| OH∙∙∙HO | 2.820 | 117.07 | −5.0 | −17.3 | −14.3 | 58.2 | 5.5 | Ry | |
| NH∙∙∙O | 2.858 | 143.47 | −39.8 | −11.2 | −14.2 | 35.6 | −40.7 | –NH2 | |
| NH∙∙∙O | 3.503 | 144.35 | −0.1 | −10 | −16.9 | 29.5 | −4 | –NH2 | |
| Zalcitabine | CH∙∙∙O * | 3.226 | 168.19 | - | - | - | - | - | Ry |
| CH∙∙∙O * | 2.726 | 95.53 | - | - | - | - | - | C(6)H/Ry | |
| CH∙∙∙O * | 2.682 | 95.91 | - | - | - | - | - | =O/Ry | |
| NH∙∙∙O | 2.977 | 166.77 | −37 | −11.6 | −13.7 | 30 | −41.2 | –NH2 | |
| NH∙∙∙O | 3.268 | 159.33 | −24.8 | −4.8 | −15.7 | 15.1 | −34.2 | –NH2 | |
| OH∙∙∙N | 2.774 | 168.64 | −73.5 | −23.3 | −26.4 | 75.3 | −62.8 | –N= | |
| Decitabine | CH∙∙∙O * | 2.687 | 98.61 | - | - | - | - | - | C(6)H/Ry |
| NH∙∙∙N | 2.873 | 166.70 | −89.2 | −22.9 | −18.7 | 104.8 | −62.9 | –NH2 | |
| OH∙∙∙O | 2.678 | 170.94 | −52.5 | −15.8 | −16.2 | 60.0 | −44.2 | =O | |
| OH∙∙∙O | 2.742 | 153.01 | −38.4 | −9.1 | −23.2 | 51.6 | −35.7 | Ry | |
| Cytosine | NH∙∙∙O | 3.022 | 166.42 | −101.4 | −27 | −17.5 | 84.8 | −90.1 | –NH2 |
| NH∙∙∙N | 2.791 | 144.49 | −41.2 | −10.8 | −5.6 | 23.5 | −42.9 | –N= | |
| NH∙∙∙O | 2.991 | 165.21 | =O | ||||||
| π···π stacking | 3.81 | - | 20.2 | −5/5 | −26.1 | 14.3 | 3.3 | - |
| Compound | RMSD_BM | ||||
|---|---|---|---|---|---|
| Total | HH | OH | NH | CH | |
| cytidine α | 0.1385 | 0.1226 | 0.1057 | 0.0989 | 0.0967 |
| cytidine, β | 0.1217 | 0.1177 | 0.1040 | 0.1029 | 0.0900 |
| 2′-deoxycytidine, α | 0.1130 | 0.1011 | 0.1089 | 0.1040 | 0.0987 |
| 2′-deoxycytidine, β | 0.1255 | 0.1062 | 0.1088 | 0.1115 | 0.0998 |
| 3′-deoxycytidine | 0.1267 | 0.1118 | 0.1088 | 0.1052 | 0.0991 |
| 2′,3′-dideoxycytidine (Zalcitabine) | 0.1165 | 0.1080 | 0.1098 | 0.1040 | 0.0955 |
| 5-aza-2′-deoxycytidine (Decitabine) | 0.1161 | 0.1046 | 0.1055 | 0.1001 | 0.0865 |
| cytosine | 0.1307 | 0.1143 | 0.1138 | 0.1056 | 0.0931 |
| Parameter | Ara-C | 2′-deoxycytidine, 1P60 | 2′-deoxycytidine, 1P61 | 2′-deoxycytidine | cytidine | Vidaza (5-azacytidine) | 3′-deoxycytidine | Zalcitabine (2′-3′-dideoxycytidine) | 3′-dehydroxy, 3,4′-didehydrocytidine | Gemcitabine, (2′, 2′-difluoro, 2′deoxycytidine), 1P62 | Gemcitabine (2′, 2′-difluoro,2′deoxycytidine) | Decitabine (5-aza-2′-deoxycytidine) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total energy | −105.083 | −101.556 | −110.763 | −99.252 | −94.384 | −99.241 | −101.853 | −96.519 | −102.211 | −96.221 | −100.881 | −96.171 |
| Protein-ligand | −120.891 | −114.636 | −114.816 | −111.000 | −105.813 | −107.250 | −109.095 | −103.851 | −101.898 | −116.96 | −121.854 | −108.103 |
| Steric | −103.105 | −100.597 | −101.318 | −97.634 | −97.468 | −95.089 | −97.641 | −94.237 | 95.288 | −102.44 | −104.982 | −93.800 |
| Hydrogen bonding | −19.582 | −14.040 | −13.498 | −15.221 | −12.397 | −12.667 | −12.498 | −10.000 | −11.310 | −14.821 | −16.872 | −14.299 |
| Mg2+-ligand | −0.149 | 0 | 0 | −0.047 | −0.300 | −0.225 | −0.176 | −0.073 | −0.313 | −0.094 | −0.153 | −0.049 |
| Binding affinity * | −25.40 | −23.07 | −23.62 | −24.58 | −21.82 | −21.30 | −22.37 | −24.18 | −22.16 | −30.76 | −31.56 | −23.62 |
| Binding affinity ** | −6.90 | −6.84 | −6.75 | −6.81 | −6.87 | −6.06 | −6.84 | −6.70 | −6.79 | −7.54 | −7.33 | −6.30 |
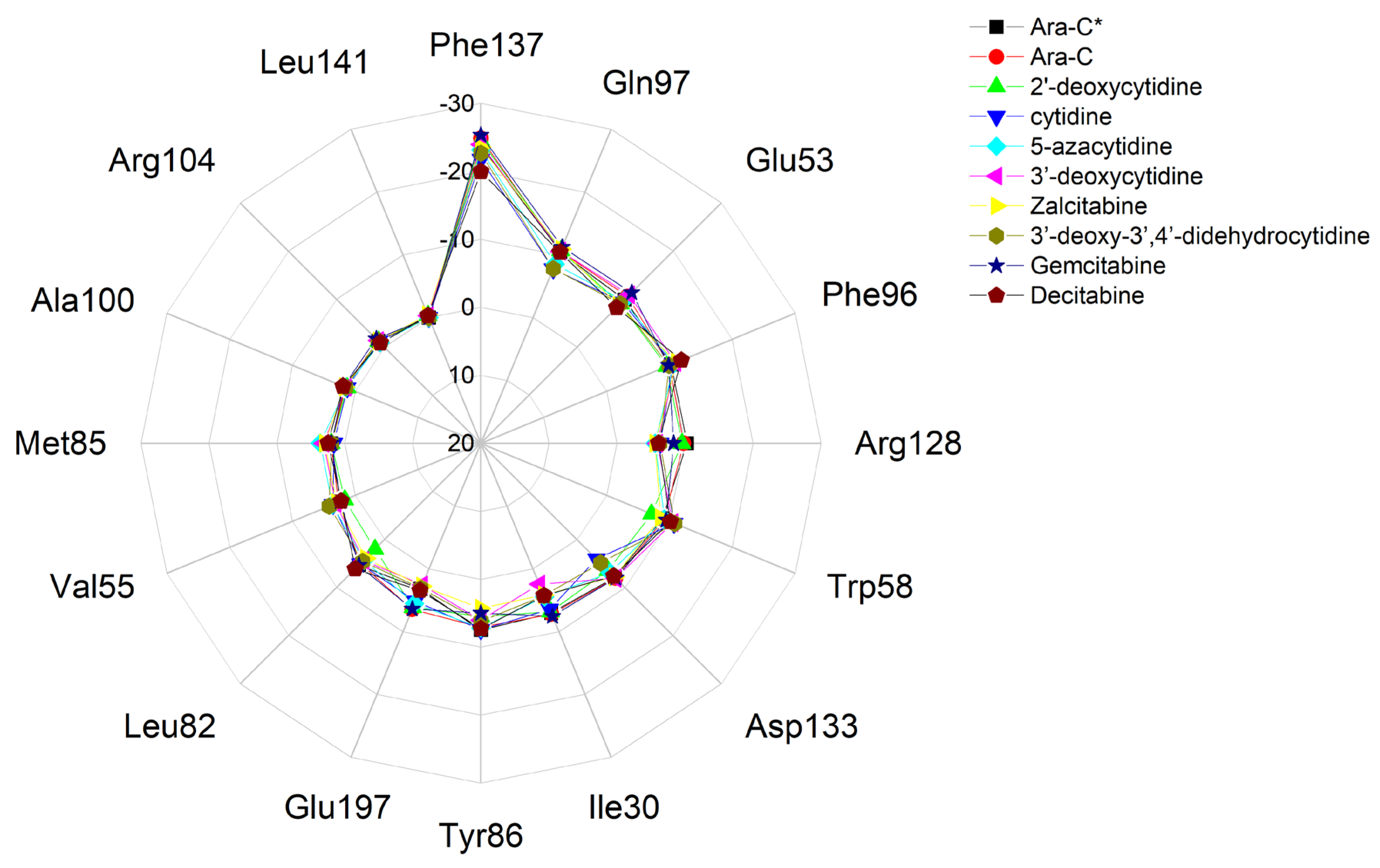
| Residue | Ara-C 15PZ | Ara-C | 2′-deoxycytidine, 1P60 | 2′-deoxycytidine, 1P61 | 2′-deoxycytidine | Cytidine | 5-azacytidine | 3′-deoxycytidine | Zalcitabine | 3′-dehydroxy, 3,4′-didehydrocytidine | Gemcitabine, 1P62 | Gemcitabine | Decitabine |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Phe137 | −23.73 | −24.86 | −22.425 | −24.102 | −24.51 | −21.75 | −23.16 | −23.97 | −23.52 | −22.668 | −26.063 | −25.343 | −19.972 |
| Gln97 | −10.46 | −10.55 | −9.412 | −10.040 | −10.67 | −7.77 | −8.51 | −10.58 | −10.91 | −7.796 | −10.135 | −11.251 | −10.409 |
| Glu53 | −9.87 | −10.53 | −8.544 | −6.823 | −9.00 | −9.34 | −9.68 | −10.89 | −8.61 | −9.036 | −10.741 | −11.348 | −8.230 |
| Phe96 | −10.30 | −10.03 | −10.662 | −10.039 | −9.60 | −10.57 | −10.71 | −10.98 | −11.17 | −9.935 | −9.887 | −9.895 | −11.919 |
| Arg128 | −10.32 | −9.83 | −8.098 | −8.338 | −9.55 | −6.34 | −5.62 | −5.95 | −5.61 | −6.320 | −6.922 | −8.340 | −6.112 |
| Trp58 | −8.89 | −9.15 | −10.177 | −10.736 | −7.17 | −10.73 | −9.18 | −10.52 | −8.59 | −10.828 | −8.875 | −9.480 | −10.151 |
| Asp133 | −7.89 | −8.05 | −7.168 | −7.856 | −6.42 | −4.07 | −6.89 | −8.08 | −7.76 | −4.878 | −7.972 | −8.040 | −7.635 |
| Ile30 | −7.03 | −7.19 | −6.473 | −5.779 | −6.80 | −6.43 | −4.45 | −2.43 | −4.16 | −4.224 | −6.875 | −7.546 | −4.279 |
| Tyr86 | −7.50 | −7.07 | −8.099 | −7.299 | −5.50 | −7.42 | −7.18 | −6.02 | −4.28 | −6.2 | −5.787 | −4.994 | −7.296 |
| Glu197 | −3.19 | −6.44 | −2.983 | −5.422 | −6.16 | −5.16 | −5.50 | −2.56 | −2.75 | −3.092 | −5.961 | −6.378 | −3.423 |
| Leu82 | −5.34 | −5.03 | −6.464 | −6.028 | −2.02 | −5.05 | −4.37 | −4.37 | −3.96 | −4.592 | −5.804 | −5.372 | −6.131 |
| Val55 | −2.40 | −2.71 | −2.405 | −3.650 | −1.65 | −3.55 | −3.62 | −3.13 | −2.95 | −4.163 | 2.626 | −2.361 | −2.248 |
| Met85 | −2.03 | −2.01 | −1.965 | −1.903 | −2.02 | −1.61 | −3.62 | −3.13 | −2.95 | −1.950 | −1.864 | −2.002 | −2.471 |
| Ala100 | −1.53 | −1.66 | −1.467 | −1.367 | −1.29 | −1.29 | −1.49 | −1.49 | −1.65 | −1.524 | −1.418 | −1.858 | −1.955 |
| Arg104 | −1.31 | −1.38 | −1.359 | −1.219 | −1.26 | −1.21 | −0.96 | −1.39 | −1.30 | −1.387 | −1.365 | −1.744 | −0.895 |
| Leu141 | 0.00 | 0.00 | 0 | 0 | −0.36 | 0.00 | 0.00 | −0.34 | −0.47 | 0 | 0 | 0 | −0.330 |
| Arg194 | 0.00 | 0.00 | −0.405 | 0 | 0.00 | −0.33 | 0.00 | 0.00 | 0.00 | 0 | 0 | 0 | 0 |
| Ile200 | −0.48 | −0.46 | 0 | −0.47 | −0.52 | −0.33 | −0.31 | 0.00 | 0.00 | 0 | −0.573 | −0.539 | −0.420 |
| Residue | Ara-C, 1P5Z | Ara-C | 2′-deoxycytidine, 1P60 | 2′-deoxycytidine, 1P61 | 2′-deoxycytidine | cytidine | 5-azacytidine | 3′-deoxycytidine | Zalcitabine | 3′-dehydro-3,4-dideoxycytitine | Gemcitabine, 1P62 | Gemcitabine | Decitabine |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RMSD_BM | - | 0.878 | - | - | 1.378 | 1.739 | 1.622 | 1.717 | 1.666 | 1.738 | - | 1.259 | 1.230 |
| RMSD_BM * | - | 0.878 | 0.913 | 1.230 | - | - | - | - | - | - | 1.786 | - | - |
| RMSD_BM ** | - | 0.878 | 1.933 | 1.660 | - | - | - | - | - | - | 1.354 | - | - |
| RMSD_BM *** | - | - | 0.989 | 0.989 | - | - | - | - | - | - | - | - | - |
| Residue | Ara-C | 2′-deoxycytidine | Cytidine | 5-azacytidine | 3′-deoxycytidine | Zalcitabine | 3′deoxy-3′,4′-didehydrocytidine | Gemcitabine | Decitabine |
|---|---|---|---|---|---|---|---|---|---|
| Phe137 | π···π | π···π | π···π | π···π | π···π | π···π | π···π | π···π | π···π |
| hydrophobic | hydrophobic | hydrophobic | hydrophobic | hydrophobic | hydrophobic | hydrophobic | hydrophobic | hydrophobic | |
| Gln97 | –NH2, –N= | –NH2, –N= | –NH2, –N= | –N= | –NH2 | –NH2, –N= | –NH2, –N= | –NH2, –N= | –NH2, –N= |
| Glu53 | CH2OH | CH2OH | CH2OH | CH2OH | CH2OH | CH2OH | CH2OH | CH2OH | CH2OH |
| Phe96 | hydrophobic | hydrophobic | hydrophobic | hydrophobic | hydrophobic | hydrophobic | hydrophobic | hydrophobic | hydrophobic |
| Arg128 | OH, CH2OH | CH2OH | - | - | - | - | - | CH2OH | CH2OH |
| Trp58 | - | - | - | - | - | - | - | - | - |
| Asp133 | NH2 | NH2 | NH2 | NH2 | NH2 | NH2 | NH2 | NH2 | NH2, |
| Ile30 | - | - | - | - | - | - | - | - | - |
| Tyr86 | OH | OH | OH | OH | OH | - | OH | OH | OH |
| Glu197 | OH | OH | OH | OH | - | - | - | OH | - |
| ϕ [°] | Ara-C | 2′-deoxycytidine 1P60 | 2′-deoxycytidine 1P61 | 2′-deoxycytidine | Cytidine | Cytidine | 5-azacytidine | 3′-deoxycytidine | Zalcitabine | 3′dehydro-3,4′-dideoxy | Gemcitabine, 1P62 | Gemcitabine | Decitabine |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| solid state | 28.83 | - | - | 44.14 | - | 18.30 | - | 35.64 | 23.29 | - | - | - | 18.13 |
| protein-ligand | 36.41 | 49.63 | 41.42 | 47.99 | 55.69 | 46.52 | 48.48 | 50.29 | 66.43 | 48.05 | 47.59 | 38.30 | 18.13 |
| Ligand | HOMO [eV] | LUMO [eV] | Gap [eV] | Χ [eV] | η [eV] | ω [eV] | S [1/eV] | ω+ [eV] | ω− [eV] | Δω [eV] | R+ [−] | R− [−] |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ara-C * | −7.490 | 0.143 | 7.633 | 3.673 | 3.816 | 1.768 | 0.262 | 4.082 | 0.408 | 4.490 | 1.000 | 1.000 |
| Ara-C | −7.321 | 0.204 | 7.525 | 3.559 | 3.762 | 1.683 | 0.266 | 3.933 | 0.374 | 4.307 | 0.964 | 0.916 |
| cytidine | −7.707 | −0.748 | 6.959 | 4.228 | 3.479 | 2.568 | 0.287 | 5.117 | 0.890 | 6.007 | 1.254 | 2.179 |
| 2′-deoxycytidine | −7.308 | 0.151 | 7.458 | 3.578 | 3.729 | 1.717 | 0.268 | 3.972 | 0.394 | 4.366 | 0.973 | 0.965 |
| 3′-deoxycytidine | −7.709 | −0.208 | 7.501 | 3.958 | 3.750 | 2.089 | 0.267 | 4.537 | 0.579 | 5.115 | 1.112 | 1.417 |
| Zalcitabine | −7.549 | 0.079 | 7.628 | 3.735 | 3.814 | 1.828 | 0.262 | 4.173 | 0.438 | 4.611 | 1.022 | 1.072 |
| 5-azacytidine | −8.406 | −0.488 | 7.917 | 4.447 | 3.959 | 2.498 | 0.253 | 5.216 | 0.769 | 5.985 | 1.278 | 1.884 |
| Gemcitabine | −7.122 | 0.107 | 7.229 | 3.507 | 3.614 | 1.702 | 0.277 | 3.907 | 0.400 | 4.307 | 0.957 | 0.979 |
| Decitabine | −7.983 | 0.187 | 8.170 | 3.898 | 4.085 | 1.859 | 0.245 | 4.319 | 0.421 | 4.740 | 1.058 | 1.032 |
| Ara-C | 2′-deoxycytidine | cytidine | Vidaza | 3′-deoxycytidine | Zalcitabine | 3′-dehydroxy, 3,4′-didehydrocytidine | Gemcitabine | Decitabine | |
|---|---|---|---|---|---|---|---|---|---|
| Biological activity | anti-leukemic (DNA, RNA) | DNA component | RNA component | anti-leukemic (RNA, DNA) | anti-virial, anticancer (DNA) | anti-retroviral | anti-virial | anti-neoplastic, anti-cancer, anti-tumor anti-virial | anti-leukemic (DNA) |
| Spectrum of activity * | AML, ALL CML | natural | natural, anti-depressant | MDS, JMML | NHL | HIV/ AIDS | metabolite | HEV, anti-cancer (pancreatic, ovarian, non-small cell lung | MDS, AML, HPV |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Latosińska, J.N.; Latosińska, M.; Seliger, J.; Žagar, V.; Apih, T. Butterfly Effect in Cytarabine: Combined NMR-NQR Experiment, Solid-State Computational Modeling, Quantitative Structure-Property Relationships and Molecular Docking Study. Pharmaceuticals 2024, 17, 445. https://doi.org/10.3390/ph17040445
Latosińska JN, Latosińska M, Seliger J, Žagar V, Apih T. Butterfly Effect in Cytarabine: Combined NMR-NQR Experiment, Solid-State Computational Modeling, Quantitative Structure-Property Relationships and Molecular Docking Study. Pharmaceuticals. 2024; 17(4):445. https://doi.org/10.3390/ph17040445
Chicago/Turabian StyleLatosińska, Jolanta Natalia, Magdalena Latosińska, Janez Seliger, Veselko Žagar, and Tomaž Apih. 2024. "Butterfly Effect in Cytarabine: Combined NMR-NQR Experiment, Solid-State Computational Modeling, Quantitative Structure-Property Relationships and Molecular Docking Study" Pharmaceuticals 17, no. 4: 445. https://doi.org/10.3390/ph17040445
APA StyleLatosińska, J. N., Latosińska, M., Seliger, J., Žagar, V., & Apih, T. (2024). Butterfly Effect in Cytarabine: Combined NMR-NQR Experiment, Solid-State Computational Modeling, Quantitative Structure-Property Relationships and Molecular Docking Study. Pharmaceuticals, 17(4), 445. https://doi.org/10.3390/ph17040445