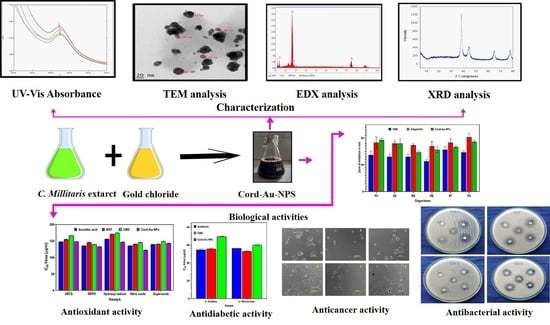
Process Optimization for the Bioinspired Synthesis of Gold Nanoparticles Using Cordyceps militaris, Its Characterization, and Assessment of Enhanced Therapeutic Efficacy
Abstract
:1. Introduction
2. Results and Discussion
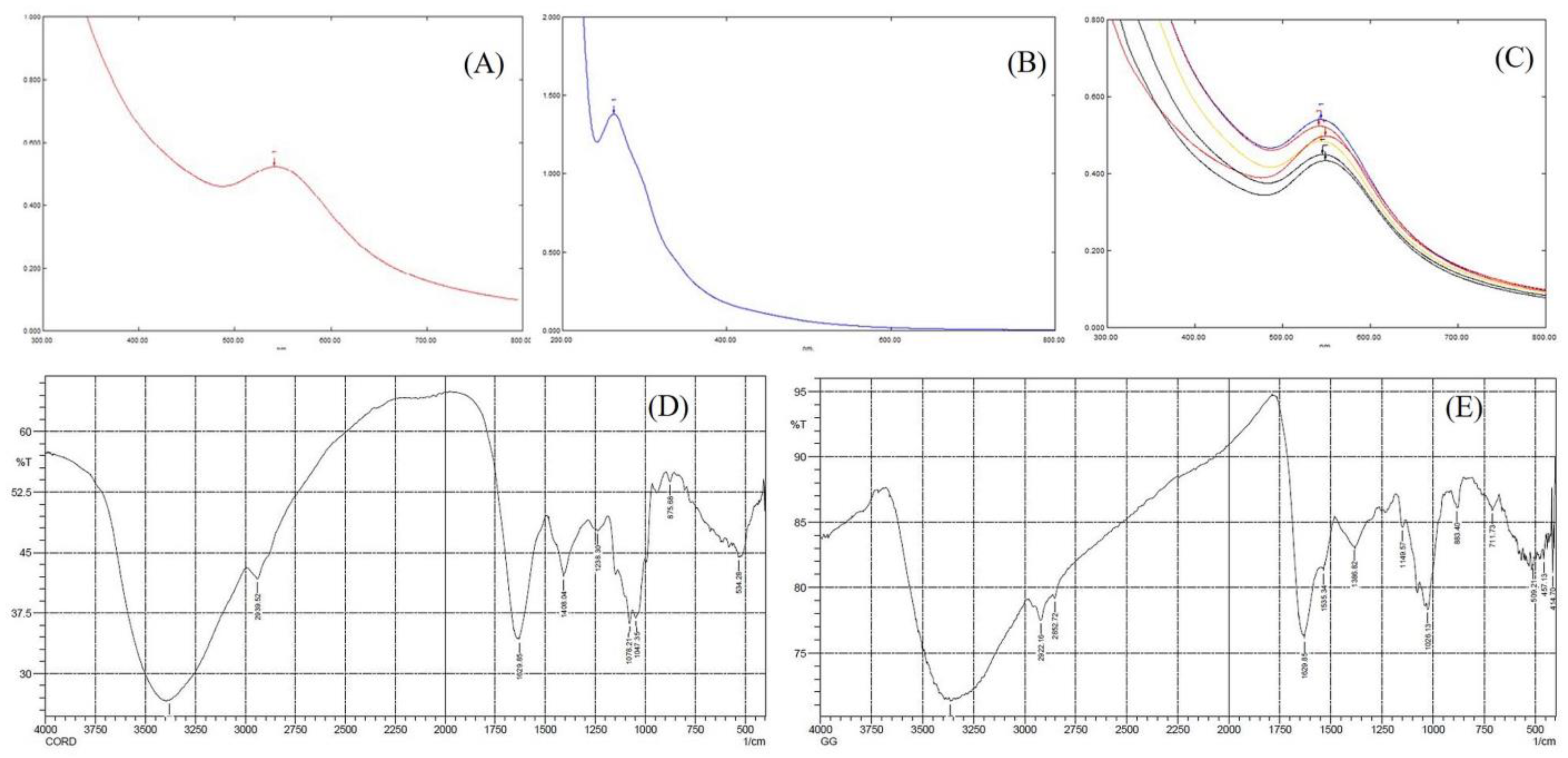
2.1. UV-Visible Spectroscopy Analysis
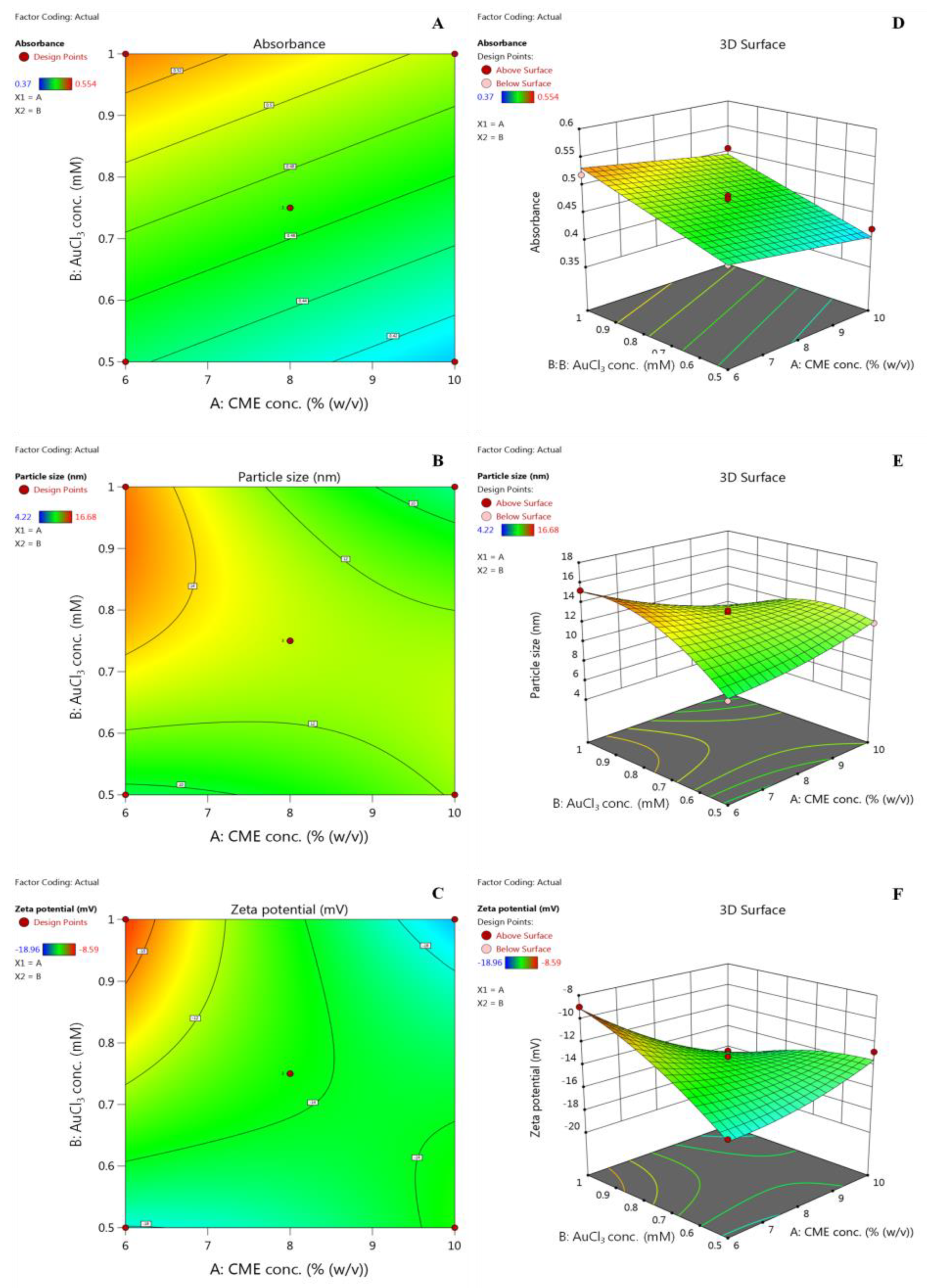
2.2. Optimization by Factorial Design Approach
Effect of Variables on Absorbance, Particle Size, and Zeta Potential
2.3. Model Validation
2.4. Fourier Transform Infrared (FTIR) Spectroscopy
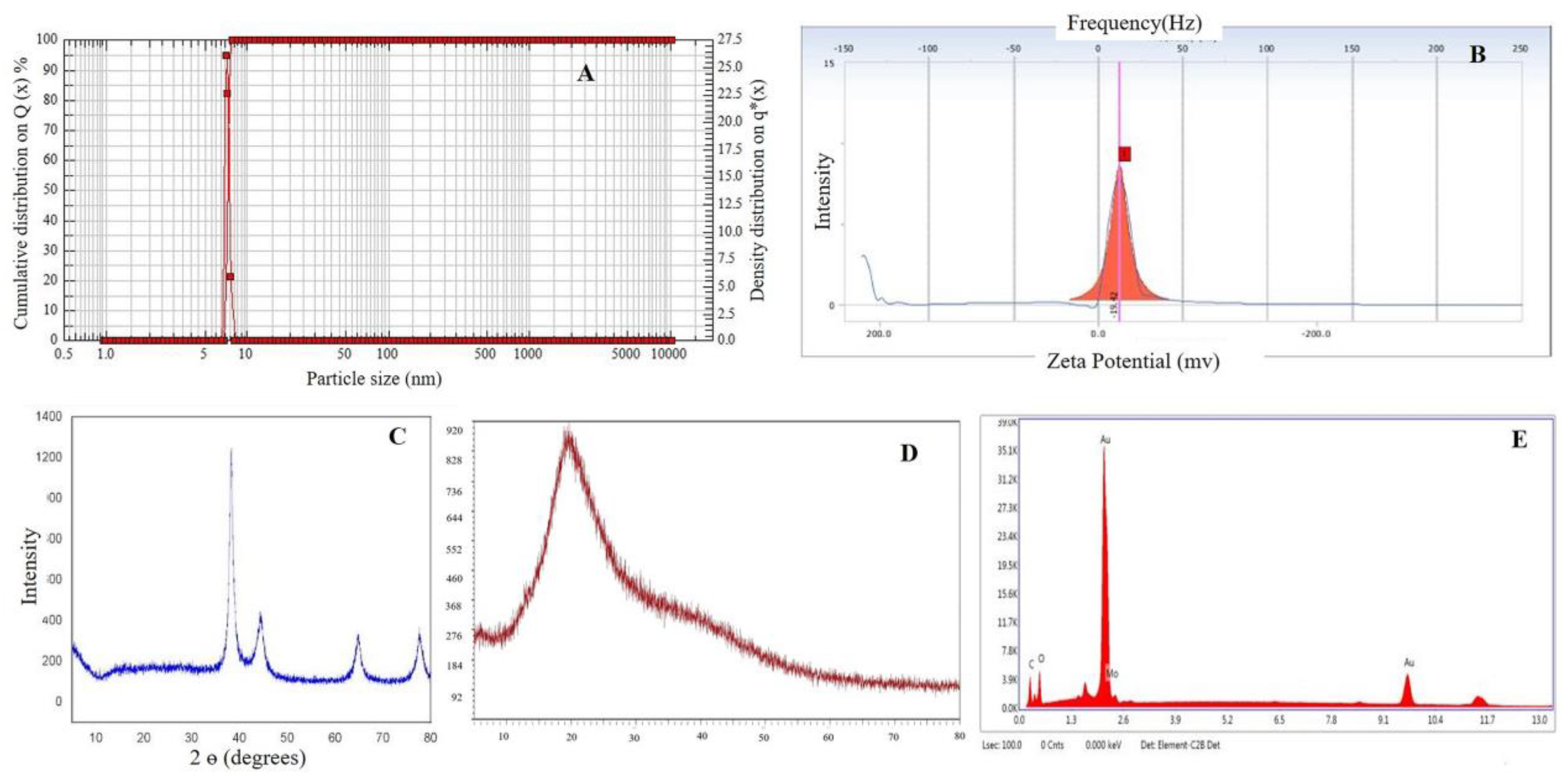
2.5. Morphological Analysis
2.6. Particle Size and Zeta Potential Analysis
2.7. X-ray Diffraction (XRD) Analysis
2.8. Energy-Dispersive X-ray Analysis
2.9. Brine Shrimp Lethality Assay
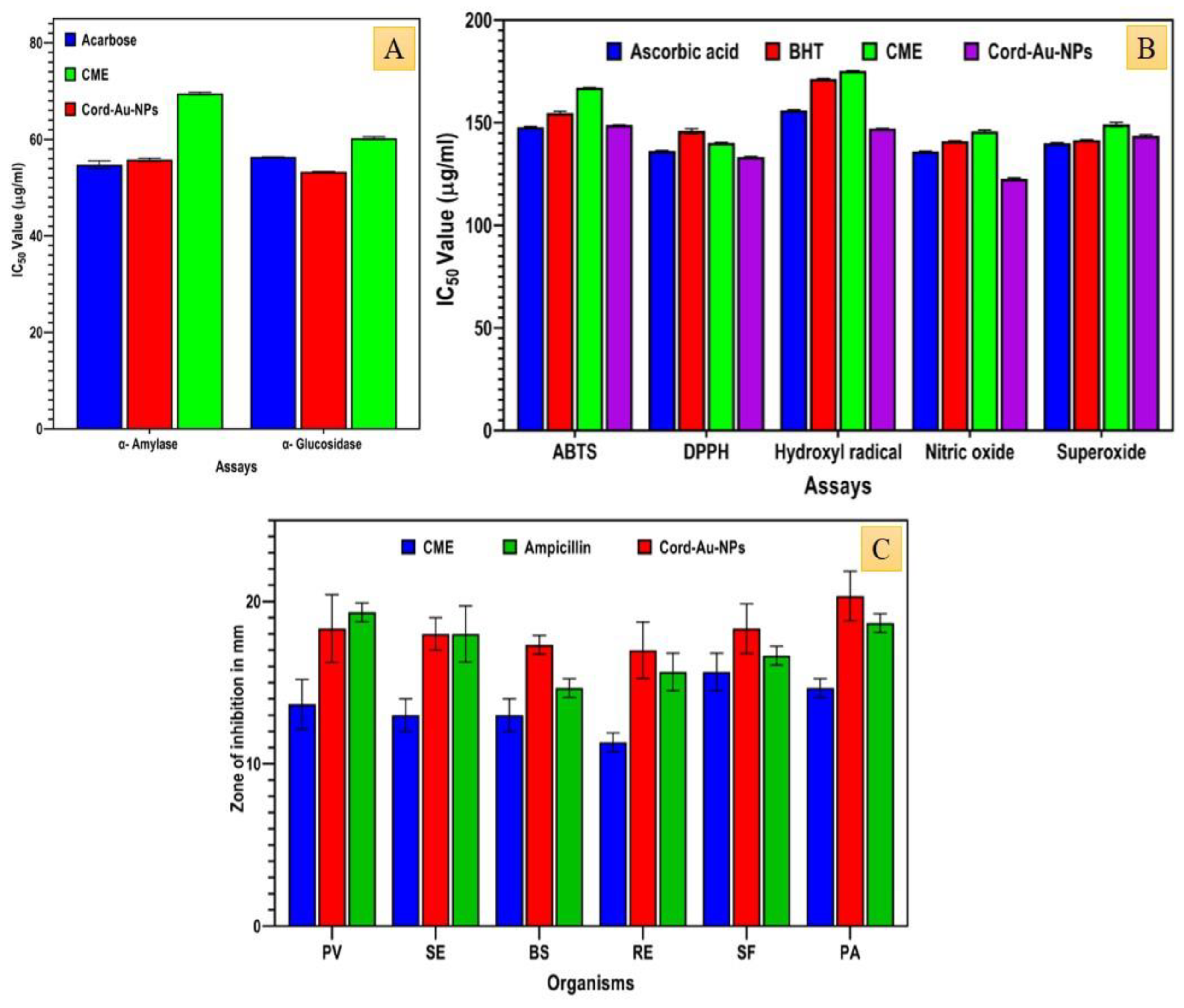
2.10. In Vitro Antidiabetic Activity
2.11. In Vitro Antioxidant Activity
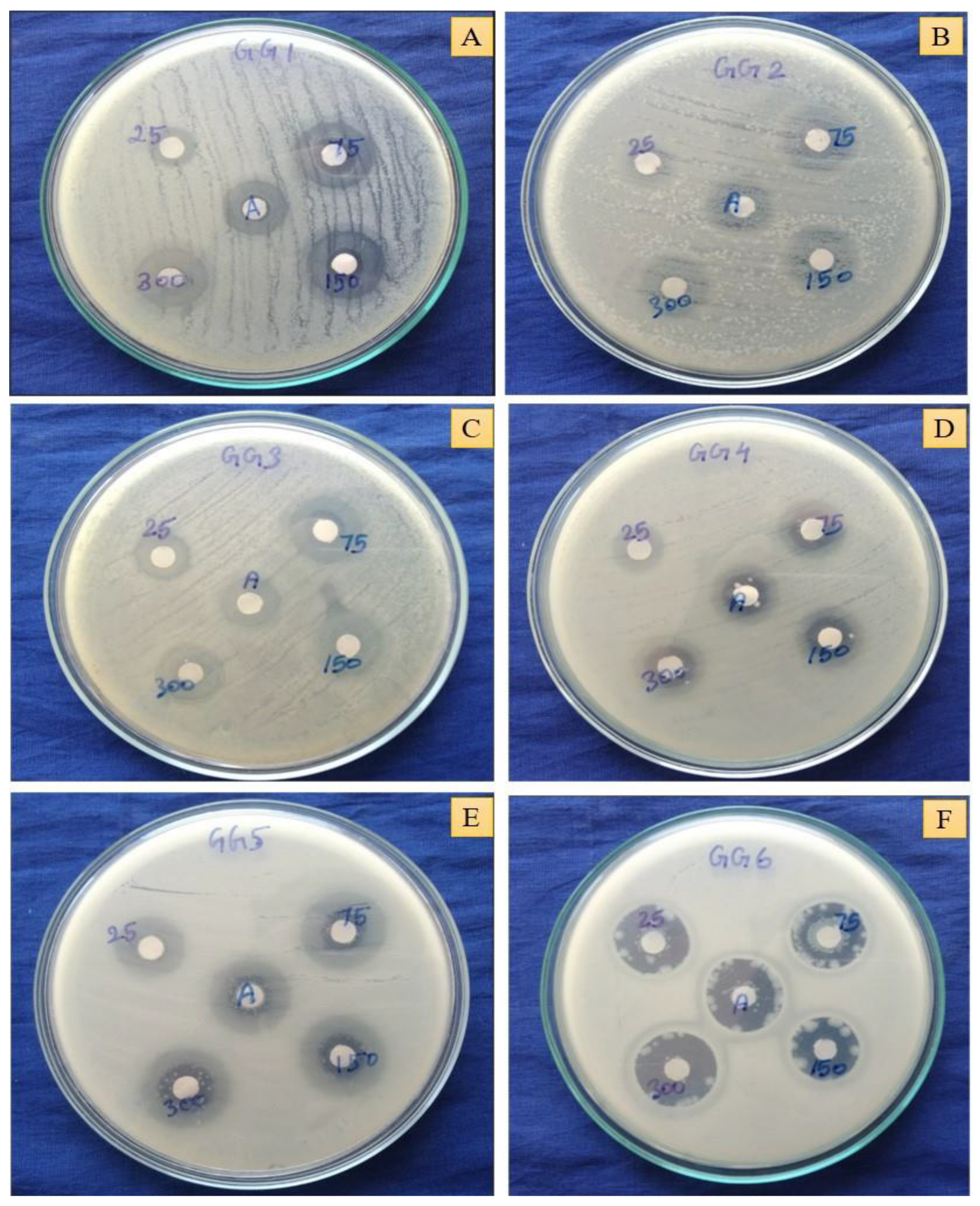
2.12. In Vitro Antibacterial Activity
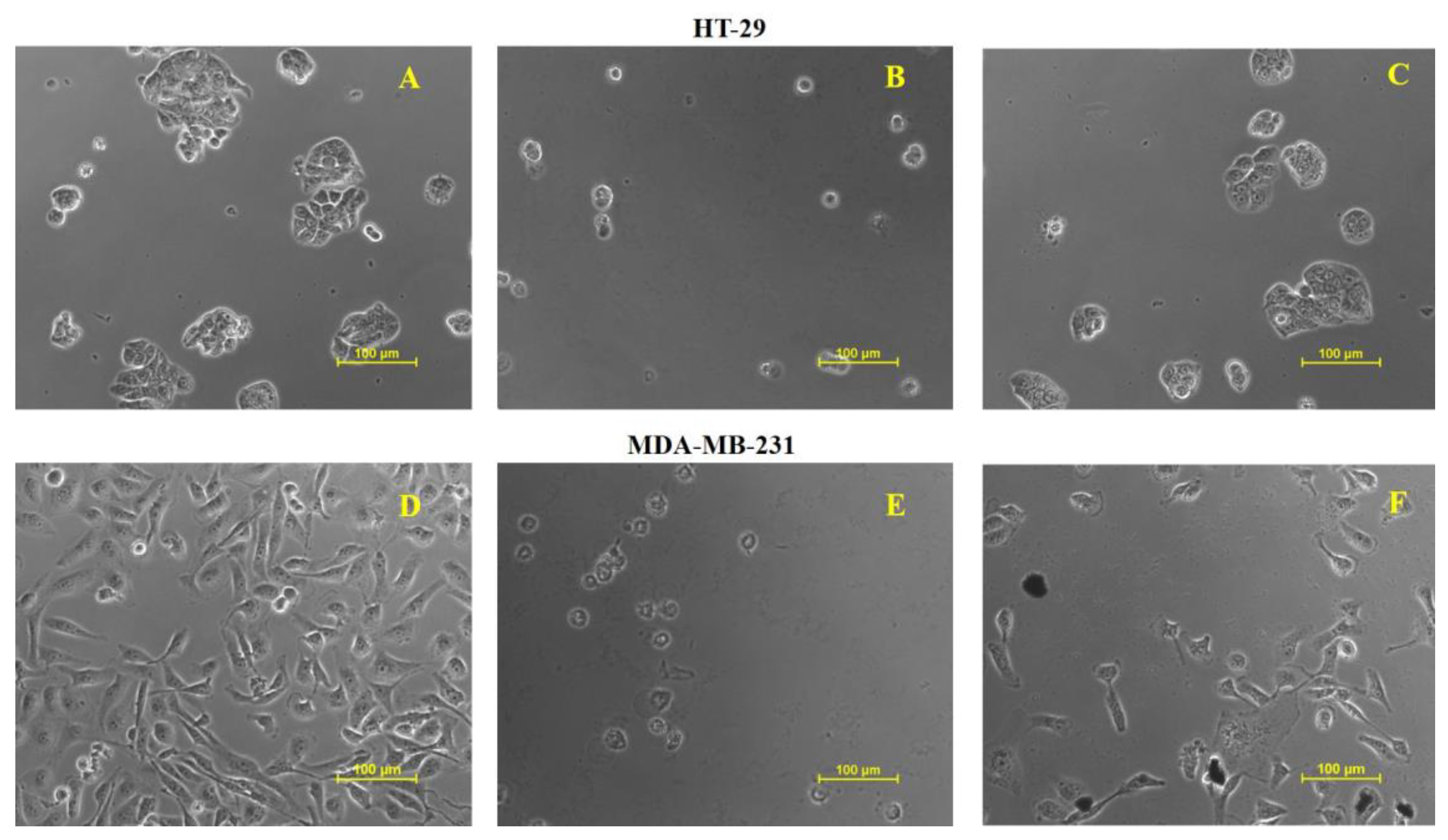
2.13. Cytotoxicity of Cord-Au-NPs
3. Materials and Methods
3.1. Chemicals
3.2. Preparation of Test Material
3.3. Cord-Au-NPs Synthesis
3.3.1. Optimization by Factorial Design Approach
3.3.2. Model Validation
3.4. Physicochemical Characterization
3.5. Therapeutic Investigation
3.5.1. Brine Shrimp Lethality Bioassay
3.5.2. Antidiabetic Activity
3.5.3. Antioxidant Activity
3.5.4. Antibacterial Activity
3.5.5. Anticancer Activity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sharma, B.; Purkayastha, D.D.; Hazra, S.; Gogoi, L.; Bhattacharjee, C.R.; Ghosh, N.N.; Rout, J. Biosynthesis of gold nanoparticles using a freshwater green alga, Prasiola crispa. Mater. Lett. 2014, 116, 94–97. [Google Scholar] [CrossRef]
- Lee, K.X.; Shameli, K.; Miyake, M.; Kuwano, N.; Khairudin, N.B.B.A.; Mohamad, S.E.B.; Yew, Y.P. Green Synthesis of Gold Nanoparticles Using Aqueous Extract of Garcinia mangostana Fruit Peels. J. Nanomater. 2016, 2016, 8489094. [Google Scholar] [CrossRef]
- Shankar, P.D.; Shobana, S.; Karuppusamy, I.; Pugazhendhi, A.; Ramkumar, V.S.; Arvindnarayan, S.; Kumar, G. A review on the biosynthesis of metallic nanoparticles (gold and silver) using bio-components of microalgae: Formation mechanism and applications. Enzym. Microb. Technol. 2016, 95, 28–44. [Google Scholar] [CrossRef] [PubMed]
- Nayak, S.; Sajankila, S.P.; Goveas, L.C.; Rao, V.C.; Mutalik, S.; Shreya, B.A. Two fold increase in synthesis of gold nanoparticles assisted by proteins and phenolic compounds in Pongamia seed cake extract: Response surface methodology approach. SN Appl. Sci. 2020, 4, 634. [Google Scholar] [CrossRef]
- Aljabali, A.A.A.; Akkam, Y.; Al Zoubi, M.S.; Al-Batayneh, K.M.; Al-Trad, B.; Alrob, O.A.; Alkilany, A.M.; Benamara, M.; Evans, D.J. Synthesis of Gold Nanoparticles Using Leaf Extract of Ziziphus zizyphus and their Antimicrobial Activity. Nanomaterials 2018, 8, 174. [Google Scholar] [CrossRef]
- Rajeshkumar, S.; Malarkodi, C.; Gnanajobitha, G.; Paulkumar, K.; Vanaja, M.; Kannan, C.; Annadurai, G. Seaweed-mediated synthesis of gold nanoparticles using Turbinaria conoides and its characterization. J. Nanostructure Chem. 2013, 3, 44. [Google Scholar] [CrossRef]
- Senapati, S.; Syed, A.; Moeez, S.; Kumar, A.; Ahmad, A. Intracellular synthesis of gold nanoparticles using alga Tetraselmis kochinensis. Mater. Lett. 2012, 79, 116–118. [Google Scholar] [CrossRef]
- Zhang, G.; Huang, Y.; Bian, Y.; Wong, J.H.; Ng, T.B.; Wang, H. Hypoglycemic activity of the fungi Cordyceps militaris, Cordyceps sinensis, Tricholoma mongolicum, and Omphalia lapidescens in streptozotocin-induced diabetic rats. Appl. Microbiol. Biotechnol. 2006, 72, 1152–1156. [Google Scholar] [CrossRef]
- Chen, X.; Wu, G.; Huang, Z. Structural analysis and antioxidant activities of polysaccharides from cultured Cordyceps militaris. Int. J. Biol. Macromol. 2013, 58, 18–22. [Google Scholar] [CrossRef]
- Jędrejko, K.J.; Lazur, J.; Muszyńska, B. Cordyceps militaris: An Overview of Its Chemical Constituents in Relation to Biological Activity. Foods 2021, 10, 2634. [Google Scholar] [CrossRef]
- Ji, Y.; Cao, Y.; Song, Y. Green synthesis of gold nanoparticles using a Cordyceps militaris extract and their antiproliferative effect in liver cancer cells (HepG2). Artif. Cells Nanomed. Biotechnol. 2019, 47, 2737–2745. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Jing, Y.; Guo, A.; Li, X.; Li, Q.; Liu, W.; Zhang, X. Biosynthesis of Platinum Nanoparticles with Cordyceps Flower Extract: Characterization, Antioxidant Activity and Antibacterial Activity. Nanomaterials 2022, 12, 1904. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liu, C.-C.; Wang, Y.-Y.; Xu, H.; Su, H.; Cheng, X. Antibacterial activities of the novel silver nanoparticles biosynthesized using Cordyceps militaris extract. Curr. Appl. Phys. 2016, 16, 969–973. [Google Scholar] [CrossRef]
- Li, J.F.; Rupa, E.J.; Hurh, J.; Huo, Y.; Chen, L.; Han, Y.; Ahn, J.C.; Park, J.K.; Lee, H.A.; Mathiyalagan, R.; et al. Cordyceps militaris fungus mediated Zinc Oxide nanoparticles for the photocatalytic degradation of Methylene blue dye. Optik 2019, 183, 691–697. [Google Scholar] [CrossRef]
- Suksiriworapong, J.; Pongprasert, N.; Bunsupa, S.; Taresco, V.; Crucitti, V.C.; Janurai, T.; Phruttiwanichakun, P.; Sakchaisri, K.; Wongrakpanich, A. CD44-Targeted Lipid Polymer Hybrid Nanoparticles Enhance Anti-Breast Cancer Effect of Cordyceps militaris Extracts. Pharmaceutics 2023, 15, 1771. [Google Scholar] [CrossRef]
- Rupa, E.J.; Li, J.F.; Arif, M.H.; Yaxi, H.; Puja, A.M.; Chan, A.J.; Hoang, V.-A.; Kaliraj, L.; Yang, D.C.; Kang, S.C. Cordyceps militaris Fungus Extracts-Mediated Nanoemulsion for Improvement Antioxidant, Antimicrobial, and Anti-Inflammatory Activities. Molecules 2020, 25, 5733. [Google Scholar] [CrossRef]
- Dias, C.; Ayyanar, M.; Amalraj, S.; Khanal, P.; Subramaniyan, V.; Das, S.; Gandhale, P.; Biswa, V.; Ali, R.; Gurav, N.; et al. Biogenic synthesis of zinc oxide nanoparticles using mushroom fungus Cordyceps militaris: Characterization and mechanistic insights of therapeutic investigation. J. Drug Deliv. Sci. Technol. 2022, 73, 103444. [Google Scholar] [CrossRef]
- Saoji, S.D.; Rarokar, N.R.; Dhore, P.W.; Dube, S.O.; Gurav, N.S.; Gurav, S.S.; Raut, N.A. Phospholipid Based Colloidal Nanocarriers for Enhanced Solubility and Therapeutic Efficacy of Withanolides. J. Drug Deliv. Sci. Technol. 2022, 70, 103251. [Google Scholar] [CrossRef]
- Moradpoor, H.; Safaei, M.; Golshah, A.; Mozaffari, H.R.; Sharifi, R.; Imani, M.M.; Mobarakeh, M.S. Green synthesis and antifungal effect of titanium dioxide nanoparticles on oral Candida albicans pathogen. Inorg. Chem. Commun. 2021, 130, 108748. [Google Scholar] [CrossRef]
- Fan, D.; Li, L.; Li, Z.; Zhang, Y.; Ma, X.; Wu, L.; Zhang, H.; Guo, F. Biosynthesis of selenium nanoparticles and their protective, antioxidative effects in streptozotocin induced diabetic rats. Sci. Technol. Adv. Mater. 2020, 21, 505–514. [Google Scholar] [CrossRef]
- Rodrigues, K.; Gurav, S.; Joshi, A.; Krishna, M.; Bhandarkar, A. Porous polymeric carrier system for modified drug release of boswellic acid. Chem. Sci. J. 2020, 11, 1–12. [Google Scholar] [CrossRef]
- Karuppiah, C.; Palanisamy, S.; Chen, S.-M.; Emmanuel, R.; Muthupandi, K.; Prakash, P. Green synthesis of gold nanoparticles and its application for the trace level determination of painter’s colic. RSC Adv. 2015, 5, 16284–16291. [Google Scholar] [CrossRef]
- Yuan, C.G.; Huo, C.; Gui, B.; Cao, W.P. Green synthesis of gold nanoparticles using Citrus maxima peel extract and their catalytic/antibacterial activities. IET Nanobiotechnol. 2017, 11, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Uthaman, S.; Kim, H.S.; Revuri, V.; Min, J.-J.; Lee, Y.-K.; Huh, K.M.; Park, I.-K. Green synthesis of bioactive polysaccharide-capped gold nanoparticles for lymph node CT imaging. Carbohydr. Polym. 2018, 181, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Dessai, S.; Ayyanar, M.; Amalraj, S.; Khanal, P.; Vijayakumar, S.; Gurav, N.; Rarokar, N.; Kalaskar, M.; Nadaf, S.; Gurav, S. Bioflavonoid mediated synthesis of TiO2 nanoparticles: Characterization and their biomedical applications. Mater. Lett. 2022, 311, 131639. [Google Scholar] [CrossRef]
- Singh, P.; Kim, Y.J.; Yang, D.C. A strategic approach for rapid synthesis of gold and silver nanoparticles by Panax ginseng leaves. Artif. Cells Nanomed. Biotechnol. 2016, 44, 1949–1957. [Google Scholar] [CrossRef]
- Veeramani, S.; Narayanan, A.P.; Yuvaraj, K.; Sivaramakrishnan, R.; Pugazhendhi, A.; Rishivarathan, I.; Jose, S.P.; Ilangovan, R. Nigella sativa flavonoids surface coated gold NPs (Au-NPs) enhancing antioxidant and anti-diabetic activity. Process. Biochem. 2021, 114, 193–202. [Google Scholar] [CrossRef]
- Guo, Y.; Jiang, N.; Zhang, L.; Yin, M. Green synthesis of gold nanoparticles from Fritillaria cirrhosa and its anti-diabetic activity on Streptozotocin induced rats. Arab. J. Chem. 2020, 13, 5096–5106. [Google Scholar] [CrossRef]
- Milanezi, F.G.; Meireles, L.M.; de Christo Scherer, M.M.; de Oliveira, J.P.; da Silva, A.R.; de Araujo, M.L.; Endringer, D.C.; Fronza, M.; Guimarães, M.C.C.; Scherer, R. Antioxidant, antimicrobial and cytotoxic activities of gold nanoparticles capped with quercetin. Saudi Pharm. J. 2019, 27, 968–974. [Google Scholar] [CrossRef]
- Zayadi, R.A.; Abu Bakar, F.; Ahmad, M.K. Elucidation of synergistic effect of eucalyptus globulus honey and Zingiber officinale in the synthesis of colloidal biogenic gold nanoparticles with antioxidant and catalytic properties. Sustain. Chem. Pharm. 2019, 13, 100156. [Google Scholar] [CrossRef]
- Sathishkumar, G.; Pradeep, K.; Vignesh, V.; Rajkuberan, C.; Jeyaraj, M.; Selvakumar, M.; Rakhi, J.; Sivaramakrishnan, S. Cannonball fruit (Couroupita guianensis, Aubl.) extract mediated synthesis of gold nanoparticles and evaluation of its antioxidant activity. J. Mol. Liq. 2016, 215, 229–236. [Google Scholar]
- Lydia, D.E.; Khusro, A.; Immanuel, P.; Esmail, G.A.; Al-Dhabi, N.A.; Arasu, M.V. Photo-activated synthesis and characterization of gold nanoparticles from Punica granatum L. seed oil: An assessment on antioxidant and anticancer properties for functional yoghurt nutraceuticals. J. Photochem. Photobiol. B Biol. 2020, 206, 111868. [Google Scholar] [CrossRef]
- Nagalingam, M.; Kalpana, V.; Devi Rajeswari, V.; Panneerselvam, A. Biosynthesis, characterization, and evaluation of bioactivities of leaf extract-mediated biocompatible gold nanoparticles from Alternanthera bettzickiana. Biotechnol. Rep. 2018, 19, e00268. [Google Scholar]
- Vinosha, M.; Palanisamy, S.; Muthukrishnan, R.; Selvam, S.; Kannapiran, E.; You, S.; Prabhu, N.M. Biogenic synthesis of gold nanoparticles from Halymenia dilatata for pharmaceutical applications: Antioxidant, anti-cancer and antibacterial activities. Process. Biochem. 2019, 85, 219–229. [Google Scholar] [CrossRef]
- Veiga, A.; Toledo, M.d.G.T.; Rossa, L.S.; Mengarda, M.; Stofella, N.C.; Oliveira, L.J.; Gonçalves, A.G.; Murakami, F.S. Colorimetric microdilution assay: Validation of a standard method for determination of MIC, IC50%, and IC90% of antimicrobial compounds. J. Microbiol. Methods 2019, 162, 50–61. [Google Scholar] [CrossRef] [PubMed]
- Hosny, M.; Fawzy, M.; Abdelfatah, A.M.; Fawzy, E.E.; Eltaweil, A.S. Comparative study on the potentialities of two halophytic species in the green synthesis of gold nanoparticles and their anticancer, antioxidant and catalytic efficiencies. Adv. Powder Technol. 2021, 32, 3220–3233. [Google Scholar] [CrossRef]

| Source | Response Y1 | Response Y2 | Response Y3 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Coeff. | Sum of Squares | F-Value | p-Value | Coeff. | Sum of Squares | F-Value | p-Value | Coeff. | Sum of Squares | F-Value | p-Value | |
| Model | 0.4689 a | 0.0274 | 93.9 | <0.0001 b | 12.86 a | 140.32 | 245.39 | <0.0001 b | −13.69 a | 90.47 | 34.89 | 0.0007 b |
| X1 - | −0.0181 | 0.0039 | 26.88 | 0.0008 | −0.9333 | 10.45 | 91.41 | 0.0002 | −1.36 | 22.09 | 42.59 | 0.0013 |
| X2 - | 0.0443 | 0.0235 | 160.93 | <0.0001 | 0.5733 | 3.94 | 34.49 | 0.002 | 0.865 | 8.98 | 17.31 | 0.0088 |
| X1X2 | −2.24 | 19.98 | 174.72 | <0.0001 | −2.61 | 27.35 | 52.75 | 0.0008 | ||||
| X12 | 0.4756 | 4.37 | 38.17 | 0.0016 | 0.581 | 6.52 | 12.57 | 0.0165 | ||||
| X22 | −1.92 | 70.92 | 620.17 | <0.0001 | −0.8252 | 13.14 | 25.35 | 0.004 | ||||
| Residual | 0.0012 | 0.5718 | 2.59 | |||||||||
| Lack of Fit | 0.0009 | 1.33 | 0.4886 c | 0.1552 | 0.2484 | 0.8586 c | 1.6 | 1.07 | 0.5175 c | |||
| Pure Error | 0.0002 | 0.4166 | 0.9978 | |||||||||
| Cor Total | 0.0286 | 140.89 | 93.06 | |||||||||
| Independent Variables | Levels | |||||||
|---|---|---|---|---|---|---|---|---|
| −α | Low (1) | Medium (0) | High (+1) | +α | ||||
| X1: CME conc. (% w/v) | 4 | 6 | 8 | 10 | 12 | |||
| X2: AuCl3 conc. (mM) | 0.25 | 0.5 | 0.75 | 1 | 1.25 | |||
| Dependent variables | Goal | |||||||
| Y1: Absorbance | Maximize | |||||||
| Y2: Particle size (nm) | Minimize | |||||||
| Y3: Zeta potential (mV) | Minimize | |||||||
| Std | Run | X1: CME Conc. (% w/v) | X2: AuCl3 Conc. (mM) | Y1: Absorbance | Y2: Particle size (nm) | Y3: Zeta potential (mV) | ||
| 11 | 1 | 8 | 0.75 | 0.461 | 13.26 | −13.24 | ||
| 10 | 2 | 8 | 0.75 | 0.476 | 13.03 | −14.65 | ||
| 9 | 3 | 8 | 0.75 | 0.482 | 12.38 | −14.02 | ||
| 7 | 4 | 8 | 0.25 | 0.37 | 4.22 | −18.96 | ||
| 4 | 5 | 10 | 1 | 0.507 | 8.96 | −16.37 | ||
| 6 | 6 | 12 | 0.75 | 0.417 | 12.89 | −14.56 | ||
| 1 | 7 | 6 | 0.5 | 0.442 | 9.32 | −15.87 | ||
| 2 | 8 | 10 | 0.5 | 0.421 | 11.98 | −12.81 | ||
| 5 | 9 | 4 | 0.75 | 0.509 | 16.68 | −8.59 | ||
| 8 | 10 | 8 | 1.25 | 0.554 | 6.21 | −15.44 | ||
| 3 | 11 | 6 | 1 | 0.519 | 15.24 | −8.97 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gawas, G.; Ayyanar, M.; Gurav, N.; Hase, D.; Murade, V.; Nadaf, S.; Khan, M.S.; Chikhale, R.; Kalaskar, M.; Gurav, S. Process Optimization for the Bioinspired Synthesis of Gold Nanoparticles Using Cordyceps militaris, Its Characterization, and Assessment of Enhanced Therapeutic Efficacy. Pharmaceuticals 2023, 16, 1311. https://doi.org/10.3390/ph16091311
Gawas G, Ayyanar M, Gurav N, Hase D, Murade V, Nadaf S, Khan MS, Chikhale R, Kalaskar M, Gurav S. Process Optimization for the Bioinspired Synthesis of Gold Nanoparticles Using Cordyceps militaris, Its Characterization, and Assessment of Enhanced Therapeutic Efficacy. Pharmaceuticals. 2023; 16(9):1311. https://doi.org/10.3390/ph16091311
Chicago/Turabian StyleGawas, Girish, Muniappan Ayyanar, Nilambari Gurav, Dinesh Hase, Vaishali Murade, Sameer Nadaf, Mohd Shahnawaz Khan, Rupesh Chikhale, Mohan Kalaskar, and Shailendra Gurav. 2023. "Process Optimization for the Bioinspired Synthesis of Gold Nanoparticles Using Cordyceps militaris, Its Characterization, and Assessment of Enhanced Therapeutic Efficacy" Pharmaceuticals 16, no. 9: 1311. https://doi.org/10.3390/ph16091311
APA StyleGawas, G., Ayyanar, M., Gurav, N., Hase, D., Murade, V., Nadaf, S., Khan, M. S., Chikhale, R., Kalaskar, M., & Gurav, S. (2023). Process Optimization for the Bioinspired Synthesis of Gold Nanoparticles Using Cordyceps militaris, Its Characterization, and Assessment of Enhanced Therapeutic Efficacy. Pharmaceuticals, 16(9), 1311. https://doi.org/10.3390/ph16091311