The Magic Methyl and Its Tricks in Drug Discovery and Development
Abstract
1. Introduction
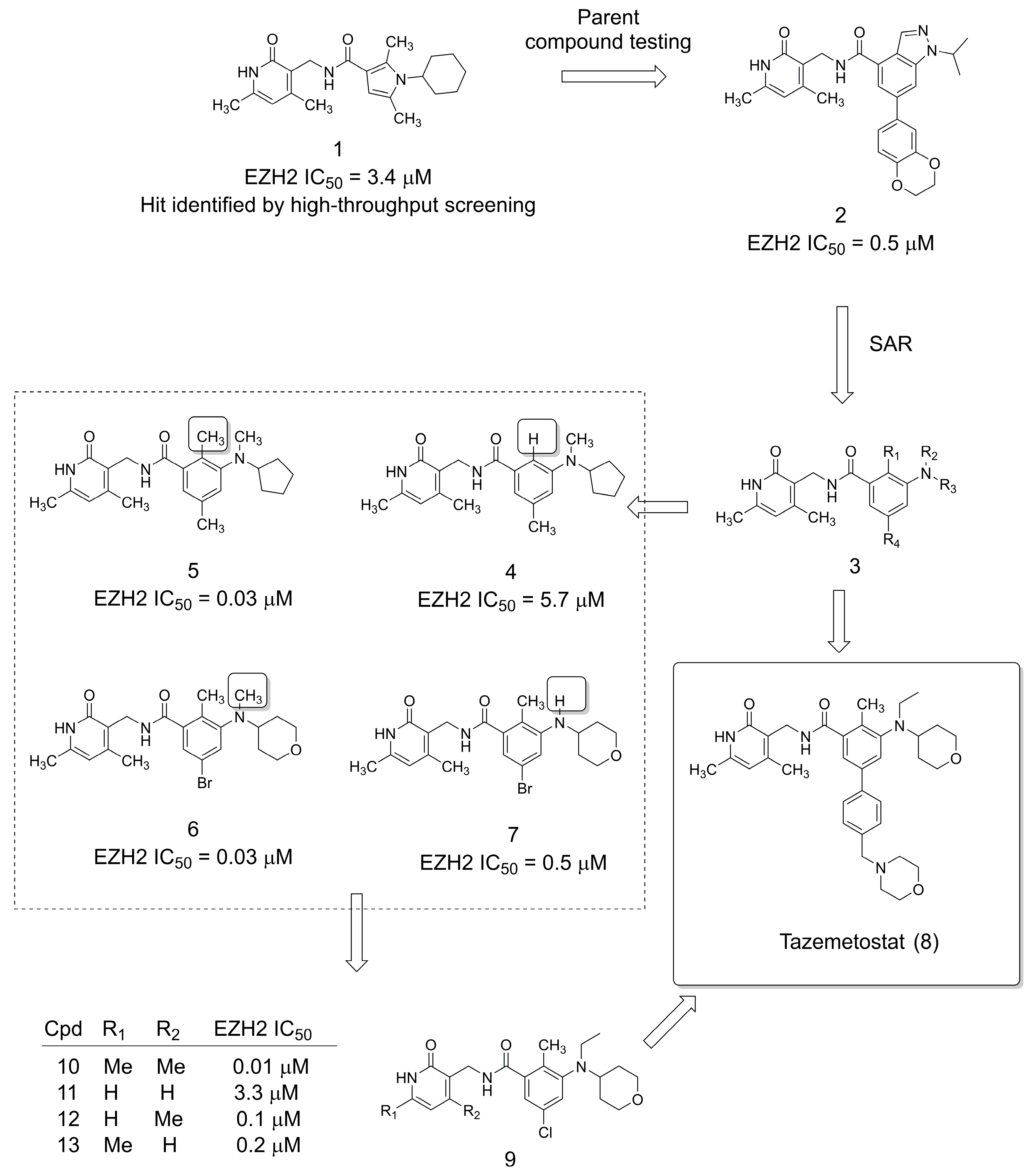
2. The Discovery of the Anticancer Drug Tazemetostat
3. The Methylation Effect in Pharmacodynamic Optimization
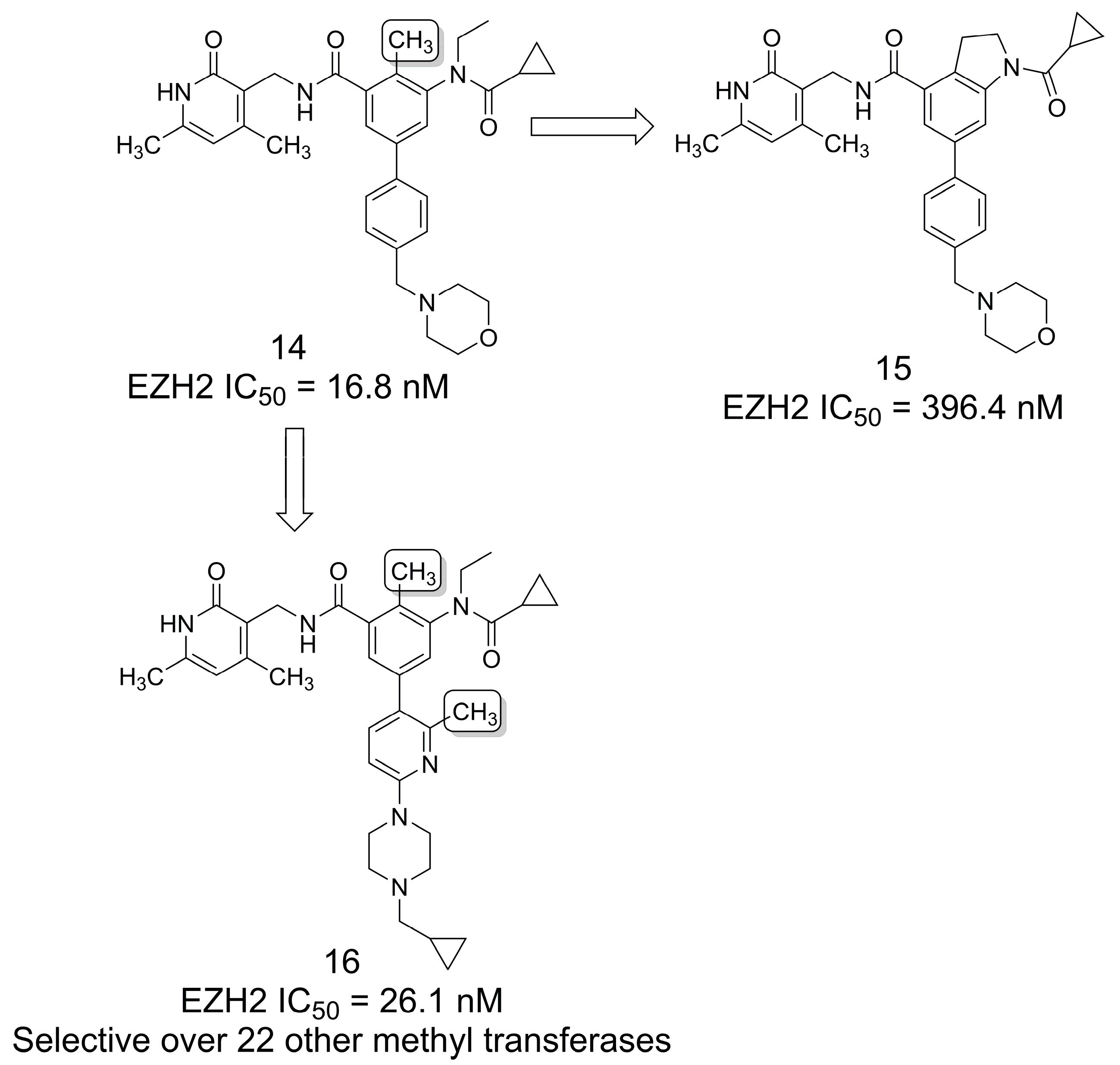
3.1. Selective EZH2 Inhibitors
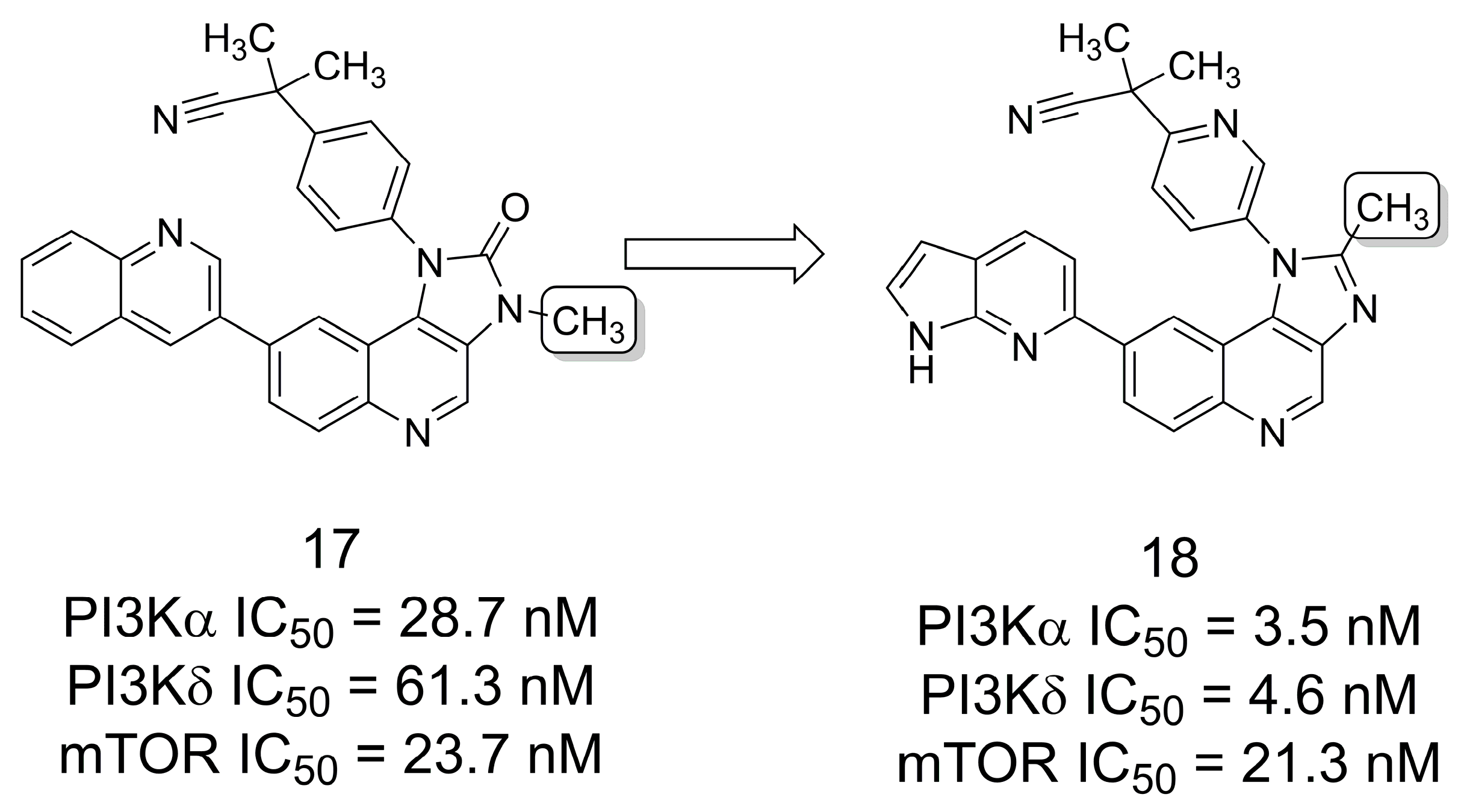
3.2. PI3K/mTOR Inhibitors
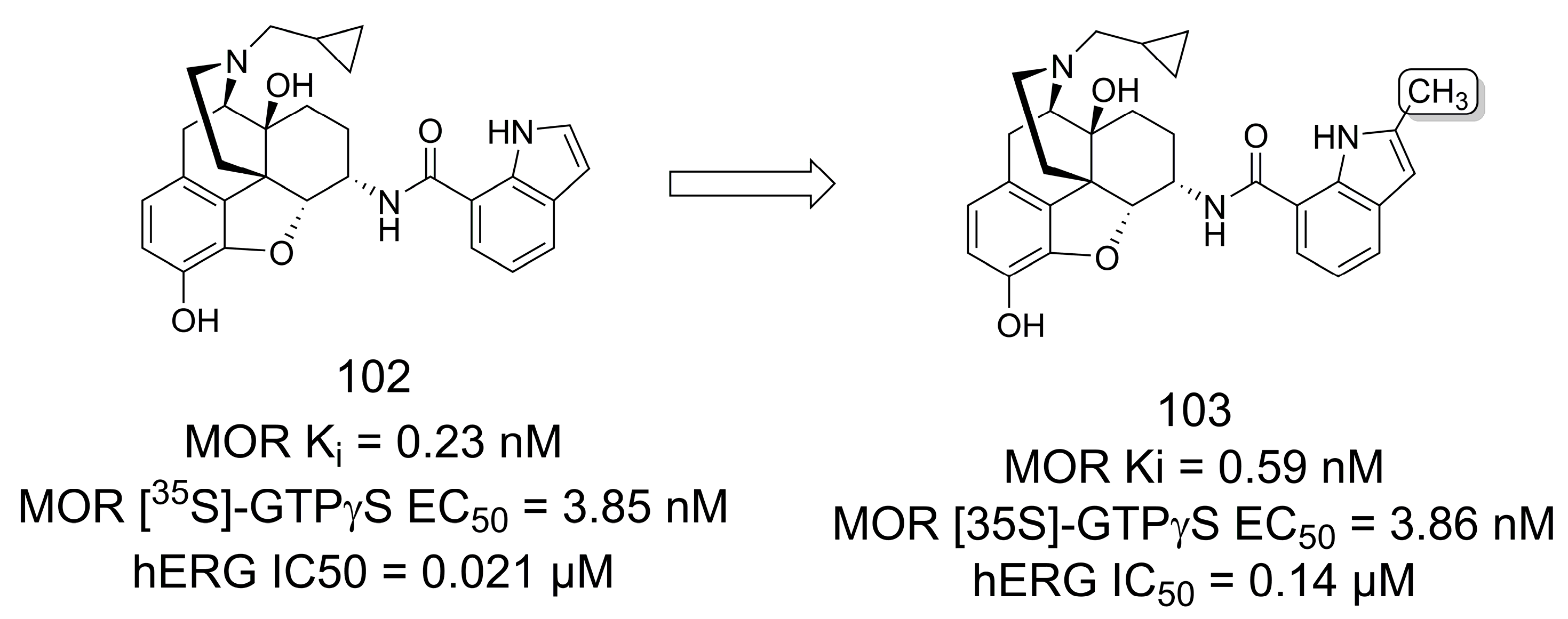
3.3. Selective κ-Opioid Receptor Antagonists
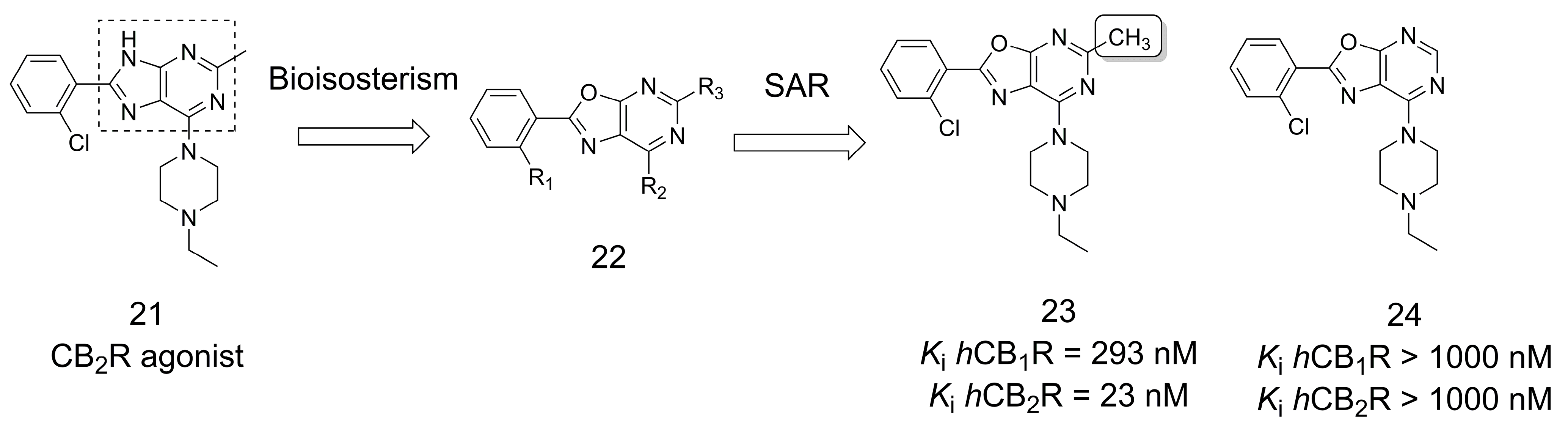
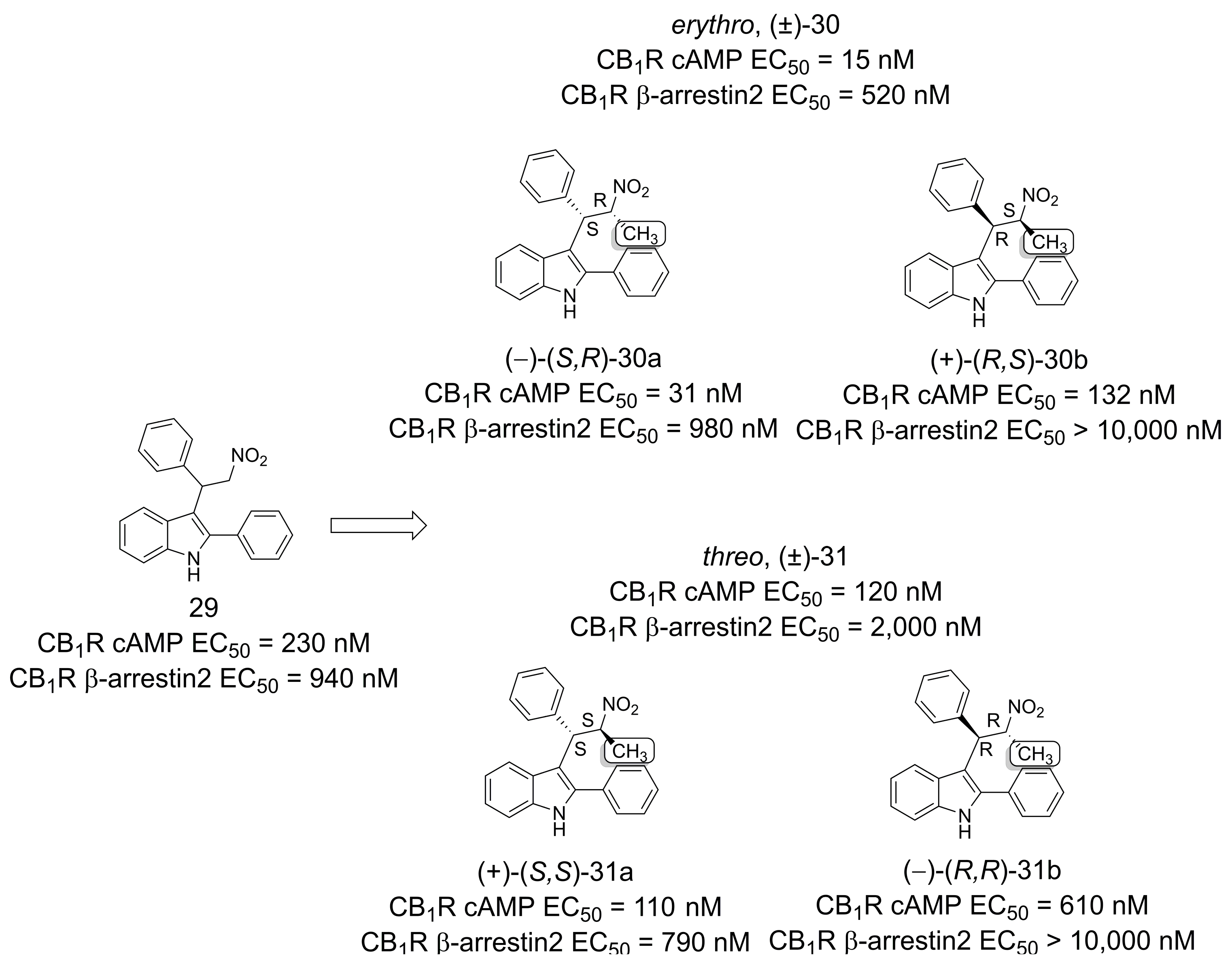
3.4. Cannabinoid Receptor Modulators
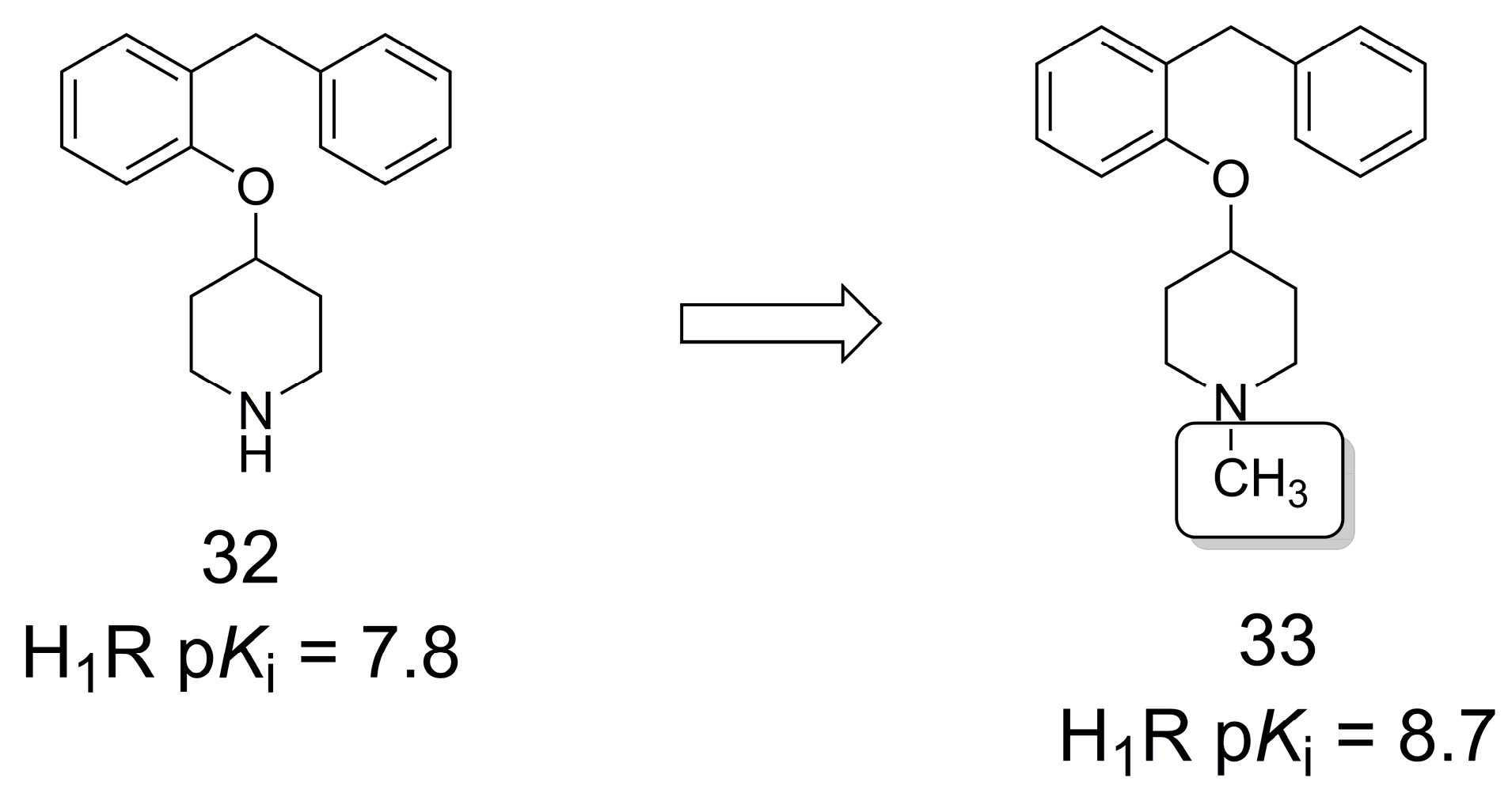
3.5. Histamine 1 Receptor Antagonists
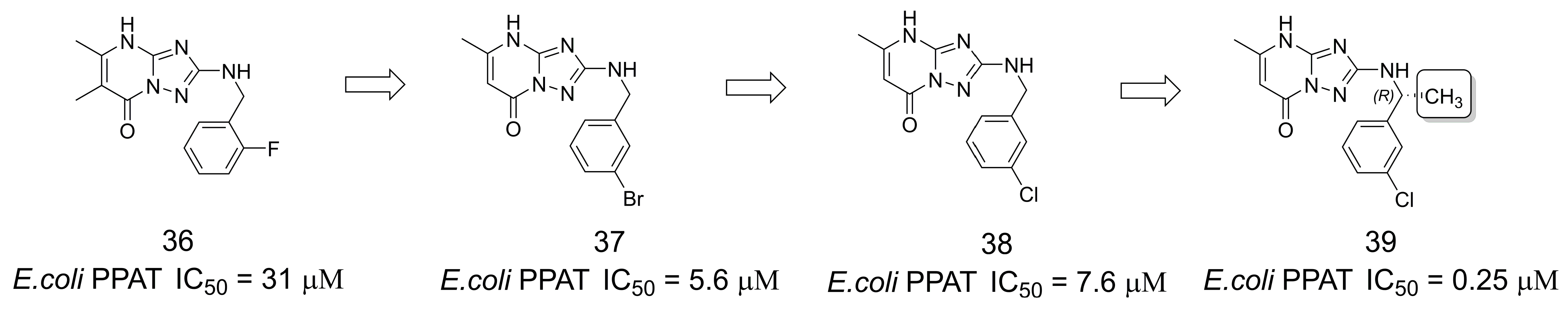
3.6. Inhibitors of Phosphopantetheine Adenylyltransferase
3.7. Genetic Depletion of the Mitotic Aurora Kinase B (AURKB)
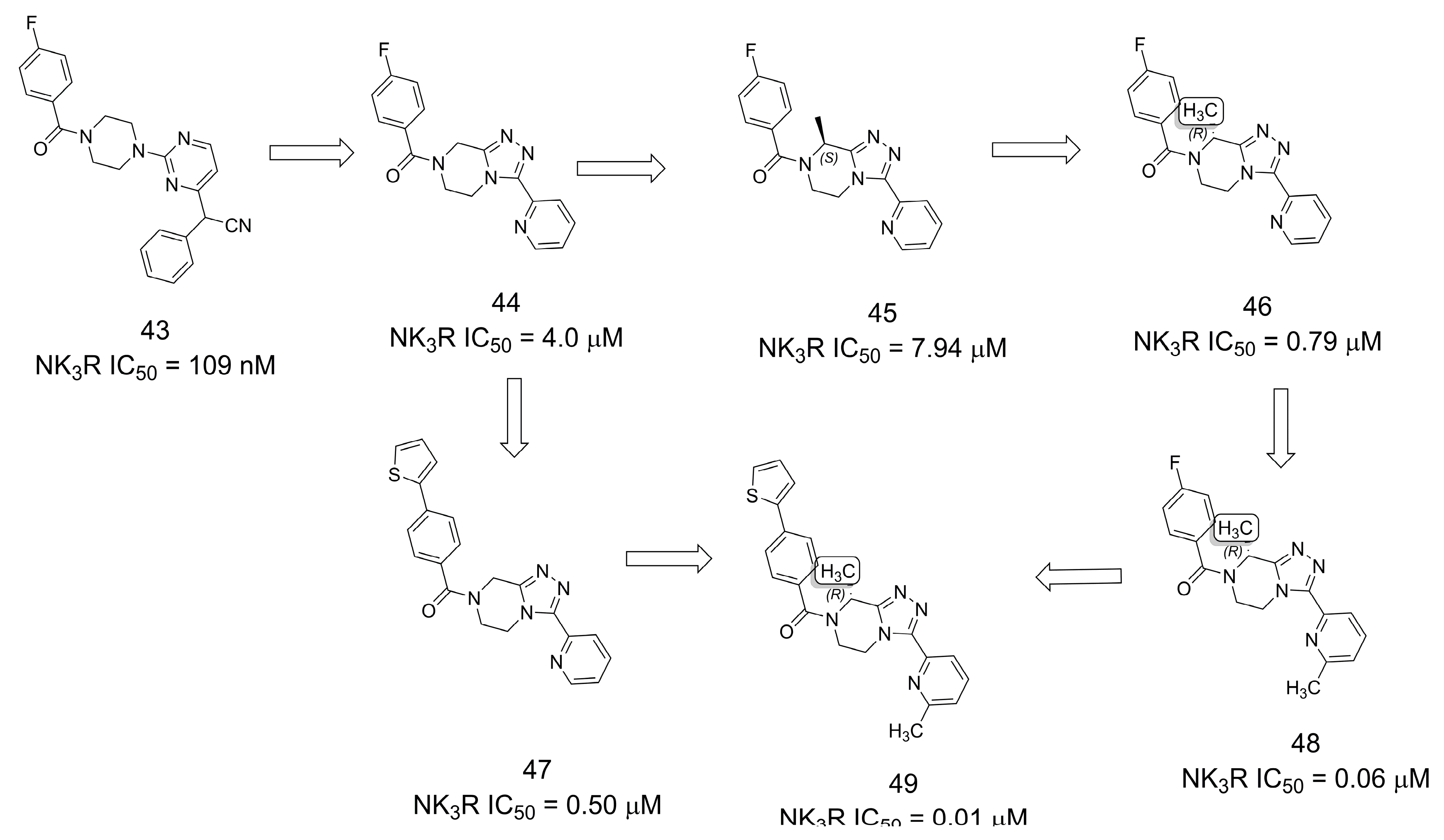
3.8. Neurokinin-3 Receptor Antagonists
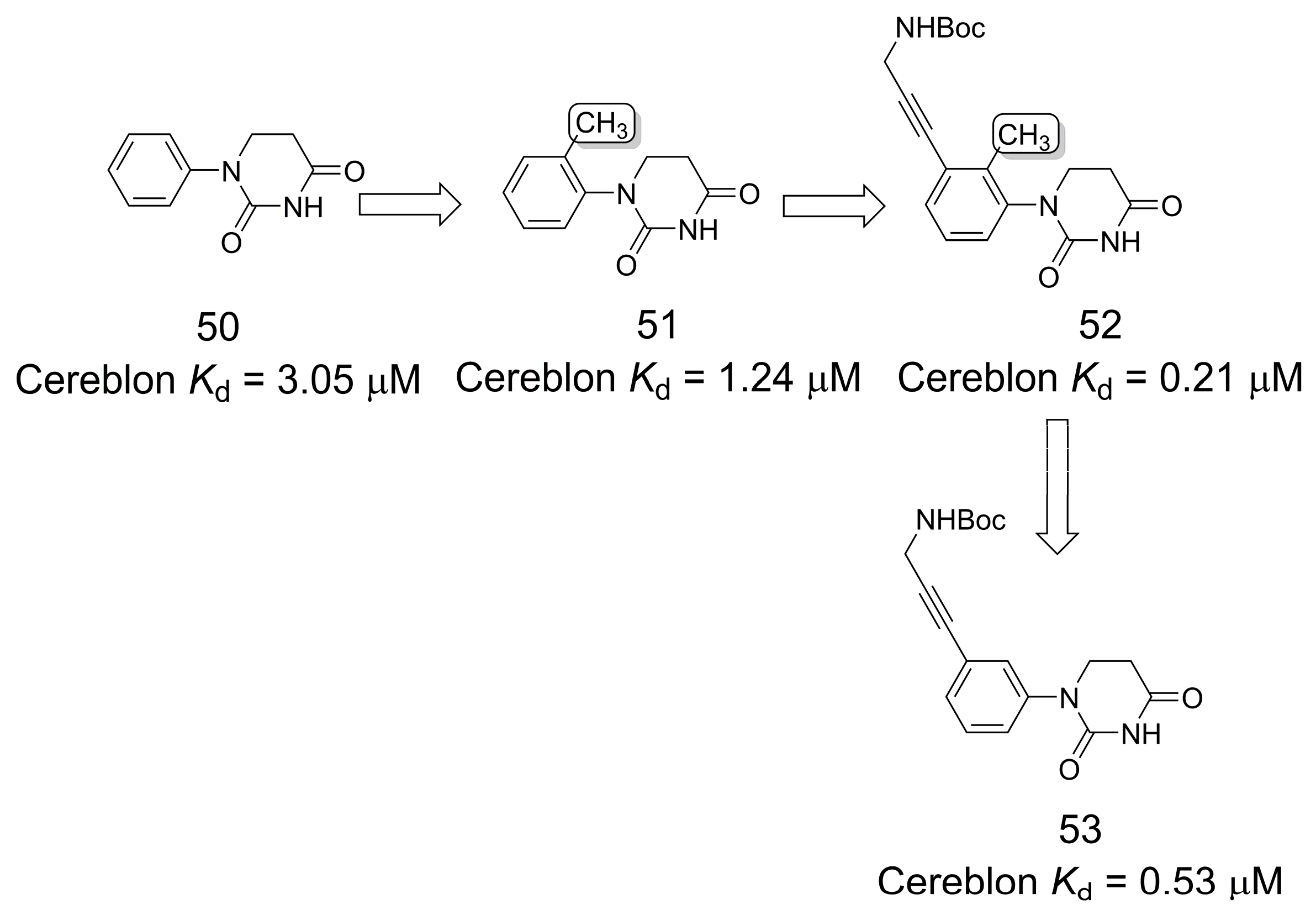
3.9. Cereblon Ligands for Targeted Protein Degradation
3.10. Putative Dual Inhibitor of Tubulin and EGFR by Phenotypic Approach
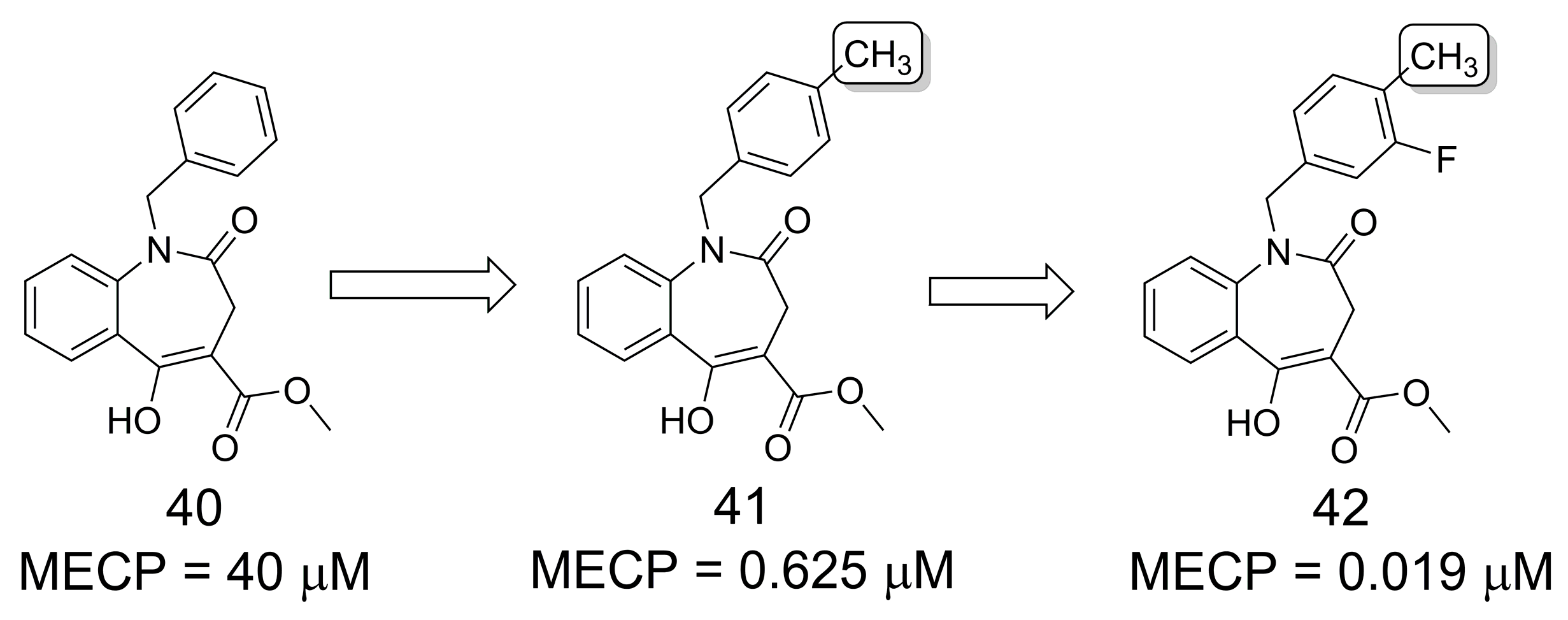
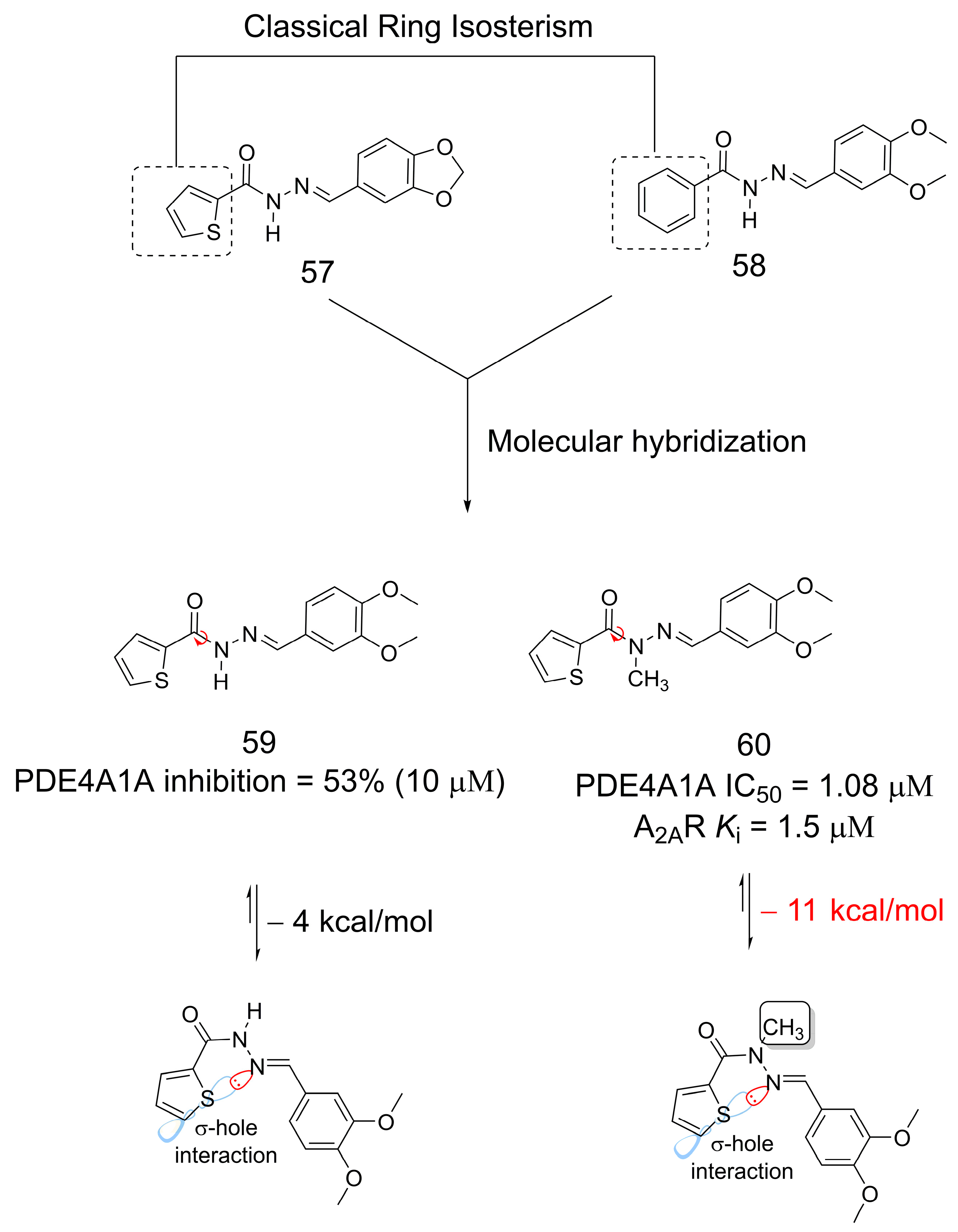
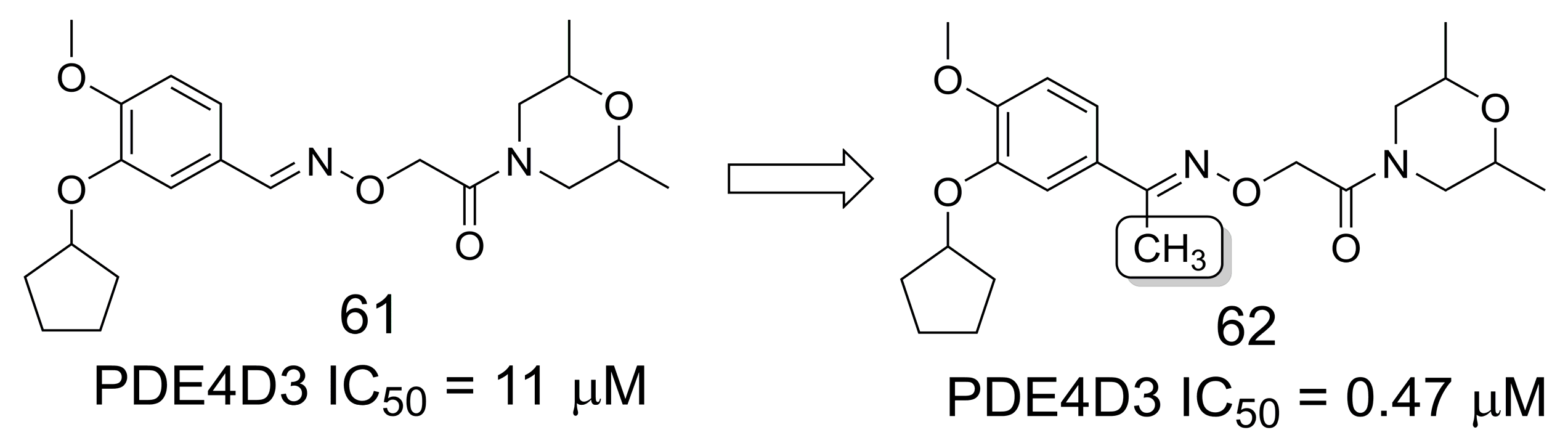
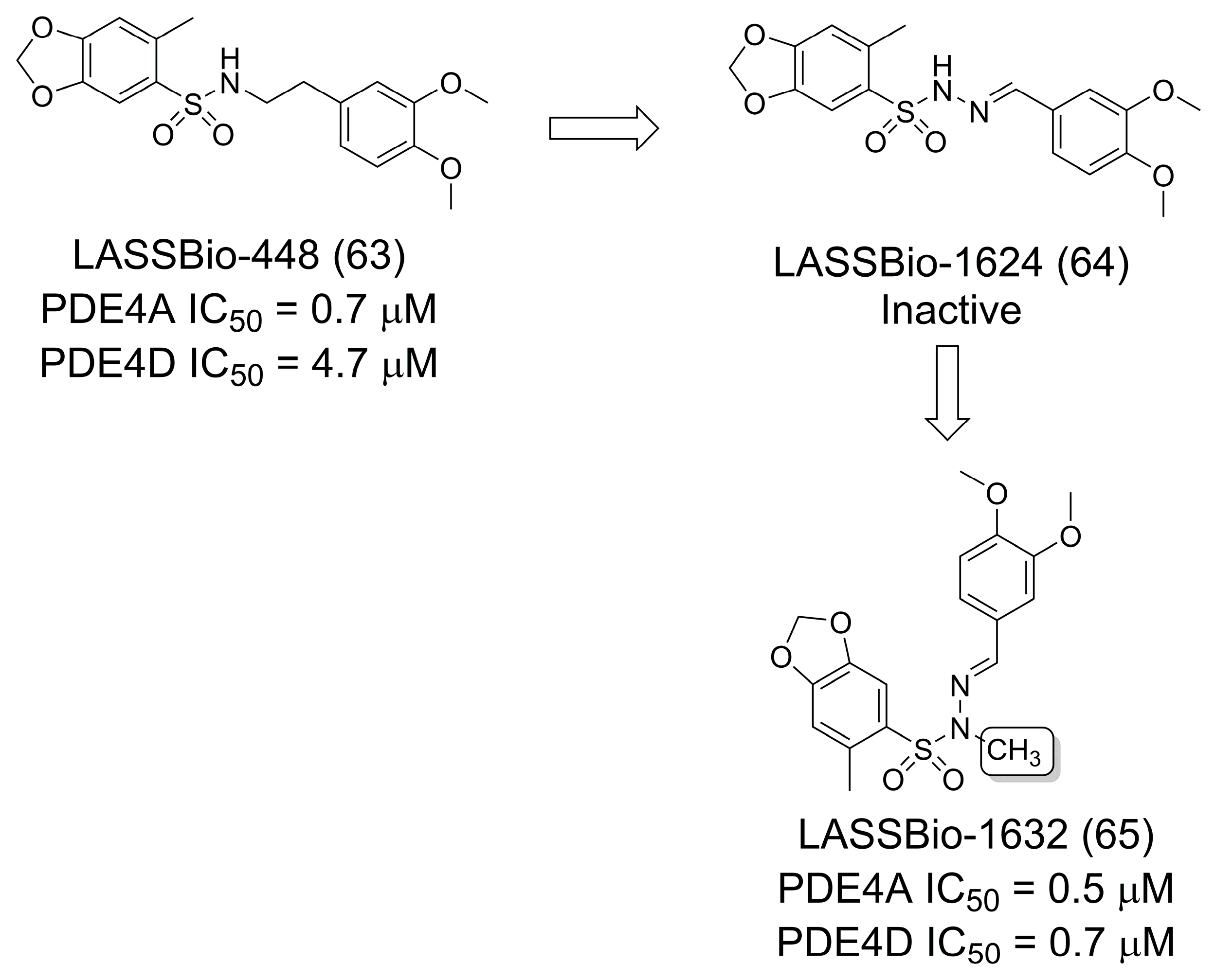
3.11. Phosphodiesterase Inhibitors
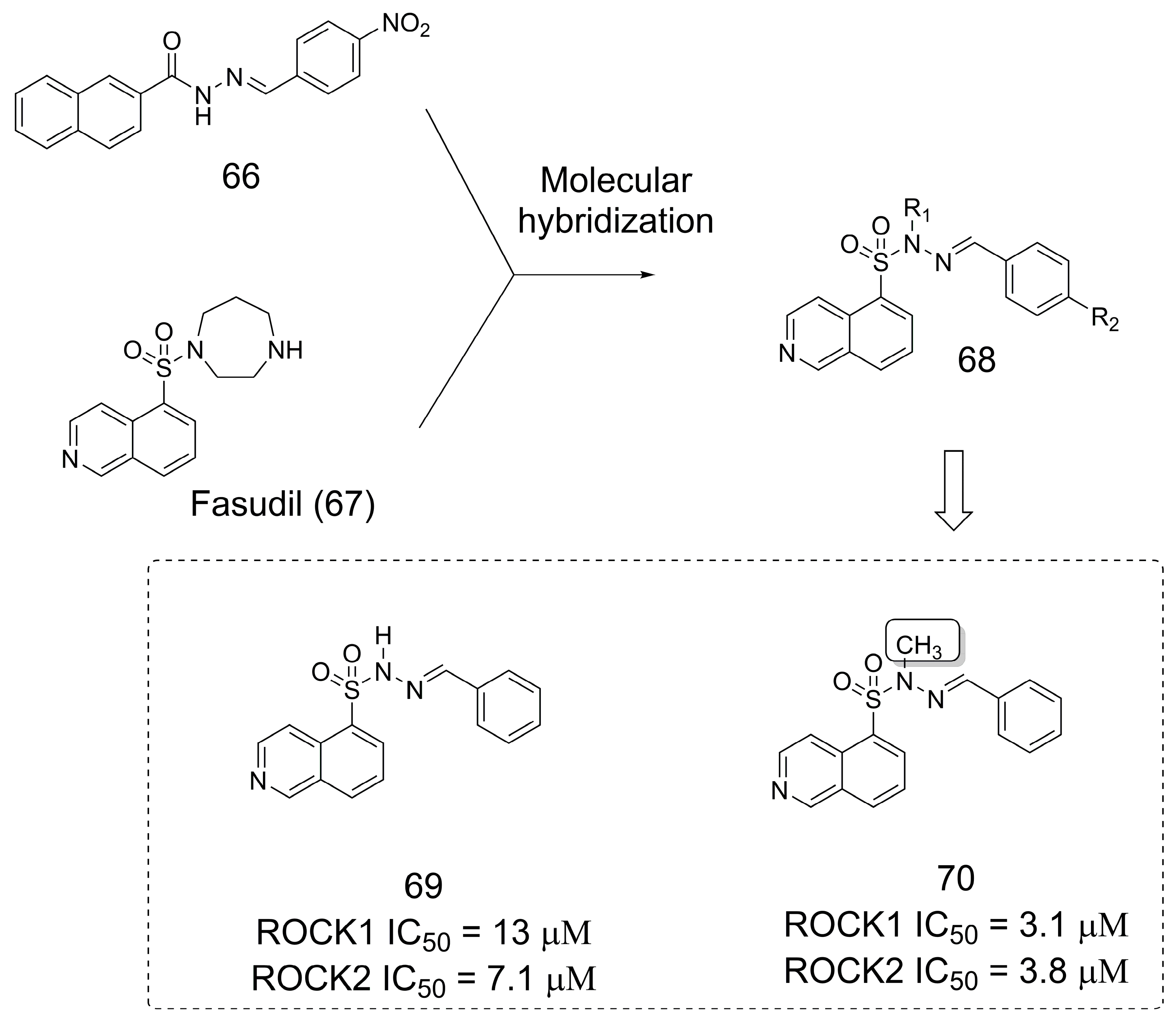
3.12. Rho-Associated Kinase (ROCK) Inhibitors
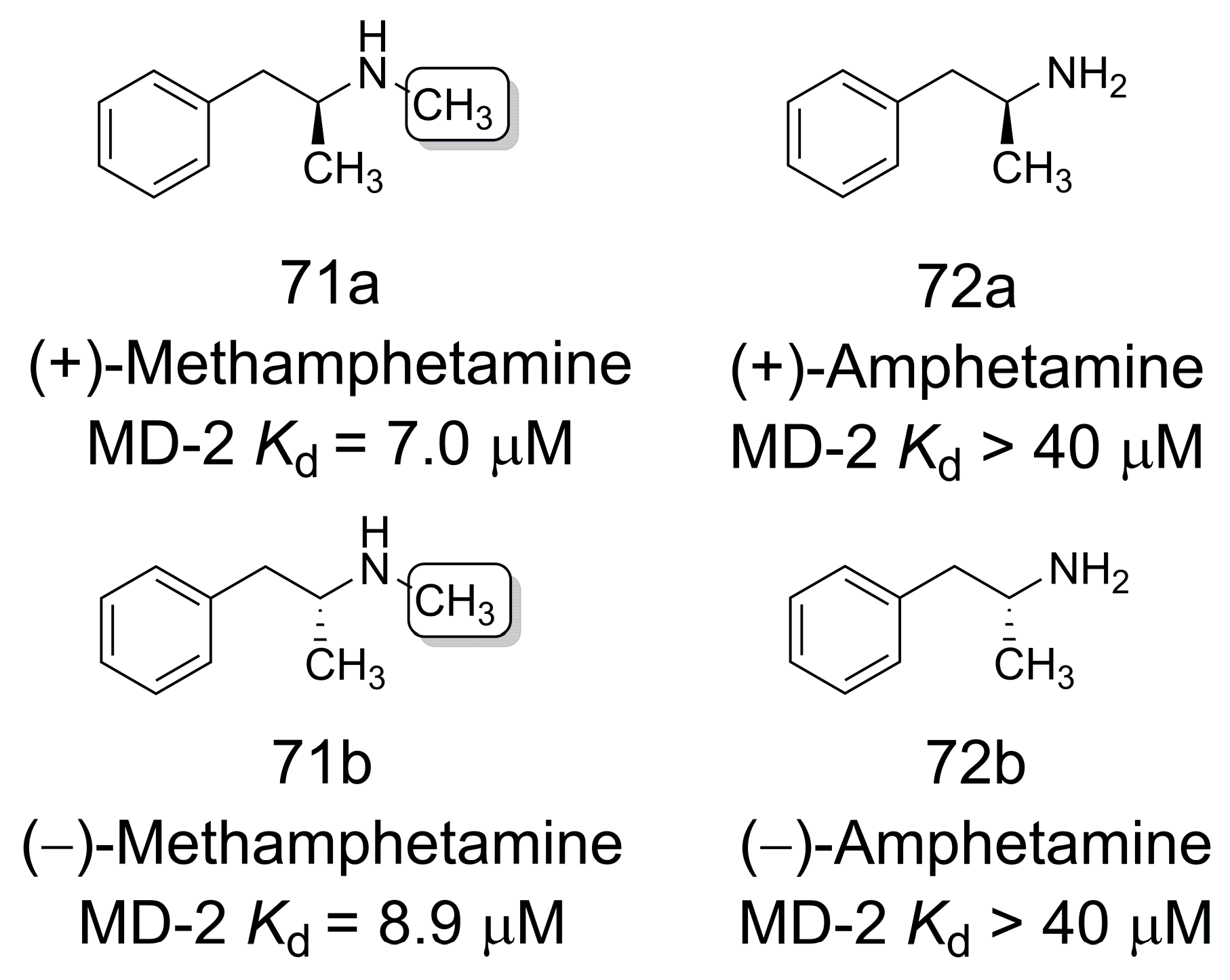
3.13. Ligands of Toll-like Receptor 4/Myeloid Differentiation Protein 2 Complex
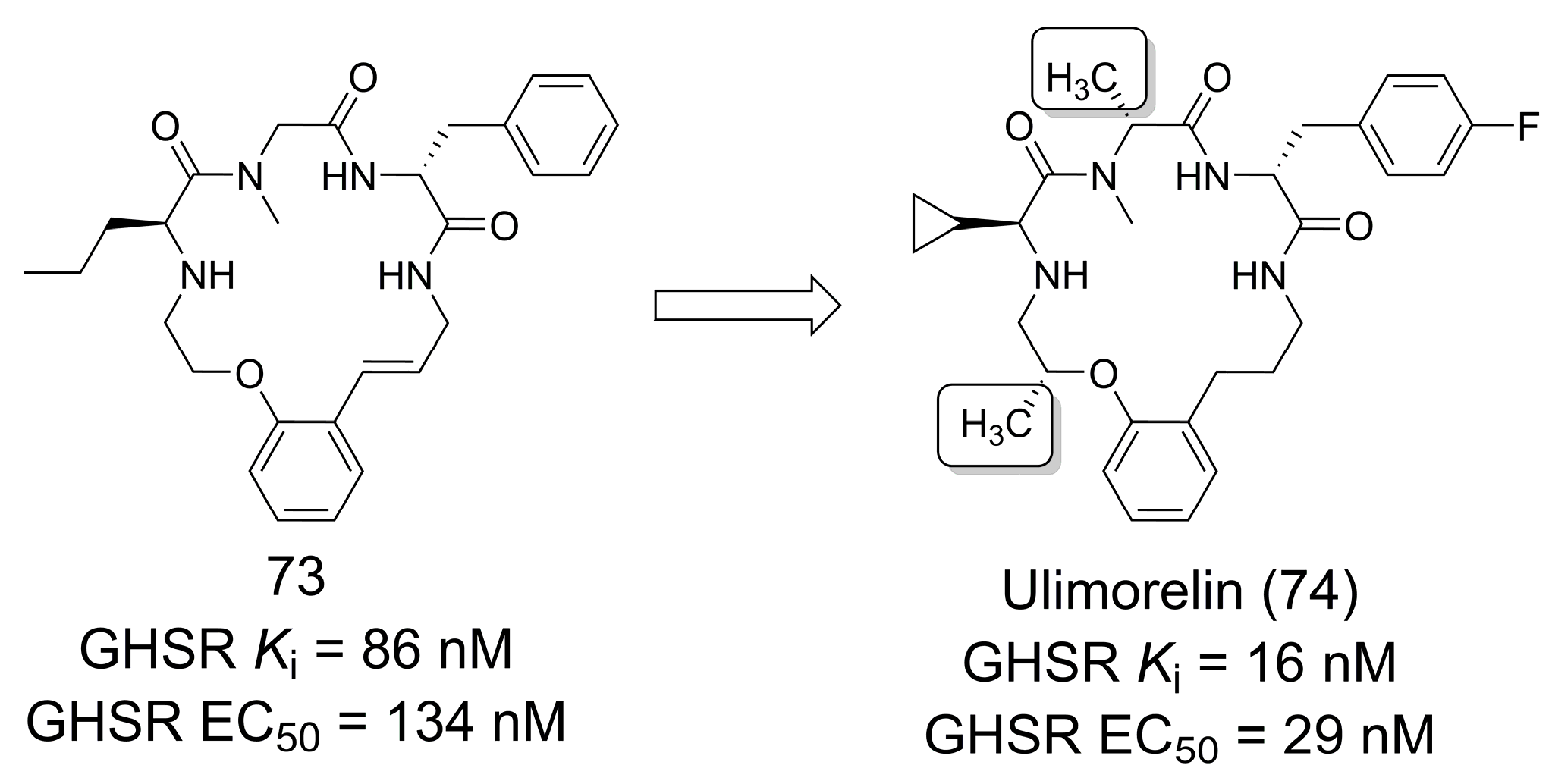
3.14. Human Ghrelin Receptor Agonist
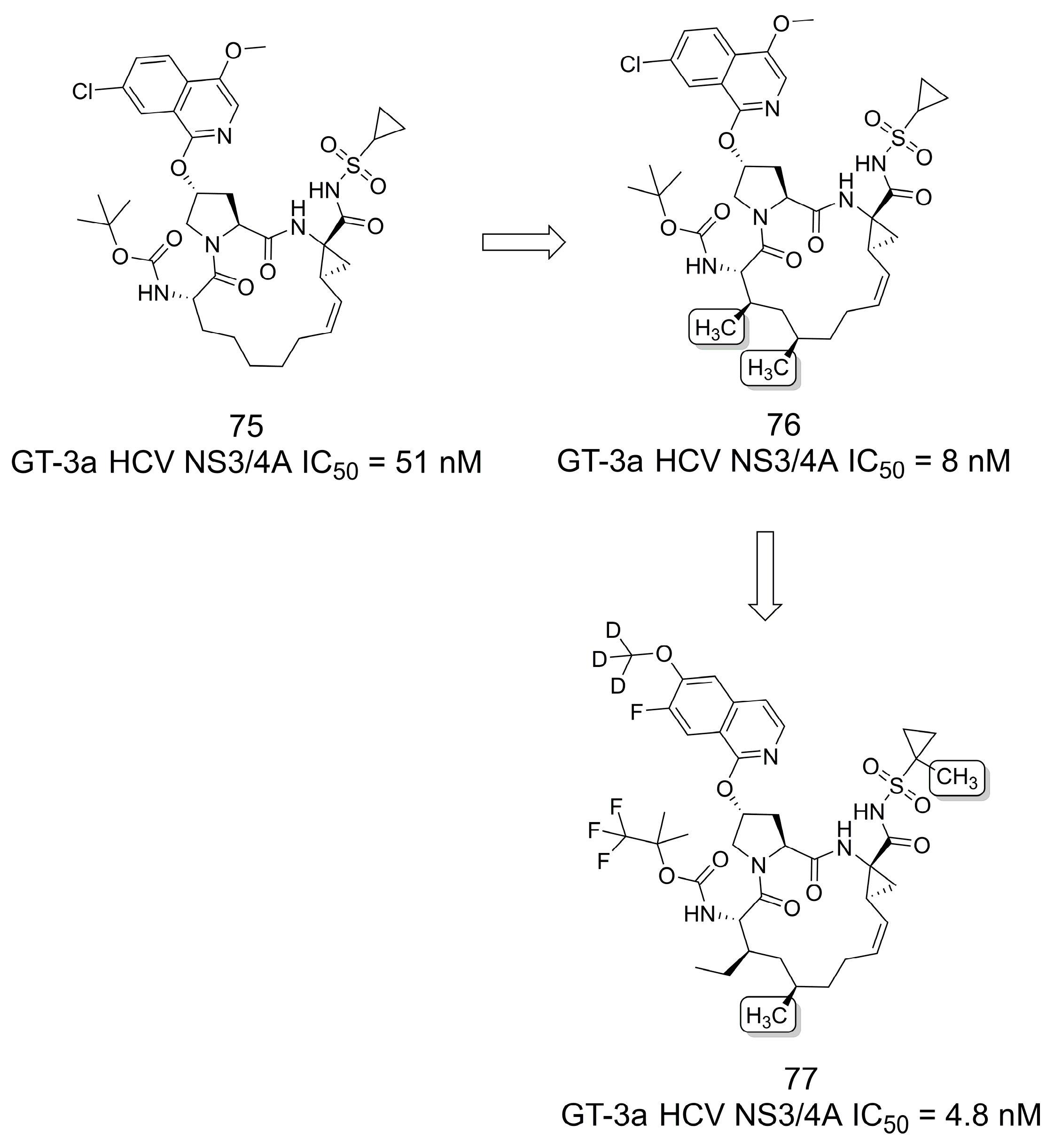
3.15. Pan-Genotype NS3/4A Protease Inhibitors
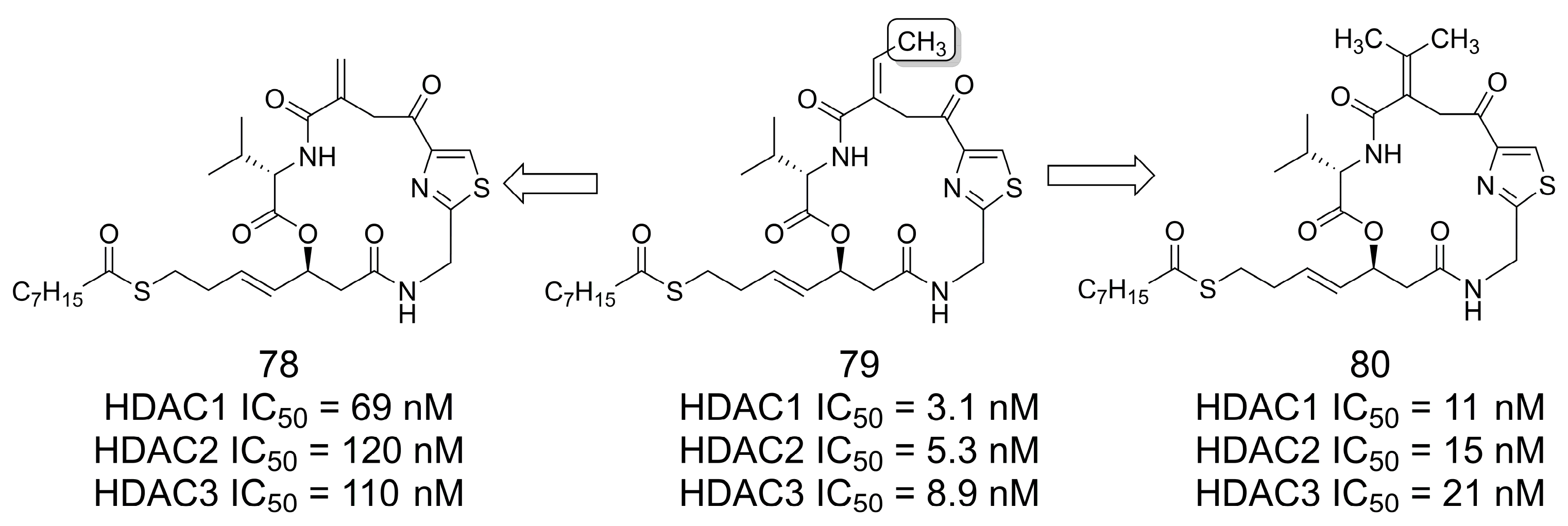
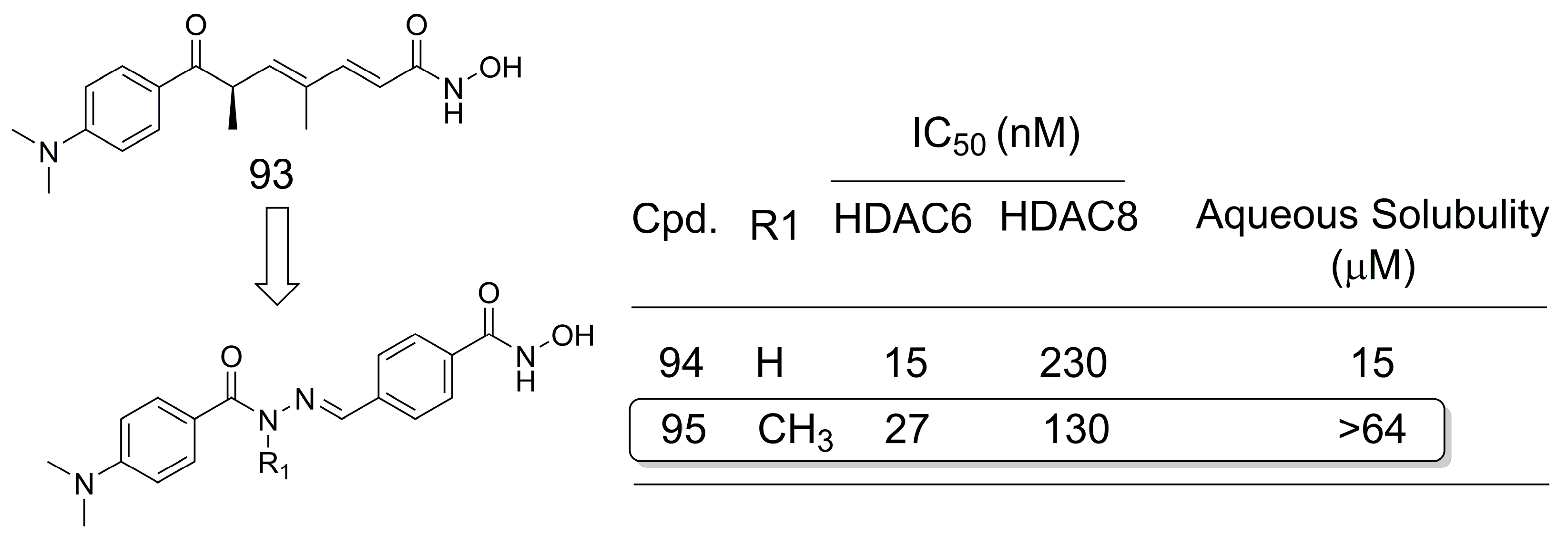
3.16. Class I Histone Deacetylase (HDAC) Inhibitors
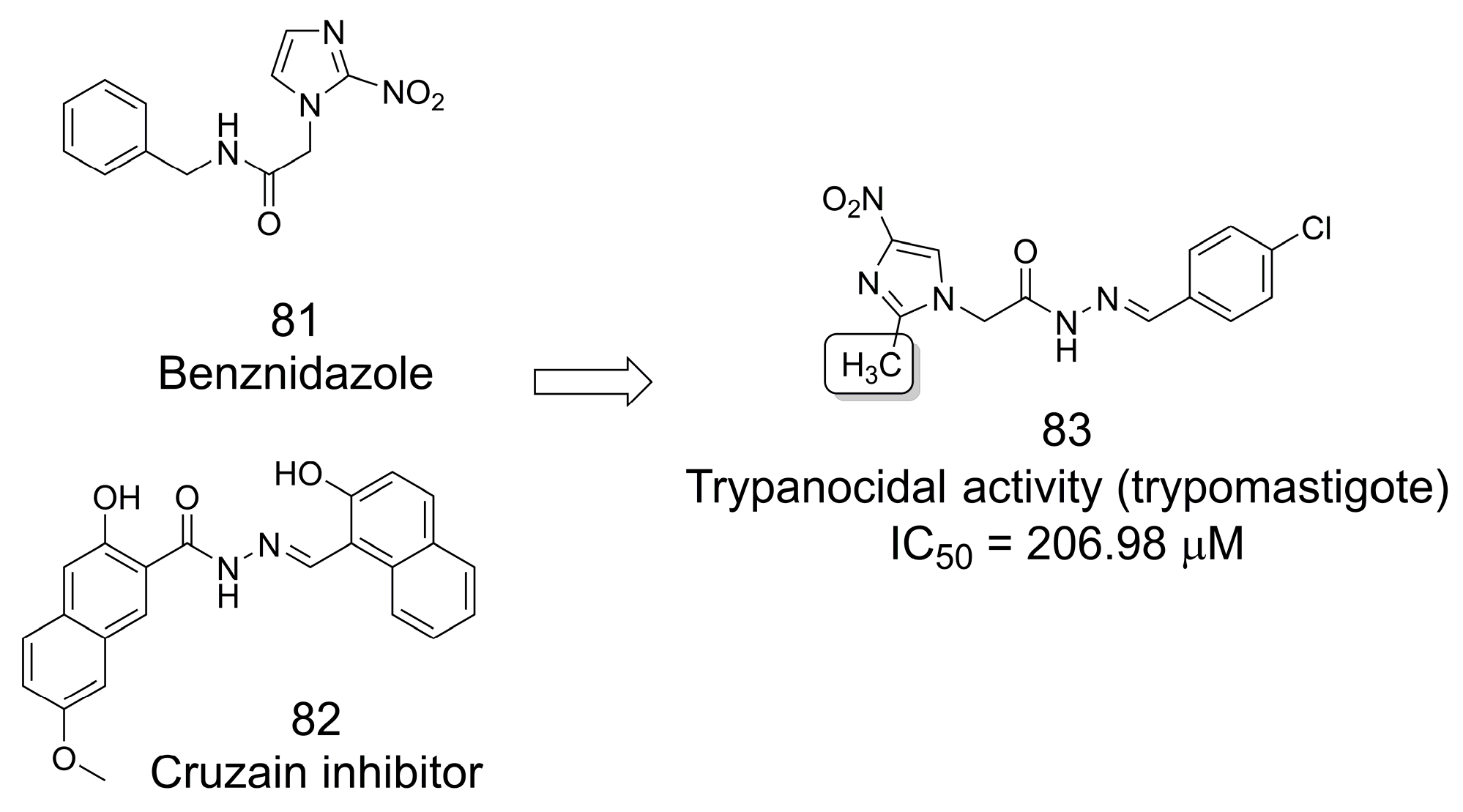
3.17. Trypanocidal Analogs of Benznidazole
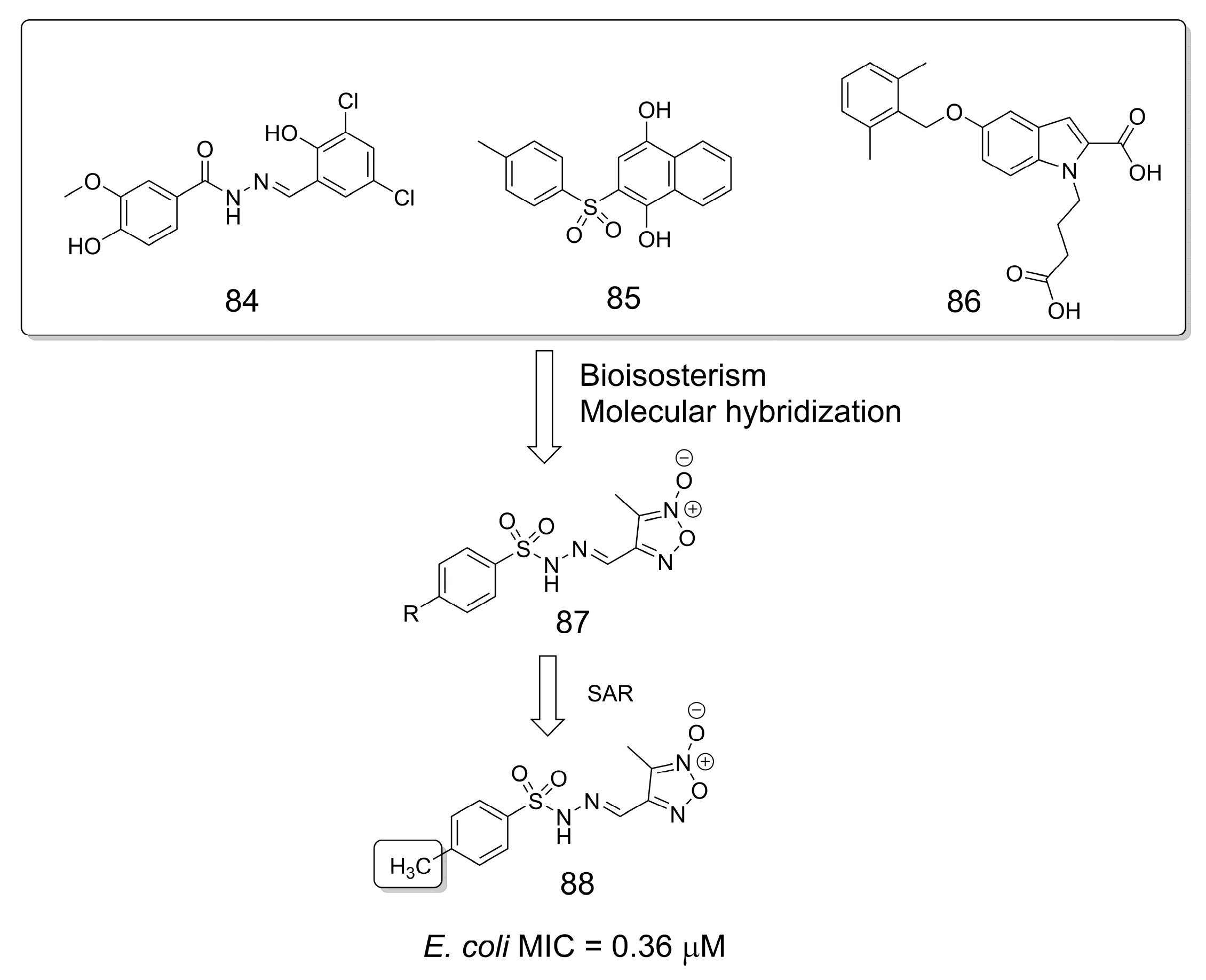
3.18. Antibacterial Agents
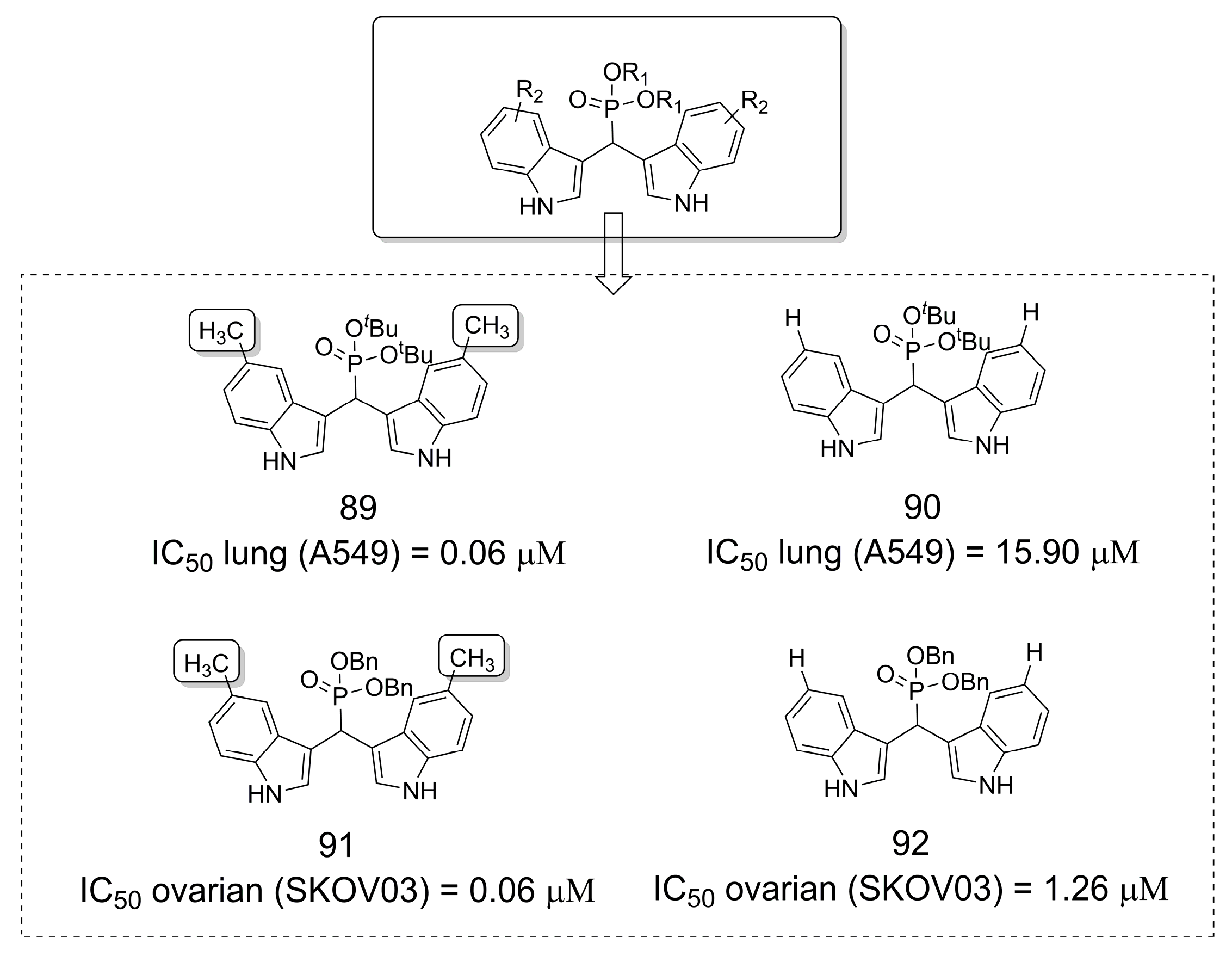
3.19. Phosphonate Derivatives as Anticancer Agents
4. The Methylation Effect in Physicochemical and Pharmacokinetic Property Optimization
4.1. Methylation Effect on Aqueous Solubility
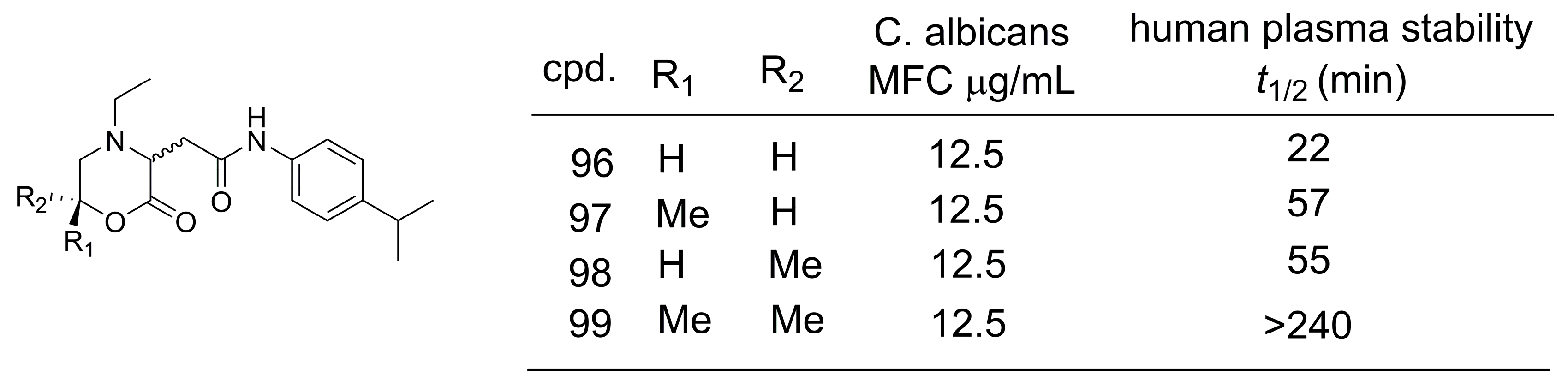
4.2. Methylation Effect on Plasma Stability
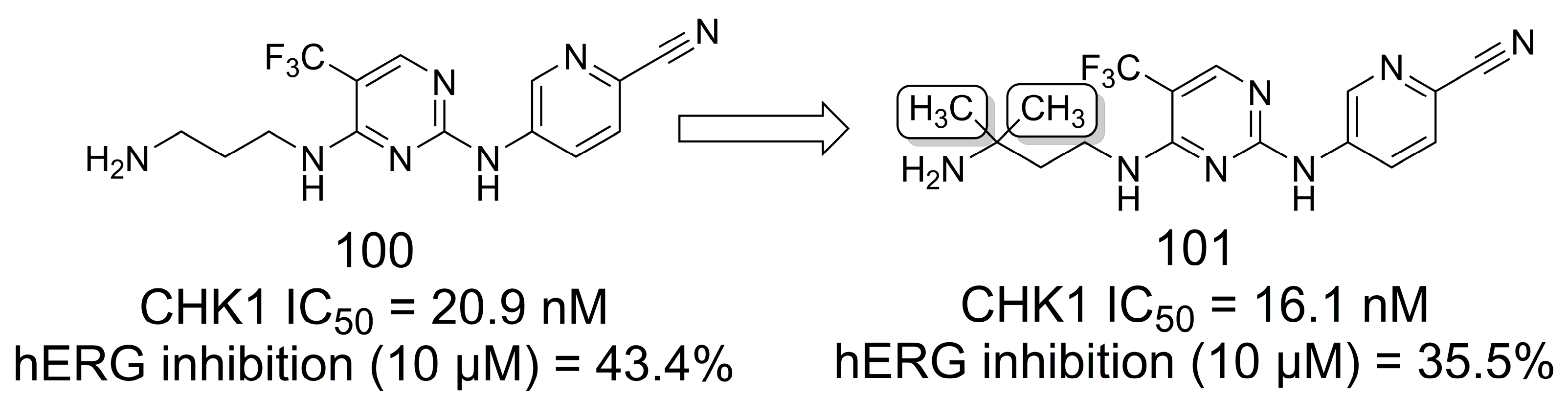
4.3. Methylation Effect on hERG Potassium Channel Inhibition
4.4. Methylation Effect on Metabolism
5. Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Barreiro, E.J.; Kümmerle, A.E.; Fraga, C.A.M. The Methylation Effect in Medicinal Chemistry. Chem. Rev. 2011, 111, 5215–5246. [Google Scholar] [CrossRef] [PubMed]
- Bissantz, C.; Kuhn, B.; Stahl, M. A Medicinal Chemist’s Guide to Molecular Interactions. J. Med. Chem. 2010, 53, 5061–5084. [Google Scholar] [CrossRef]
- Klebe, G. Applying Thermodynamic Profiling in Lead Finding and Optimization. Nat. Rev. Drug. Discov. 2015, 14, 95–110. [Google Scholar] [CrossRef]
- Zheng, Y.; Tice, C.M.; Singh, S.B. Conformational Control in Structure-Based Drug Design. Bioorg. Med. Chem. Lett. 2017, 27, 2825–2837. [Google Scholar] [CrossRef] [PubMed]
- de Sena, M.; Pinheiro, P.; Rodrigues, D.A.; do Couto Maia, R.; Thota, S.; Fraga, C.A.M. The Use of Conformational Restriction in Medicinal Chemistry. Curr. Top. Med. Chem. 2019, 19, 1712–1733. [Google Scholar] [CrossRef]
- Patani, G.A.; LaVoie, E.J. Bioisosterism: A Rational Approach in Drug Design. Chem. Rev. 1996, 96, 3147–3176. [Google Scholar] [CrossRef] [PubMed]
- Lima, L.; Barreiro, E. Bioisosterism: A Useful Strategy for Molecular Modification and Drug Design. Curr. Med. Chem. 2005, 12, 23–49. [Google Scholar] [CrossRef]
- Lima, L.M.; Barreiro, E.J. Beyond Bioisosterism: New Concepts in Drug Discovery. In Comprehensive Medicinal Chemistry III; Elsevier: Amsterdam, The Netherlands, 2017; pp. 186–210. [Google Scholar]
- Lima, L.M.; Alves, M.A.; do Amaral, D.N. Homologation: A Versatile Molecular Modification Strategy to Drug Discovery. Curr. Top. Med. Chem. 2019, 19, 1734–1750. [Google Scholar] [CrossRef]
- Ishikawa, M.; Hashimoto, Y. Improvement in Aqueous Solubility in Small Molecule Drug Discovery Programs by Disruption of Molecular Planarity and Symmetry. J. Med. Chem. 2011, 54, 1539–1554. [Google Scholar] [CrossRef]
- Ishikawa, M.; Hashimoto, Y. Improving the Water-Solubility of Compounds by Molecular Modification to Disrupt Crystal Packing. In The Practice of Medicinal Chemistry; Elsevier: Amsterdam, The Netherlands, 2015; pp. 747–765. [Google Scholar]
- Schönherr, H.; Cernak, T. Profound Methyl Effects in Drug Discovery and a Call for New C—H Methylation Reactions. Angew. Chem. Int. Ed. 2013, 52, 12256–12267. [Google Scholar] [CrossRef]
- Aynetdinova, D.; Callens, M.C.; Hicks, H.B.; Poh, C.Y.X.; Shennan, B.D.A.; Boyd, A.M.; Lim, Z.H.; Leitch, J.A.; Dixon, D.J. Installing the “Magic Methyl”—C–H Methylation in Synthesis. Chem. Soc. Rev. 2021, 50, 5517–5563. [Google Scholar] [CrossRef]
- Leung, C.S.; Leung, S.S.F.; Tirado-Rives, J.; Jorgensen, W.L. Methyl Effects on Protein–Ligand Binding. J. Med. Chem. 2012, 55, 4489–4500. [Google Scholar] [CrossRef] [PubMed]
- FDA Granted Accelerated Approval to Tazemetostat for Follicular Lymphoma. Available online: https://www.fda.gov/drugs/fda-granted-accelerated-approval-tazemetostat-follicular-lymphoma (accessed on 12 August 2023).
- Kuntz, K.W.; Campbell, J.E.; Keilhack, H.; Pollock, R.M.; Knutson, S.K.; Porter-Scott, M.; Richon, V.M.; Sneeringer, C.J.; Wigle, T.J.; Allain, C.J.; et al. The Importance of Being Me: Magic Methyls, Methyltransferase Inhibitors, and the Discovery of Tazemetostat. J. Med. Chem. 2016, 59, 1556–1564. [Google Scholar] [CrossRef] [PubMed]
- Makita, S.; Tobinai, K. Targeting EZH2 with Tazemetostat. Lancet Oncol. 2018, 19, 586–587. [Google Scholar] [CrossRef]
- Kaniskan, H.Ü.; Martini, M.L.; Jin, J. Inhibitors of Protein Methyltransferases and Demethylases. Chem. Rev. 2018, 118, 989–1068. [Google Scholar] [CrossRef]
- Zhou, B.; Liang, X.; Mei, H.; Qi, S.; Jiang, Z.; Wang, A.; Zou, F.; Liu, Q.; Liu, J.; Wang, W.; et al. Discovery of IHMT-EZH2-115 as a Potent and Selective Enhancer of Zeste Homolog 2 (EZH2) Inhibitor for the Treatment of B-Cell Lymphomas. J. Med. Chem. 2021, 64, 15170–15188. [Google Scholar] [CrossRef]
- Yang, J.; Liu, Y.; Lan, S.; Yu, S.; Ma, X.; Luo, D.; Shan, H.; Zhong, X.; Yan, G.; Li, R. Discovery of 2-Methyl-2-(4-(2-Methyl-8-(1 H -Pyrrolo[2,3- b ]Pyridin-6-Yl)-1 H -Naphtho[1,2- d ]Imidazol-1-Yl)Phenyl)Propanenitrile as a Novel PI3K/MTOR Inhibitor with Enhanced Antitumor Efficacy In Vitro and In Vivo. J. Med. Chem. 2022, 65, 12781–12801. [Google Scholar] [CrossRef]
- Liu, T.-J.; Koul, D.; LaFortune, T.; Tiao, N.; Shen, R.J.; Maira, S.-M.; Garcia-Echevrria, C.; Yung, W.K.A. NVP-BEZ235, a Novel Dual Phosphatidylinositol 3-Kinase/Mammalian Target of Rapamycin Inhibitor, Elicits Multifaceted Antitumor Activities in Human Gliomas. Mol. Cancer Ther. 2009, 8, 2204–2210. [Google Scholar] [CrossRef]
- Rodrigues, D.A.; Sagrillo, F.S.; Fraga, C.A.M. Duvelisib: A 2018 Novel FDA-Approved Small Molecule Inhibiting Phosphoinositide 3-Kinases. Pharmaceuticals 2019, 12, 69. [Google Scholar] [CrossRef]
- Zhang, Y.; Kwok-Shing Ng, P.; Kucherlapati, M.; Chen, F.; Liu, Y.; Tsang, Y.H.; de Velasco, G.; Jeong, K.J.; Akbani, R.; Hadjipanayis, A.; et al. A Pan-Cancer Proteogenomic Atlas of PI3K/AKT/MTOR Pathway Alterations. Cancer Cell 2017, 31, 820–832. [Google Scholar] [CrossRef] [PubMed]
- Carroll, F.I.; Carlezon, W.A. Development of κ Opioid Receptor Antagonists. J. Med. Chem. 2013, 56, 2178–2195. [Google Scholar] [CrossRef]
- Kormos, C.M.; Ondachi, P.W.; Runyon, S.P.; Thomas, J.B.; Mascarella, S.W.; Decker, A.M.; Navarro, H.A.; Carroll, F.I. Simple Tetrahydroisoquinolines Are Potent and Selective Kappa Opioid Receptor Antagonists. ACS Med. Chem. Lett. 2017, 8, 742–745. [Google Scholar] [CrossRef]
- Ondachi, P.W.; Kormos, C.M.; Runyon, S.P.; Thomas, J.B.; Mascarella, S.W.; Decker, A.M.; Navarro, H.A.; Fennell, T.R.; Snyder, R.W.; Carroll, F.I. Potent and Selective Tetrahydroisoquinoline Kappa Opioid Receptor Antagonists of Lead Compound (3 R)-7-Hydroxy- N -[(1 S)-2-Methyl-1-(Piperidin-1-Ylmethyl)Propyl]-1,2,3,4-Tetrahydroisoquinoline-3-Carboxamide (PDTic). J. Med. Chem. 2018, 61, 7525–7545. [Google Scholar] [CrossRef]
- Aizpurua-Olaizola, O.; Elezgarai, I.; Rico-Barrio, I.; Zarandona, I.; Etxebarria, N.; Usobiaga, A. Targeting the Endocannabinoid System: Future Therapeutic Strategies. Drug Discov. Today 2017, 22, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Pennant, N.M.; Hinton, C.V. The Evolution of Cannabinoid Receptors in Cancer. WIREs Mech. Dis. 2023, 15, e1602. [Google Scholar] [CrossRef] [PubMed]
- Tuo, W.; Bollier, M.; Leleu-Chavain, N.; Lemaire, L.; Barczyk, A.; Dezitter, X.; Klupsch, F.; Szczepanski, F.; Spencer, J.; Chavatte, P.; et al. Development of Novel Oxazolo[5,4-d]Pyrimidines as Competitive CB2 Neutral Antagonists Based on Scaffold Hopping. Eur. J. Med. Chem. 2018, 146, 68–78. [Google Scholar] [CrossRef]
- Hollinshead, S.P.; Tidwell, M.W.; Palmer, J.; Guidetti, R.; Sanderson, A.; Johnson, M.P.; Chambers, M.G.; Oskins, J.; Stratford, R.; Astles, P.C. Selective Cannabinoid Receptor Type 2 (CB2) Agonists: Optimization of a Series of Purines Leading to the Identification of a Clinical Candidate for the Treatment of Osteoarthritic Pain. J. Med. Chem. 2013, 56, 5722–5733. [Google Scholar] [CrossRef] [PubMed]
- Mugnaini, C.; Rabbito, A.; Brizzi, A.; Palombi, N.; Petrosino, S.; Verde, R.; Di Marzo, V.; Ligresti, A.; Corelli, F. Synthesis of Novel 2-(1-Adamantanylcarboxamido)Thiophene Derivatives. Selective Cannabinoid Type 2 (CB2) Receptor Agonists as Potential Agents for the Treatment of Skin Inflammatory Disease. Eur. J. Med. Chem. 2019, 161, 239–251. [Google Scholar] [CrossRef]
- Garai, S.; Leo, L.M.; Szczesniak, A.-M.; Hurst, D.P.; Schaffer, P.C.; Zagzoog, A.; Black, T.; Deschamps, J.R.; Miess, E.; Schulz, S.; et al. Discovery of a Biased Allosteric Modulator for Cannabinoid 1 Receptor: Preclinical Anti-Glaucoma Efficacy. J. Med. Chem. 2021, 64, 8104–8126. [Google Scholar] [CrossRef]
- Laprairie, R.B.; Kulkarni, P.M.; Deschamps, J.R.; Kelly, M.E.M.; Janero, D.R.; Cascio, M.G.; Stevenson, L.A.; Pertwee, R.G.; Kenakin, T.P.; Denovan-Wright, E.M.; et al. Enantiospecific Allosteric Modulation of Cannabinoid 1 Receptor. ACS Chem. Neurosci. 2017, 8, 1188–1203. [Google Scholar] [CrossRef]
- Erlanson, D.A.; Fesik, S.W.; Hubbard, R.E.; Jahnke, W.; Jhoti, H. Twenty Years on: The Impact of Fragments on Drug Discovery. Nat. Rev. Drug Discov. 2016, 15, 605–619. [Google Scholar] [CrossRef]
- Rees, D.C.; Congreve, M.; Murray, C.W.; Carr, R. Fragment-Based Lead Discovery. Nat. Rev. Drug Discov. 2004, 3, 660–672. [Google Scholar] [CrossRef]
- de Graaf, C.; Kooistra, A.J.; Vischer, H.F.; Katritch, V.; Kuijer, M.; Shiroishi, M.; Iwata, S.; Shimamura, T.; Stevens, R.C.; de Esch, I.J.P.; et al. Crystal Structure-Based Virtual Screening for Fragment-like Ligands of the Human Histamine H 1 Receptor. J. Med. Chem. 2011, 54, 8195–8206. [Google Scholar] [CrossRef]
- Kuhne, S.; Kooistra, A.J.; Bosma, R.; Bortolato, A.; Wijtmans, M.; Vischer, H.F.; Mason, J.S.; de Graaf, C.; de Esch, I.J.P.; Leurs, R. Identification of Ligand Binding Hot Spots of the Histamine H 1 Receptor Following Structure-Based Fragment Optimization. J. Med. Chem. 2016, 59, 9047–9061. [Google Scholar] [CrossRef]
- Moreau, R.J.; Skepper, C.K.; Appleton, B.A.; Blechschmidt, A.; Balibar, C.J.; Benton, B.M.; Drumm, J.E.; Feng, B.Y.; Geng, M.; Li, C.; et al. Fragment-Based Drug Discovery of Inhibitors of Phosphopantetheine Adenylyltransferase from Gram-Negative Bacteria. J. Med. Chem. 2018, 61, 3309–3324. [Google Scholar] [CrossRef] [PubMed]
- Nigg, E.A. Mitotic Kinases as Regulators of Cell Division and Its Checkpoints. Nat. Rev. Mol. Cell Biol. 2001, 2, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Shi, Q.; Choudhry, N.; Li, H.; Yang, C.; Kalashova, J.; Yan, Z.; Li, J.; Reddy, M.C.; Gopala, S.G.; et al. Discovery and Optimization of Seven-Membered Lactam-Based Compounds to Phenocopy the Inhibition of the Aurora Kinase B. ACS Med. Chem. Lett. 2022, 13, 1091–1098. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yan, Z.; Li, H.; Shi, Q.; Huang, L.; Nimishetti, N.; Allen, T.D.; Yang, D.; Zhang, J. A High-Content Screen for Anti-Mitosis and Polyploidy-Induction Identifies an Unknown Activity of Two Benzophenanthridine Alkaloids from Corydalis Longicalcarata. Phytochem. Lett. 2021, 41, 180–185. [Google Scholar] [CrossRef]
- Hoveyda, H.R.; Fraser, G.L.; Roy, M.-O.; Dutheuil, G.; Batt, F.; El Bousmaqui, M.; Korac, J.; Lenoir, F.; Lapin, A.; Noël, S.; et al. Discovery and Optimization of Novel Antagonists to the Human Neurokinin-3 Receptor for the Treatment of Sex-Hormone Disorders (Part I). J. Med. Chem. 2015, 58, 3060–3082. [Google Scholar] [CrossRef]
- Cieślak, M.; Słowianek, M. Cereblon-Recruiting PROTACs: Will New Drugs Have to Face Old Challenges? Pharmaceutics 2023, 15, 812. [Google Scholar] [CrossRef] [PubMed]
- Boichenko, I.; Bär, K.; Deiss, S.; Heim, C.; Albrecht, R.; Lupas, A.N.; Hernandez Alvarez, B.; Hartmann, M.D. Chemical Ligand Space of Cereblon. ACS Omega 2018, 3, 11163–11171. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.; Li, C.; Tang, H.; Tandon, I.; Liao, J.; Roberts, B.L.; Zhao, Y.; Tang, W. Development of Substituted Phenyl Dihydrouracil as the Novel Achiral Cereblon Ligands for Targeted Protein Degradation. J. Med. Chem. 2023, 66, 2904–2917. [Google Scholar] [CrossRef]
- Barbosa, G.; Gelves, L.G.V.; Costa, C.M.X.; Franco, L.S.; de Lima, J.A.L.; Aparecida-Silva, C.; Teixeira, J.D.; Mermelstein, C.d.S.; Barreiro, E.J.; Lima, L.M. Discovery of Putative Dual Inhibitor of Tubulin and EGFR by Phenotypic Approach on LASSBio-1586 Homologs. Pharmaceuticals 2022, 15, 913. [Google Scholar] [CrossRef] [PubMed]
- Viegas-Junior, C.; Danuello, A.; da Silva Bolzani, V.; Barreiro, E.J.; Fraga, C.A.M. Manssour Fraga Molecular Hybridization: A Useful Tool in the Design of New Drug Prototypes. Curr. Med. Chem. 2007, 14, 1829–1852. [Google Scholar] [CrossRef]
- Leal, C.M.; Pereira, S.L.; Kümmerle, A.E.; Leal, D.M.; Tesch, R.; de Sant’Anna, C.M.R.; Fraga, C.A.M.; Barreiro, E.J.; Sudo, R.T.; Zapata-Sudo, G. Antihypertensive Profile of 2-Thienyl-3,4-Methylenedioxybenzoylhydrazone Is Mediated by Activation of the A2A Adenosine Receptor. Eur. J. Med. Chem. 2012, 55, 49–57. [Google Scholar] [CrossRef]
- Kümmerle, A.E.; Schmitt, M.; Cardozo, S.V.S.; Lugnier, C.; Villa, P.; Lopes, A.B.; Romeiro, N.C.; Justiniano, H.; Martins, M.A.; Fraga, C.A.M.; et al. Design, Synthesis, and Pharmacological Evaluation of N -Acylhydrazones and Novel Conformationally Constrained Compounds as Selective and Potent Orally Active Phosphodiesterase-4 Inhibitors. J. Med. Chem. 2012, 55, 7525–7545. [Google Scholar] [CrossRef] [PubMed]
- Bastos, I.; Pinheiro, P.; Costa, F.; Rocha, M.; Sant’Anna, C.; Braz, D.; Souza, E.; Martins, M.; Barreiro, E.; Ferreira, F.; et al. Design, Synthesis, Experimental and Theoretical Characterization of a New Multitarget 2-Thienyl-N-Acylhydrazone Derivative. Pharmaceuticals 2018, 11, 119. [Google Scholar] [CrossRef]
- Pinheiro, P.d.S.M.; Rodrigues, D.A.; Alves, M.A.; Tinoco, L.W.; Ferreira, G.B.; de Sant’Anna, C.M.R.; Fraga, C.A.M. Theoretical and Experimental Characterization of 1,4-N⋯S σ-Hole Intramolecular Interactions in Bioactive N -Acylhydrazone Derivatives. New J. Chem. 2018, 42, 497–505. [Google Scholar] [CrossRef]
- Brullo, C.; Rapetti, F.; Abbate, S.; Prosdocimi, T.; Torretta, A.; Semrau, M.; Massa, M.; Alfei, S.; Storici, P.; Parisini, E.; et al. Design, Synthesis, Biological Evaluation and Structural Characterization of Novel GEBR Library PDE4D Inhibitors. Eur. J. Med. Chem. 2021, 223, 113638. [Google Scholar] [CrossRef]
- Nunes, I.K.d.C.; de Souza, E.T.; Martins, I.R.R.; Barbosa, G.; Moraes Junior, M.O.d.; Medeiros, M.d.M.; Silva, S.W.D.; Balliano, T.L.; da Silva, B.A.; Silva, P.M.R.; et al. Discovery of Sulfonyl Hydrazone Derivative as a New Selective PDE4A and PDE4D Inhibitor by Lead-Optimization Approach on the Prototype LASSBio-448: In Vitro and in Vivo Preclinical Studies. Eur. J. Med. Chem. 2020, 204, 112492. [Google Scholar] [CrossRef]
- Nunes, I.K.d.C.; de Souza, E.T.; Cardozo, S.V.S.; Carvalho, V.d.F.; Romeiro, N.C.; Silva, P.M.R.e.; Martins, M.A.; Barreiro, E.J.; Lima, L.M. Synthesis, Pharmacological Profile and Docking Studies of New Sulfonamides Designed as Phosphodiesterase-4 Inhibitors. PLoS ONE 2016, 11, e0162895. [Google Scholar] [CrossRef]
- Tolomeu, H.V.; Fraga, C.A.M. The Outcomes of Small-Molecule Kinase Inhibitors and the Role of ROCK2 as a Molecular Target for the Treatment of Alzheimer’s Disease. CNS Neurol. Disord Drug Targets 2022, 21, 188–205. [Google Scholar] [CrossRef]
- Montagnoli, T.L.; de Oliveira, D.R.; Fraga, C.A.M. Therapeutic Perspectives on ROCK Inhibition for Cerebral Cavernous Malformations. Kinases Phosphatases 2023, 1, 72–96. [Google Scholar] [CrossRef]
- De Oliveira, R.G.; Guerra, F.S.; Mermelstein, C.d.S.; Fernandes, P.D.; Bastos, I.T.d.S.; Costa, F.N.; Barroso, R.C.R.; Ferreira, F.F.; Fraga, C.A.M. Synthesis and Pharmacological Evaluation of Novel Isoquinoline N -Sulphonylhydrazones Designed as ROCK Inhibitors. J. Enzym. Inhib. Med. Chem. 2018, 33, 1181–1193. [Google Scholar] [CrossRef]
- Jacobs, M.; Hayakawa, K.; Swenson, L.; Bellon, S.; Fleming, M.; Taslimi, P.; Doran, J. The Structure of Dimeric ROCK I Reveals the Mechanism for Ligand Selectivity. J. Biol. Chem. 2006, 281, 260–268. [Google Scholar] [CrossRef]
- Avila, C.M.; Lopes, A.B.; Gonçalves, A.S.; da Silva, L.L.; Romeiro, N.C.; Miranda, A.L.P.; Sant’Anna, C.M.R.; Barreiro, E.J.; Fraga, C.A.M. Structure-Based Design and Biological Profile of (E)-N-(4-Nitrobenzylidene)-2-Naphthohydrazide, a Novel Small Molecule Inhibitor of IκB Kinase-β. Eur. J. Med. Chem. 2011, 46, 1245–1253. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, Y.; Wang, H.; Li, H.; Zhang, T.; Peng, Y.; Wang, X. Exploring Methamphetamine Nonenantioselectively Targeting Toll-like Receptor 4/Myeloid Differentiation Protein 2 by in Silico Simulations and Wet-Lab Techniques. J. Chem. Inf. Model. 2020, 60, 1607–1613. [Google Scholar] [CrossRef]
- Bachtell, R.; Hutchinson, M.; Wang, X.; Rice, K.; Maier, S.; Watkins, L. Targeting the Toll of Drug Abuse: The Translational Potential of Toll-Like Receptor 4. CNS Neurol. Disord Drug Targets 2015, 14, 692–699. [Google Scholar] [CrossRef]
- Wang, X.; Northcutt, A.L.; Cochran, T.A.; Zhang, X.; Fabisiak, T.J.; Haas, M.E.; Amat, J.; Li, H.; Rice, K.C.; Maier, S.F.; et al. Methamphetamine Activates Toll-Like Receptor 4 to Induce Central Immune Signaling within the Ventral Tegmental Area and Contributes to Extracellular Dopamine Increase in the Nucleus Accumbens Shell. ACS Chem. Neurosci. 2019, 10, 3622–3634. [Google Scholar] [CrossRef]
- Räder, A.F.B.; Weinmüller, M.; Reichart, F.; Schumacher-Klinger, A.; Merzbach, S.; Gilon, C.; Hoffman, A.; Kessler, H. Orally Active Peptides: Is There a Magic Bullet? Angew. Chem. Int. Ed. 2018, 57, 14414–14438. [Google Scholar] [CrossRef]
- Vinogradov, A.A.; Yin, Y.; Suga, H. Macrocyclic Peptides as Drug Candidates: Recent Progress and Remaining Challenges. J. Am. Chem. Soc. 2019, 141, 4167–4181. [Google Scholar] [CrossRef]
- Hoveyda, H.R.; Marsault, E.; Gagnon, R.; Mathieu, A.P.; Vézina, M.; Landry, A.; Wang, Z.; Benakli, K.; Beaubien, S.; Saint-Louis, C.; et al. Optimization of the Potency and Pharmacokinetic Properties of a Macrocyclic Ghrelin Receptor Agonist (Part I): Development of Ulimorelin (TZP-101) from Hit to Clinic. J. Med. Chem. 2011, 54, 8305–8320. [Google Scholar] [CrossRef]
- Sun, L.-Q.; Mull, E.; D’Andrea, S.; Zheng, B.; Hiebert, S.; Gillis, E.; Bowsher, M.; Kandhasamy, S.; Baratam, V.R.; Puttaswamy, S.; et al. Discovery of BMS-986144, a Third-Generation, Pan-Genotype NS3/4A Protease Inhibitor for the Treatment of Hepatitis C Virus Infection. J. Med. Chem. 2020, 63, 14740–14760. [Google Scholar] [CrossRef]
- Vermehren, J.; Park, J.S.; Jacobson, I.M.; Zeuzem, S. Challenges and Perspectives of Direct Antivirals for the Treatment of Hepatitis C Virus Infection. J. Hepatol. 2018, 69, 1178–1187. [Google Scholar] [CrossRef]
- Zhang, K.; Yao, Y.; Tu, Z.; Liao, C.; Wang, Z.; Qiu, Y.; Chen, D.; Hamilton, D.J.; Li, Z.; Jiang, S. Discovery of Class I Histone Deacetylase Inhibitors Based on Romidpesin with Promising Selectivity for Cancer Cells. Future Med. Chem. 2020, 12, 311–323. [Google Scholar] [CrossRef]
- Yao, Y.; Tu, Z.; Liao, C.; Wang, Z.; Li, S.; Yao, H.; Li, Z.; Jiang, S. Discovery of Novel Class I Histone Deacetylase Inhibitors with Promising in Vitro and in Vivo Antitumor Activities. J. Med. Chem. 2015, 58, 7672–7680. [Google Scholar] [CrossRef]
- de Alcântara, G.P.; Barbosa, J.M.C.; Salomão, K.; de Castro, S.L.; Wardell, J.L.; Low, J.N.; Wardell, S.M.S.V.; Carvalho, S.A.; da Silva, E.F.; Fraga, C.A.M. New 4-Nitro-Imidazole-N-Glycinyl-Hydrazones Designed as Trypanocidal Analogues of Benznidazole. Lett. Drug Des. Discov. 2023, 20, 488–497. [Google Scholar] [CrossRef]
- Wang, X.-L.; Zhang, Y.-B.; Tang, J.-F.; Yang, Y.-S.; Chen, R.-Q.; Zhang, F.; Zhu, H.-L. Design, Synthesis and Antibacterial Activities of Vanillic Acylhydrazone Derivatives as Potential β-Ketoacyl-Acyl Carrier Protein Synthase III (FabH) Inhibitors. Eur. J. Med. Chem. 2012, 57, 373–382. [Google Scholar] [CrossRef]
- Segretti, N.D.; Serafim, R.A.M.; Segretti, M.C.F.; Miyata, M.; Coelho, F.R.; Augusto, O.; Ferreira, E.I. New Antibacterial Agents: Hybrid Bioisoster Derivatives as Potential E. Coli FabH Inhibitors. Bioorg. Med. Chem. Lett. 2016, 26, 3988–3993. [Google Scholar] [CrossRef]
- Maestro, A.; Martín-Encinas, E.; Alonso, C.; Martinez de Marigorta, E.; Rubiales, G.; Vicario, J.; Palacios, F. Synthesis of Novel Antiproliferative Hybrid Bis-(3-Indolyl)Methane Phosphonate Derivatives. Eur. J. Med. Chem. 2018, 158, 874–883. [Google Scholar] [CrossRef]
- Rodrigues, D.A.; Ferreira-Silva, G.À.; Ferreira, A.C.S.; Fernandes, R.A.; Kwee, J.K.; Sant’Anna, C.M.R.; Ionta, M.; Fraga, C.A.M. Design, Synthesis, and Pharmacological Evaluation of Novel N -Acylhydrazone Derivatives as Potent Histone Deacetylase 6/8 Dual Inhibitors. J. Med. Chem. 2016, 59, 655–670. [Google Scholar] [CrossRef]
- Kümmerle, A.E.; Raimundo, J.M.; Leal, C.M.; da Silva, G.S.; Balliano, T.L.; Pereira, M.A.; de Simone, C.A.; Sudo, R.T.; Zapata-Sudo, G.; Fraga, C.A.M. Studies towards the Identification of Putative Bioactive Conformation of Potent Vasodilator Arylidene N-Acylhydrazone Derivatives. Eur. J. Med. Chem. 2009, 44, 4004–4009. [Google Scholar] [CrossRef] [PubMed]
- Bardiot, D.; Thevissen, K.; De Brucker, K.; Peeters, A.; Cos, P.; Taborda, C.P.; McNaughton, M.; Maes, L.; Chaltin, P.; Cammue, B.P.A.; et al. 2-(2-Oxo-Morpholin-3-Yl)-Acetamide Derivatives as Broad-Spectrum Antifungal Agents. J. Med. Chem. 2015, 58, 1502–1512. [Google Scholar] [CrossRef] [PubMed]
- Jin, T.; Xu, L.; Wang, P.; Hu, X.; Zhang, R.; Wu, Z.; Du, W.; Kan, W.; Li, K.; Wang, C.; et al. Discovery and Development of a Potent, Selective, and Orally Bioavailable CHK1 Inhibitor Candidate: 5-((4-((3-Amino-3-Methylbutyl)Amino)-5-(Trifluoromethyl)Pyrimidin-2-Yl)Amino)Picolinonitrile. J. Med. Chem. 2021, 64, 15069–15090. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Pagare, P.P.; Li, M.; Neel, L.T.; Mendez, R.E.; Gillespie, J.C.; Stevens, D.L.; Dewey, W.L.; Selley, D.E.; Zhang, Y. Structural Alterations of the “Address” Moiety of NAN Leading to the Discovery of a Novel Opioid Receptor Modulator with Reduced HERG Toxicity. J. Med. Chem. 2023, 66, 577–595. [Google Scholar] [CrossRef]
- Obeng, S.; Jali, A.; Zheng, Y.; Wang, H.; Schwienteck, K.L.; Chen, C.; Stevens, D.L.; Akbarali, H.I.; Dewey, W.L.; Banks, M.L.; et al. Characterization of 17-Cyclopropylmethyl-3,14β-Dihydroxy-4,5α-Epoxy-6α-(Indole-7-Carboxamido)Morphinan (NAN) as a Novel Opioid Receptor Modulator for Opioid Use Disorder Treatment. ACS Chem. Neurosci. 2019, 10, 2518–2532. [Google Scholar] [CrossRef]
- Liu, K.; Li, D.; Zheng, W.; Shi, M.; Chen, Y.; Tang, M.; Yang, T.; Zhao, M.; Deng, D.; Zhang, C.; et al. Discovery, Optimization, and Evaluation of Quinazolinone Derivatives with Novel Linkers as Orally Efficacious Phosphoinositide-3-Kinase Delta Inhibitors for Treatment of Inflammatory Diseases. J. Med. Chem. 2021, 64, 8951–8970. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pinheiro, P.d.S.M.; Franco, L.S.; Fraga, C.A.M. The Magic Methyl and Its Tricks in Drug Discovery and Development. Pharmaceuticals 2023, 16, 1157. https://doi.org/10.3390/ph16081157
Pinheiro PdSM, Franco LS, Fraga CAM. The Magic Methyl and Its Tricks in Drug Discovery and Development. Pharmaceuticals. 2023; 16(8):1157. https://doi.org/10.3390/ph16081157
Chicago/Turabian StylePinheiro, Pedro de Sena Murteira, Lucas Silva Franco, and Carlos Alberto Manssour Fraga. 2023. "The Magic Methyl and Its Tricks in Drug Discovery and Development" Pharmaceuticals 16, no. 8: 1157. https://doi.org/10.3390/ph16081157
APA StylePinheiro, P. d. S. M., Franco, L. S., & Fraga, C. A. M. (2023). The Magic Methyl and Its Tricks in Drug Discovery and Development. Pharmaceuticals, 16(8), 1157. https://doi.org/10.3390/ph16081157








