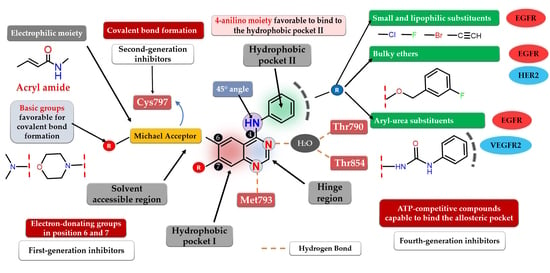
Structure–Activity Relationship Studies Based on Quinazoline Derivatives as EGFR Kinase Inhibitors (2017–Present)
Abstract
1. Introduction
2. Epidermal Growth Factor Receptor (EGFR)
2.1. EGFR Implications in Cancer
2.2. EGFR Inhibition in Cancer
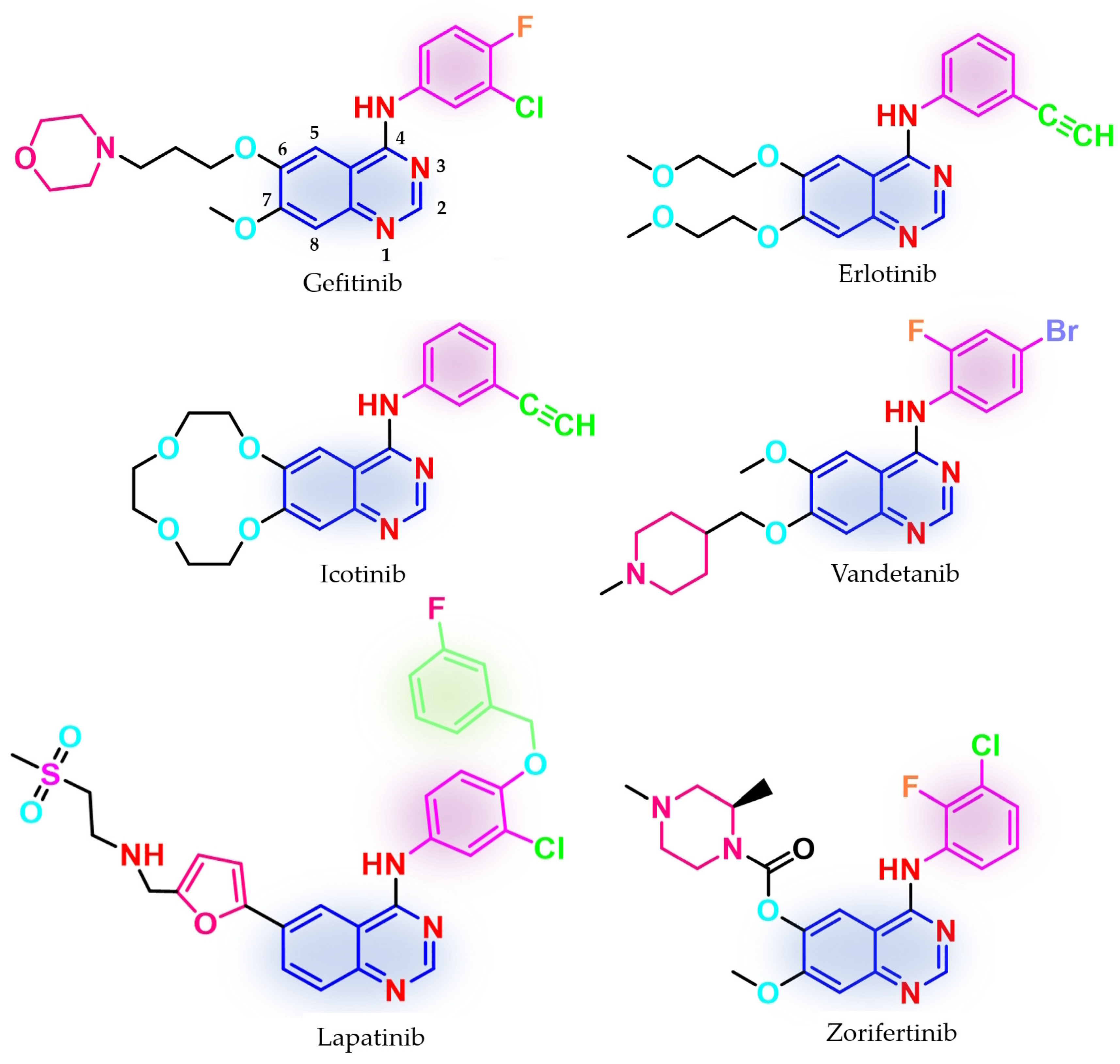
3. First Generation of EGFR TKIs
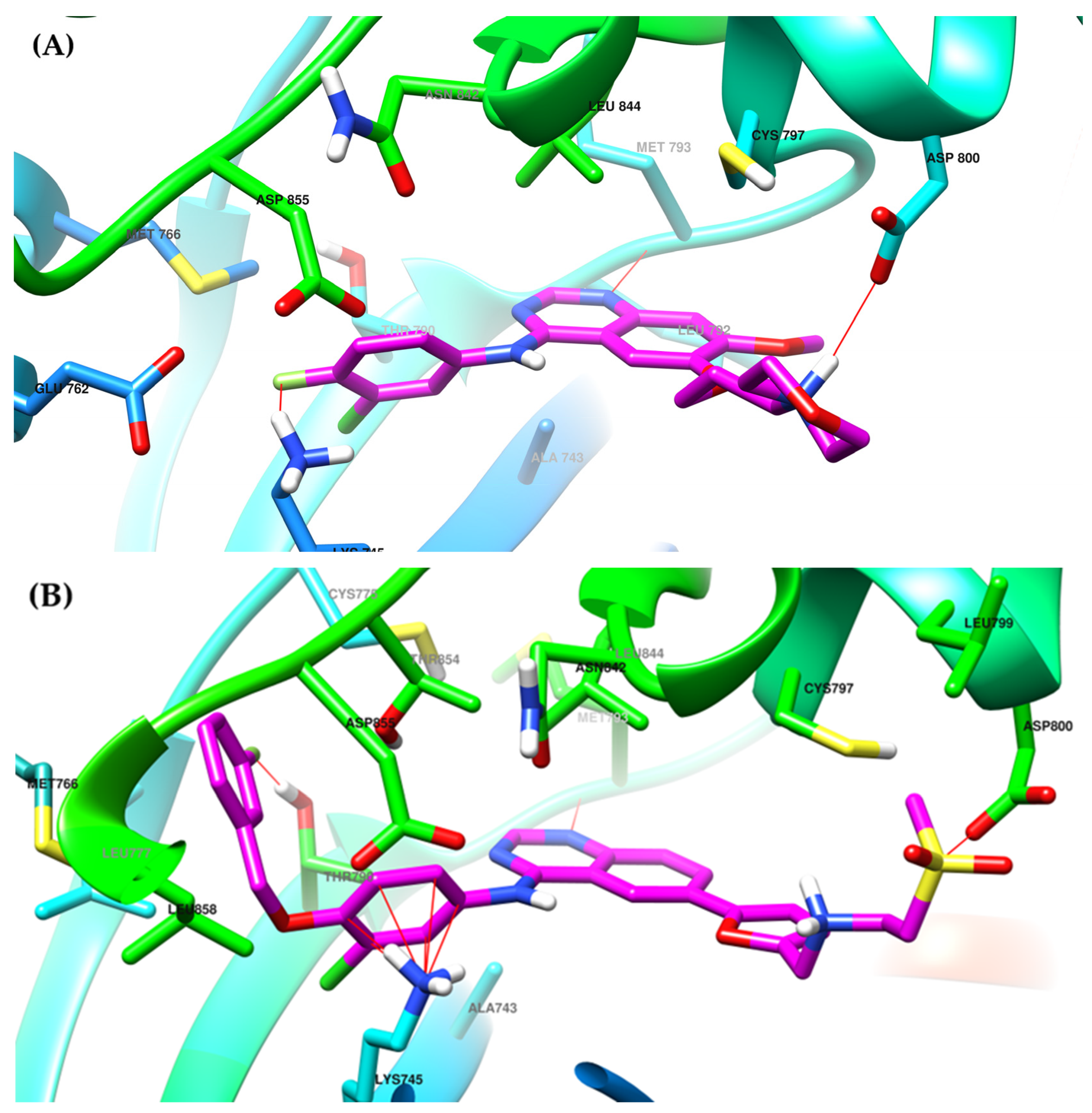
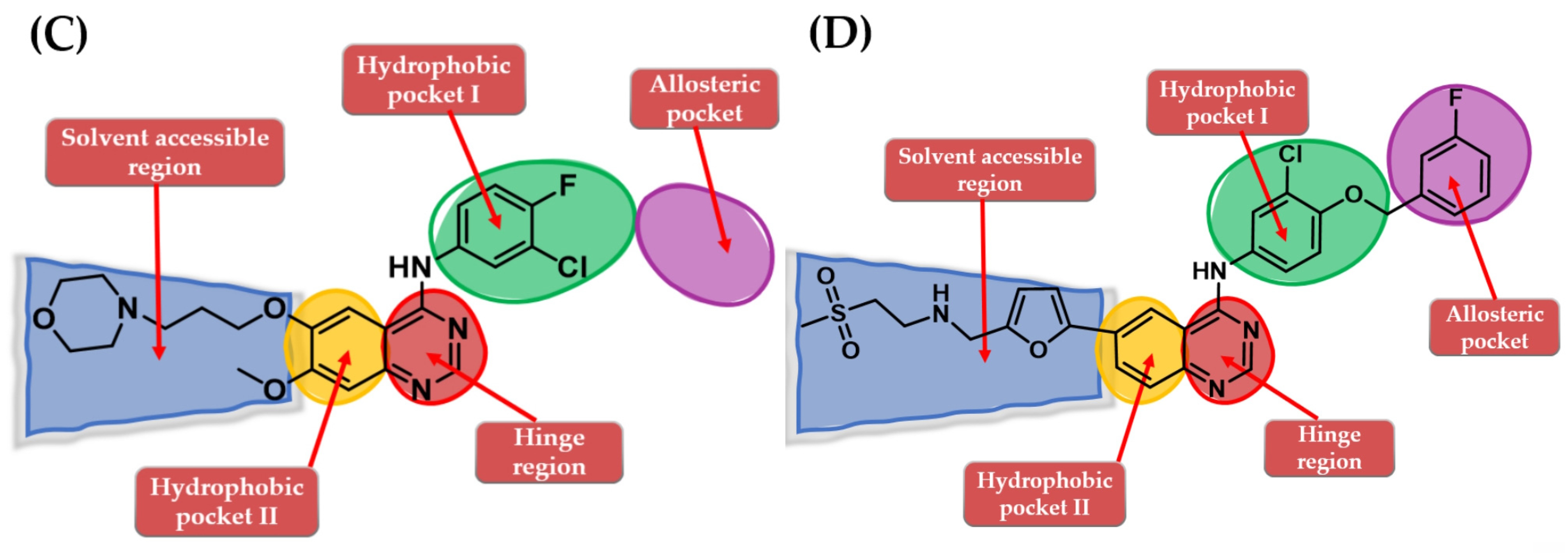
4. Novel First-Generation Quinazoline EGFR TKIs (2017–Present)
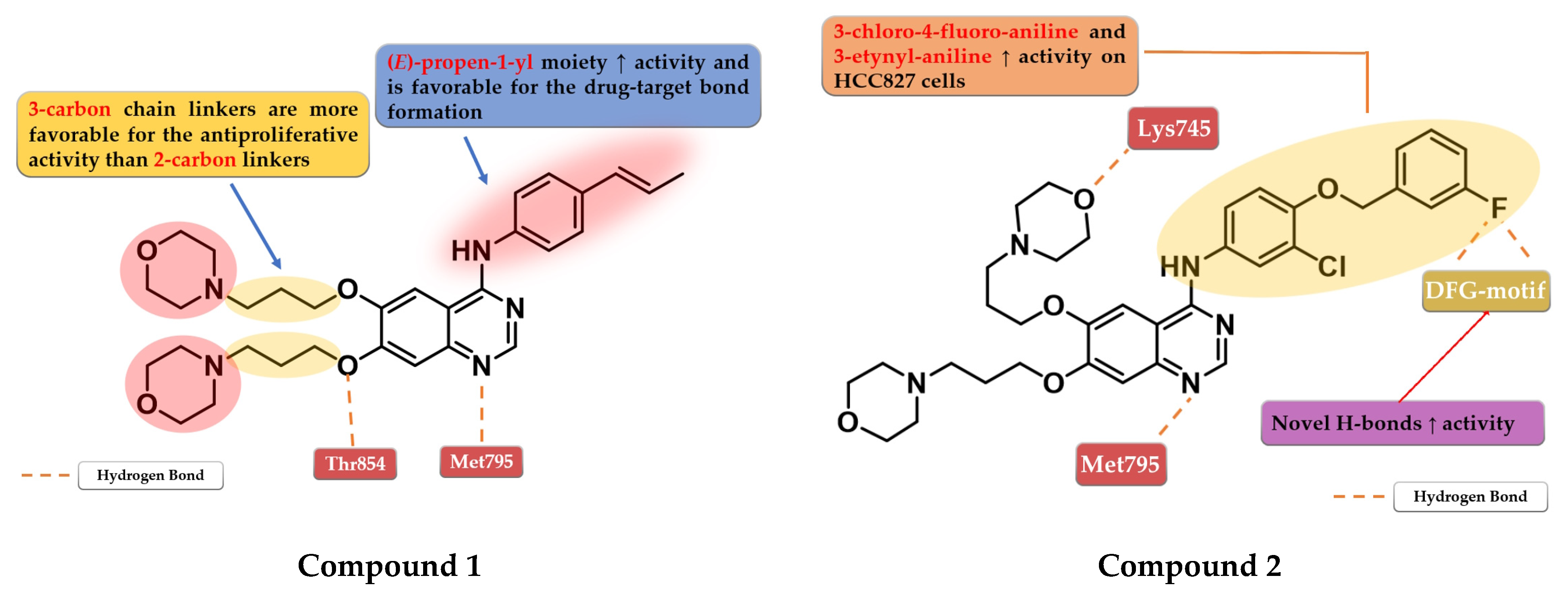
4.1. 6,7-Dimorpholinoalkoxy-4-anilino-quinazolines
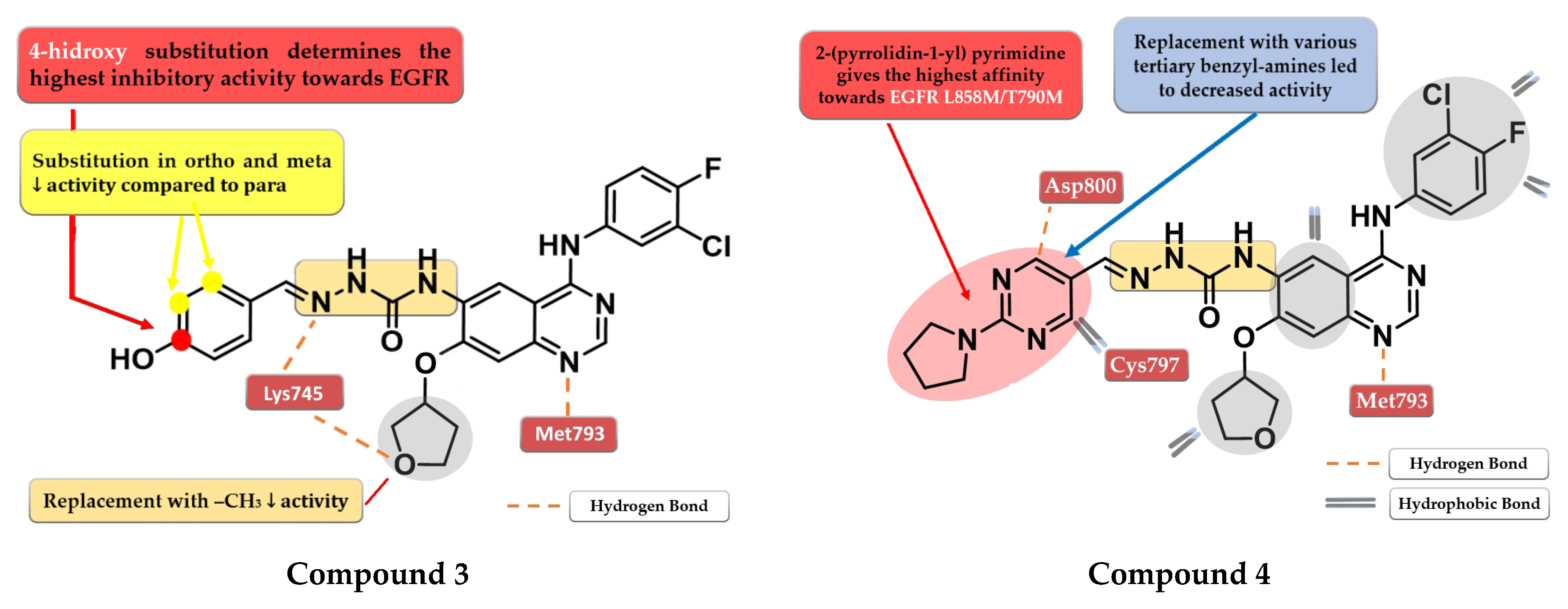
4.2. 6-Aryl-semicarbazone-4-anilino-quinazoline Derivatives
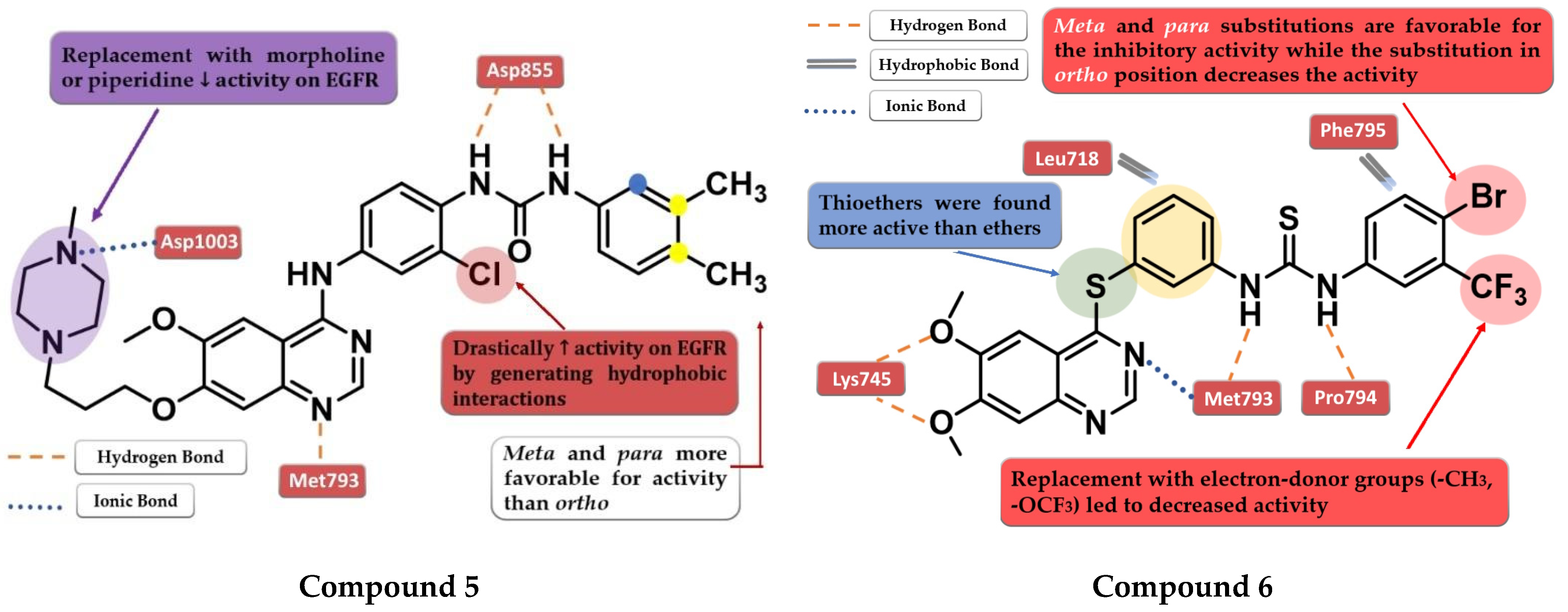
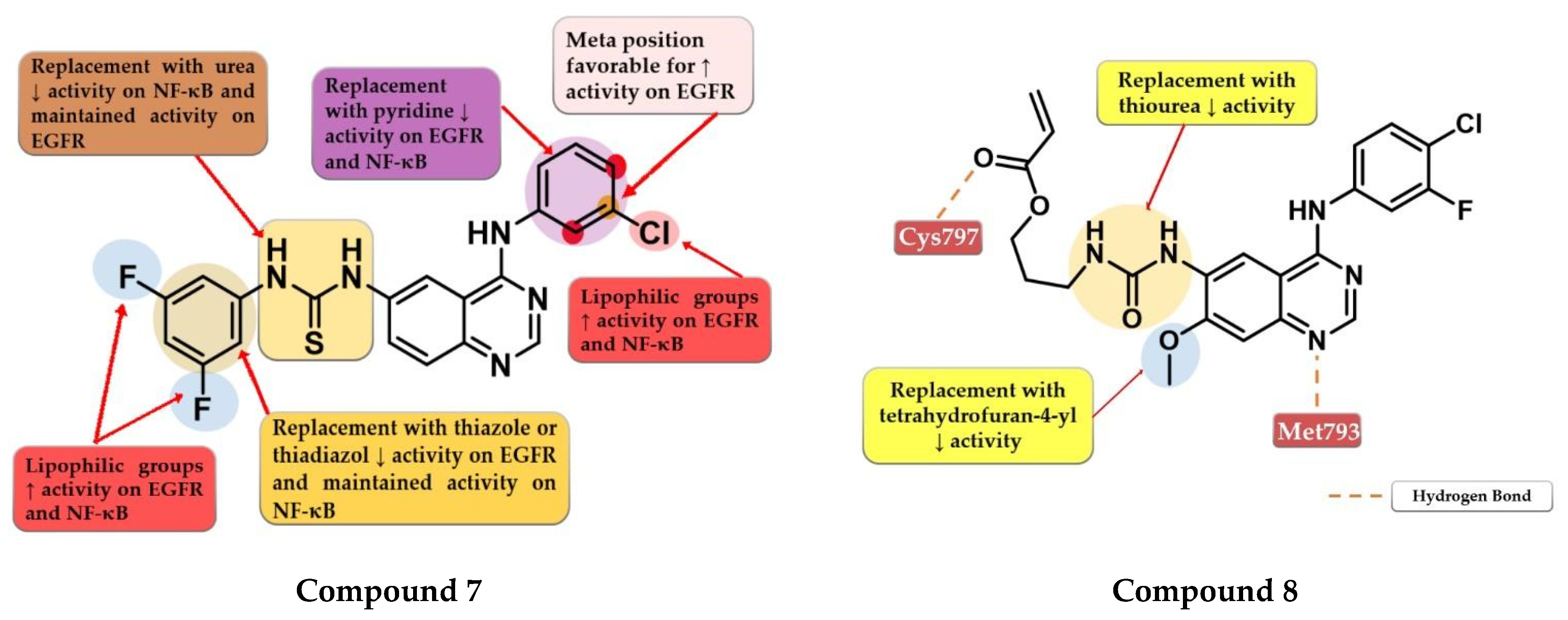
4.3. Disubstituted-Urea/Thiourea-Linked 4-Amino-quinazoline Derivatives
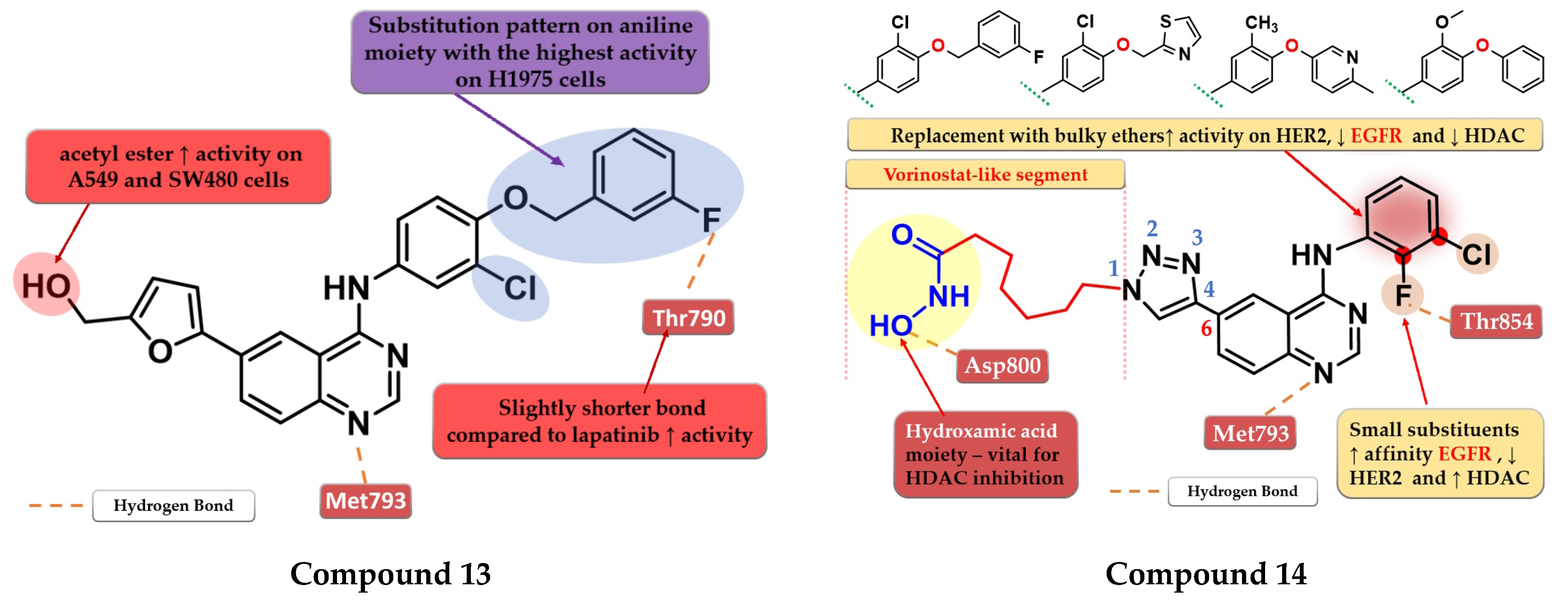
4.4. 6-Substituted-amide-4-amino-quinazoline Derivatives
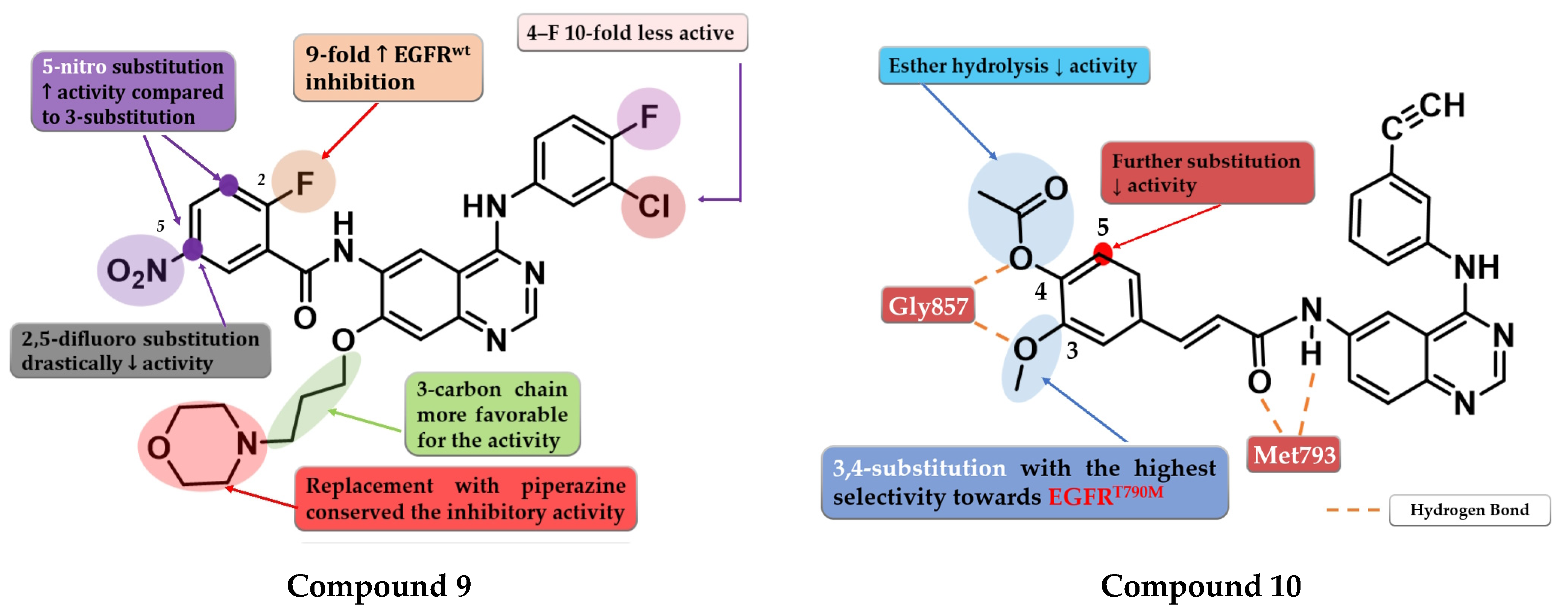
4.5. 6.7-Dimethoxy-4-amino-quinazoline Derivatives
4.6. 6-Heteroaryl-4-amino-quinazoline Derivatives
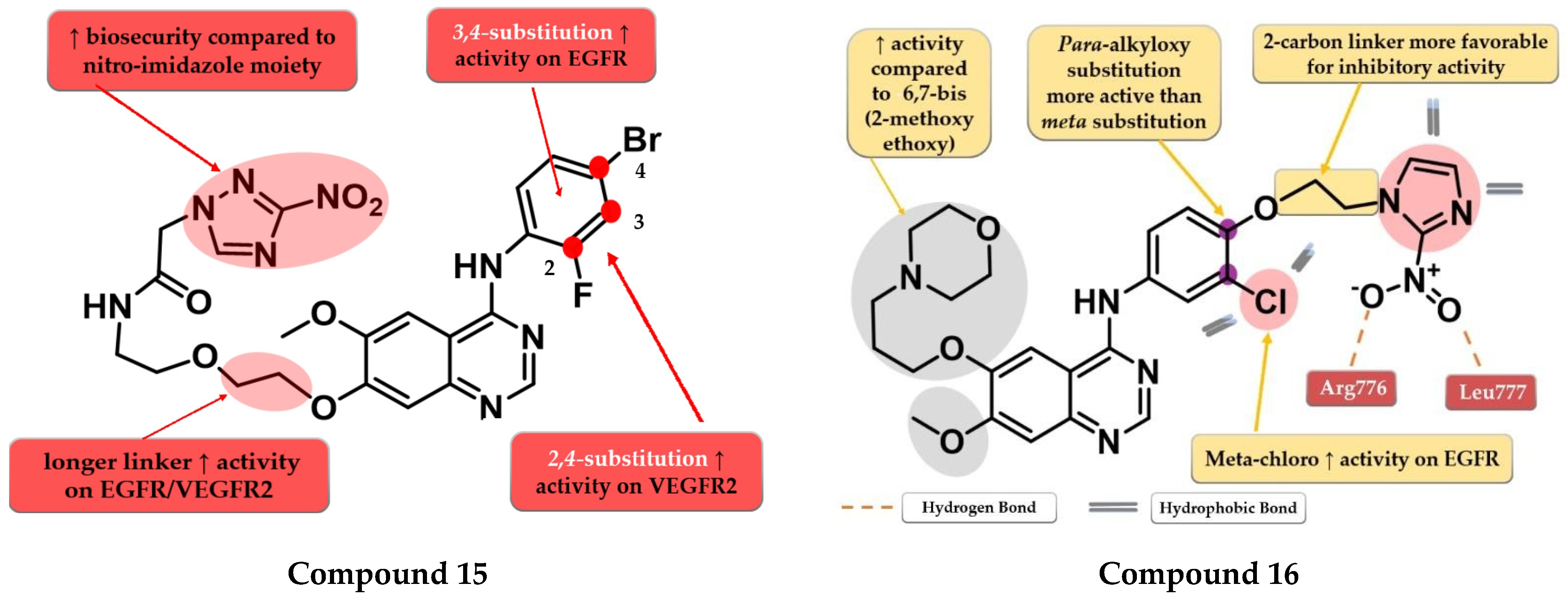
4.7. Nitro-Substituted-Azole Linked 4-Amino-quinazoline Derivatives
4.8. Fused 2,3-Dihydro-[1,4] Dioxino-[2,3-f] Quinazoline Derivatives
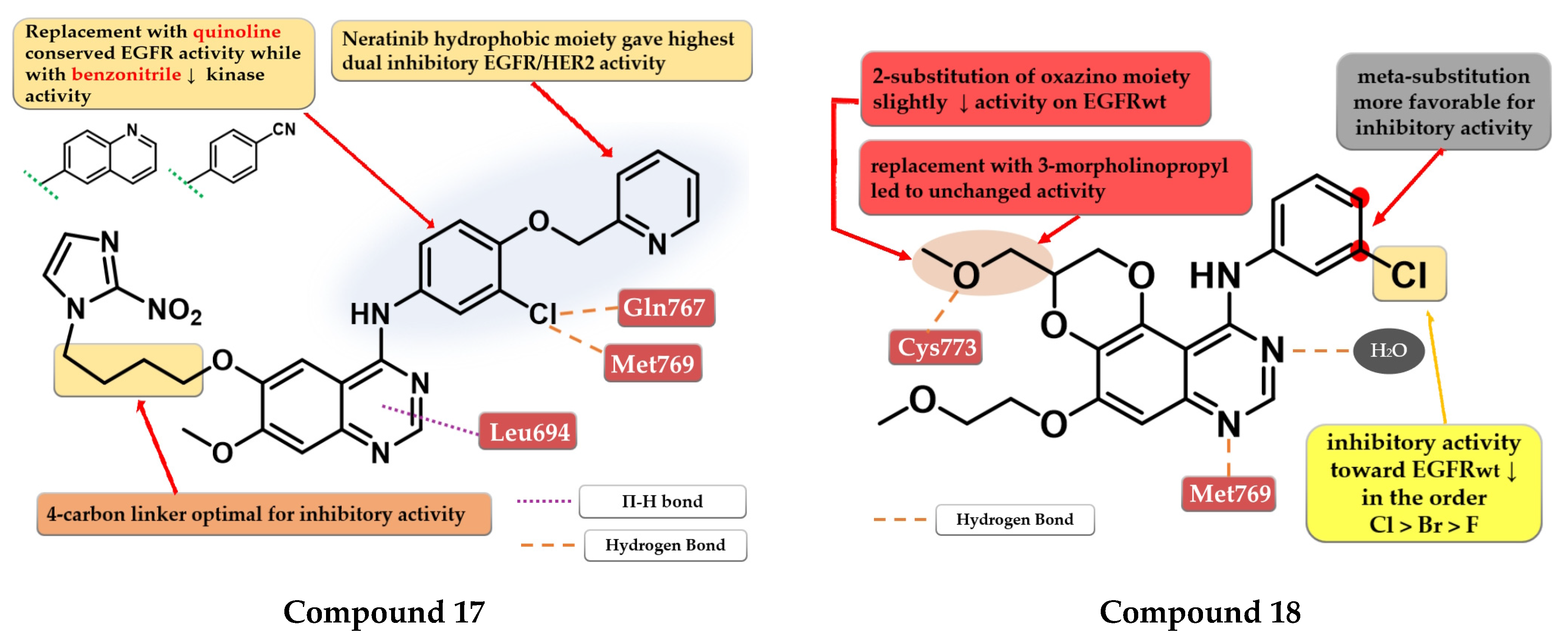
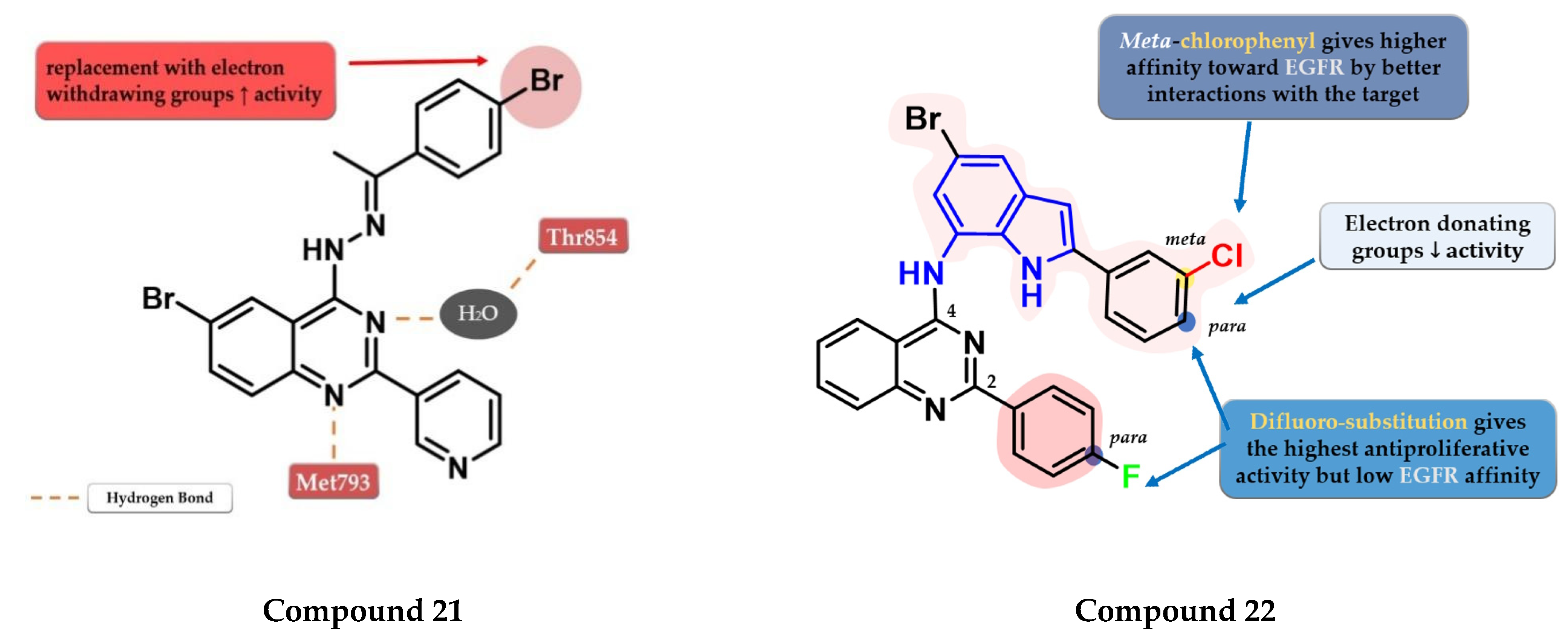
4.9. (2-Bromo-phenyl)-4-amino-quinazoline Derivatives
4.10. 2-Aryl-4-Substituted Quinazoline Derivatives
4.11. Sulfonamide Linked Quinazoline Derivatives
4.12. (4-Cyano-phenyl)-4-amino-quinazoline Derivatives
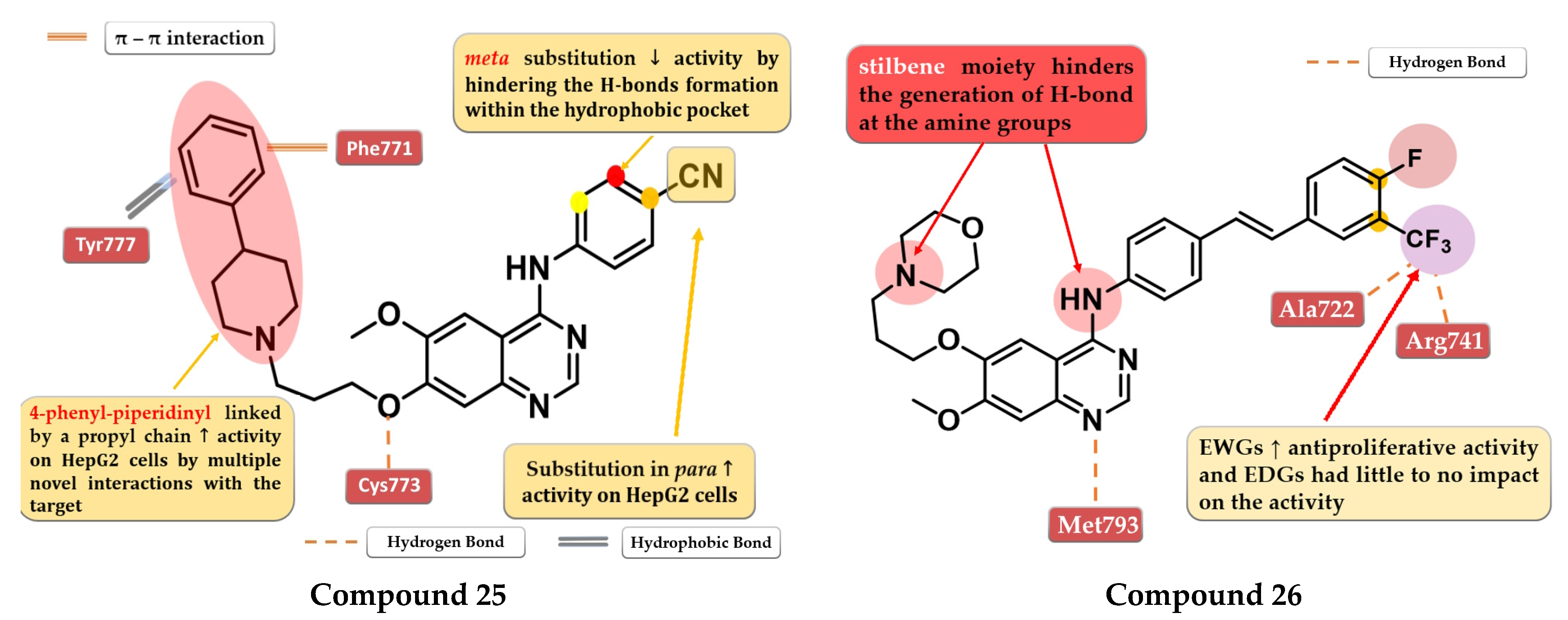
4.13. 6,7-Dialkoxy-4-stilbenyl-amino-quinazolines
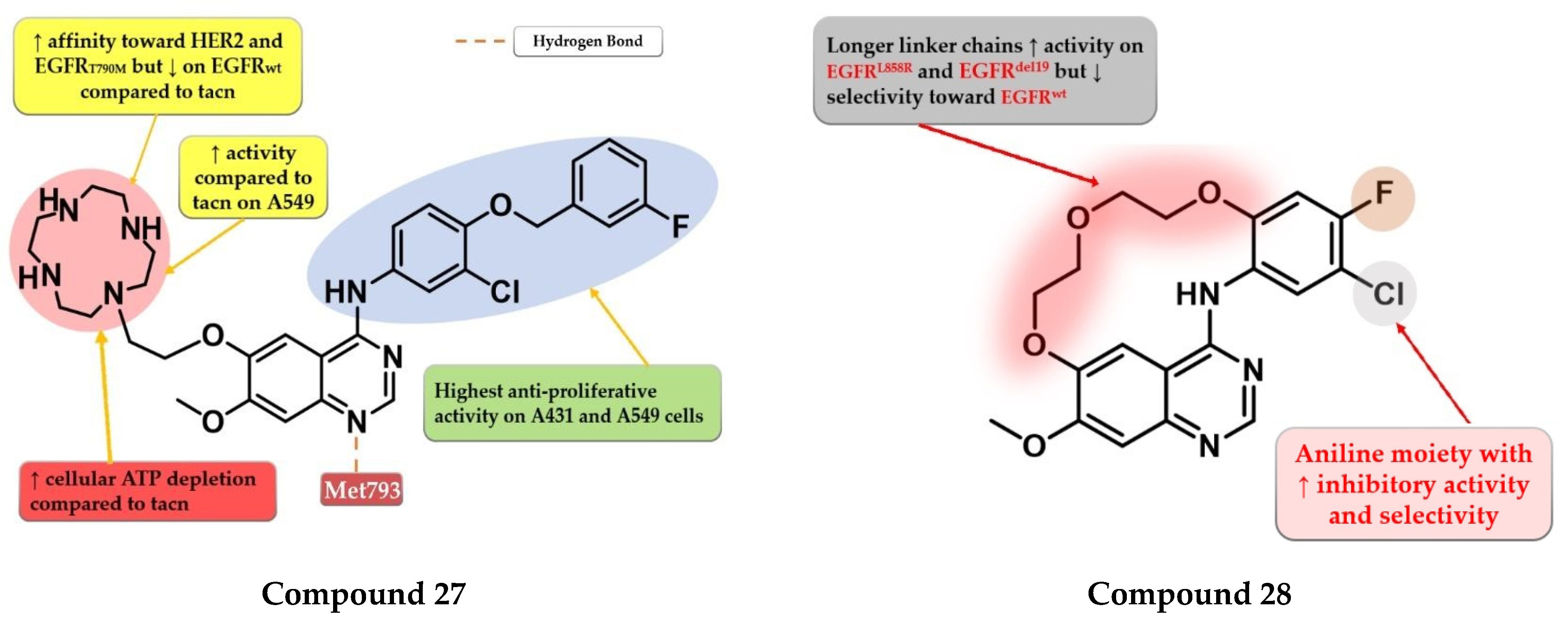
4.14. Macrocyclic Quinazoline Derivatives
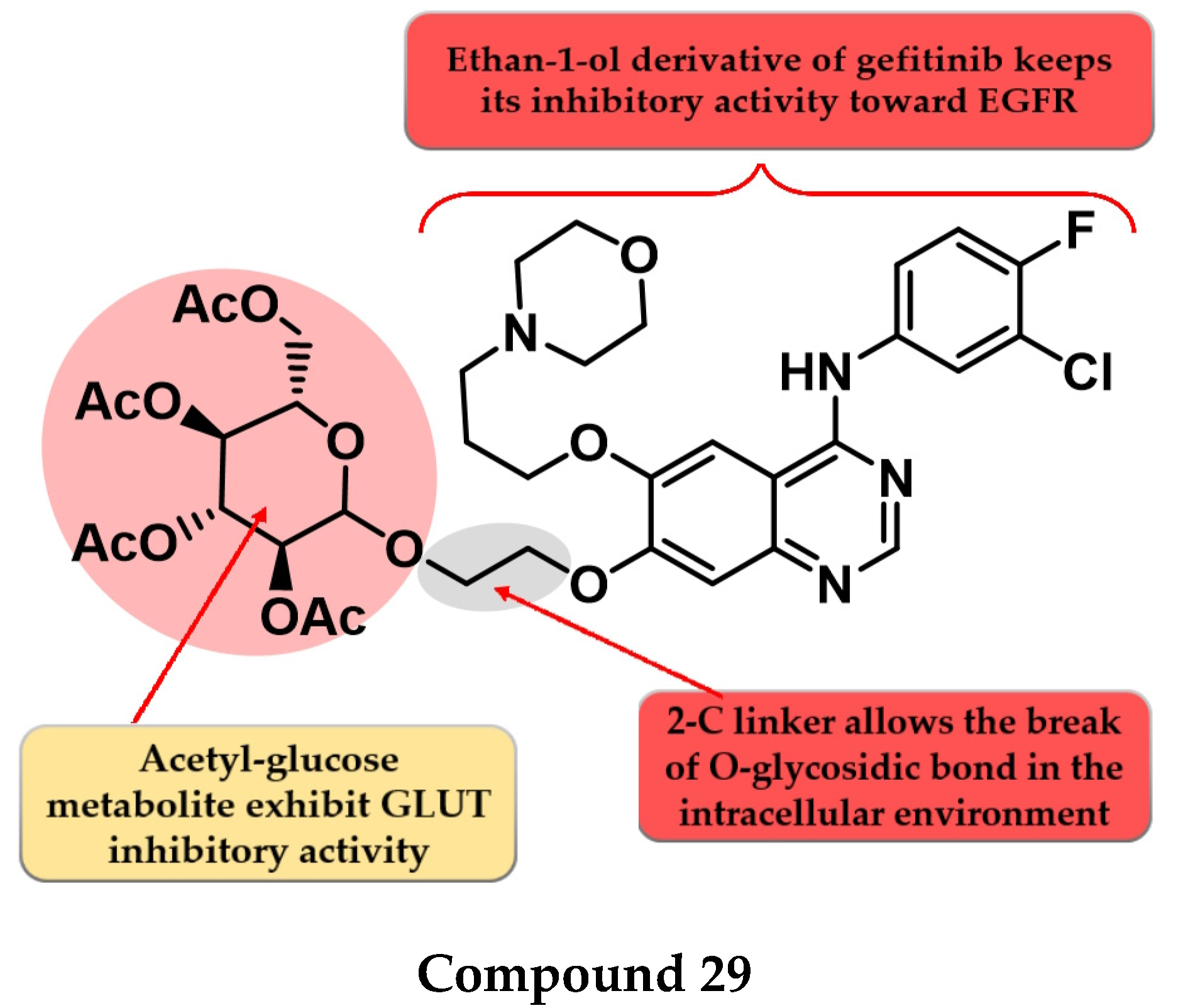
4.15. Acetyl Glucose-Modified 4-Anilino-quinazoline Derivatives
5. Second Generation of EGFR TKIs
6. Novel Second-Generation Quinazoline EGFR TKIs (2017–Present)
6.1. α-Chlorofluoro Acetamide Derivatives
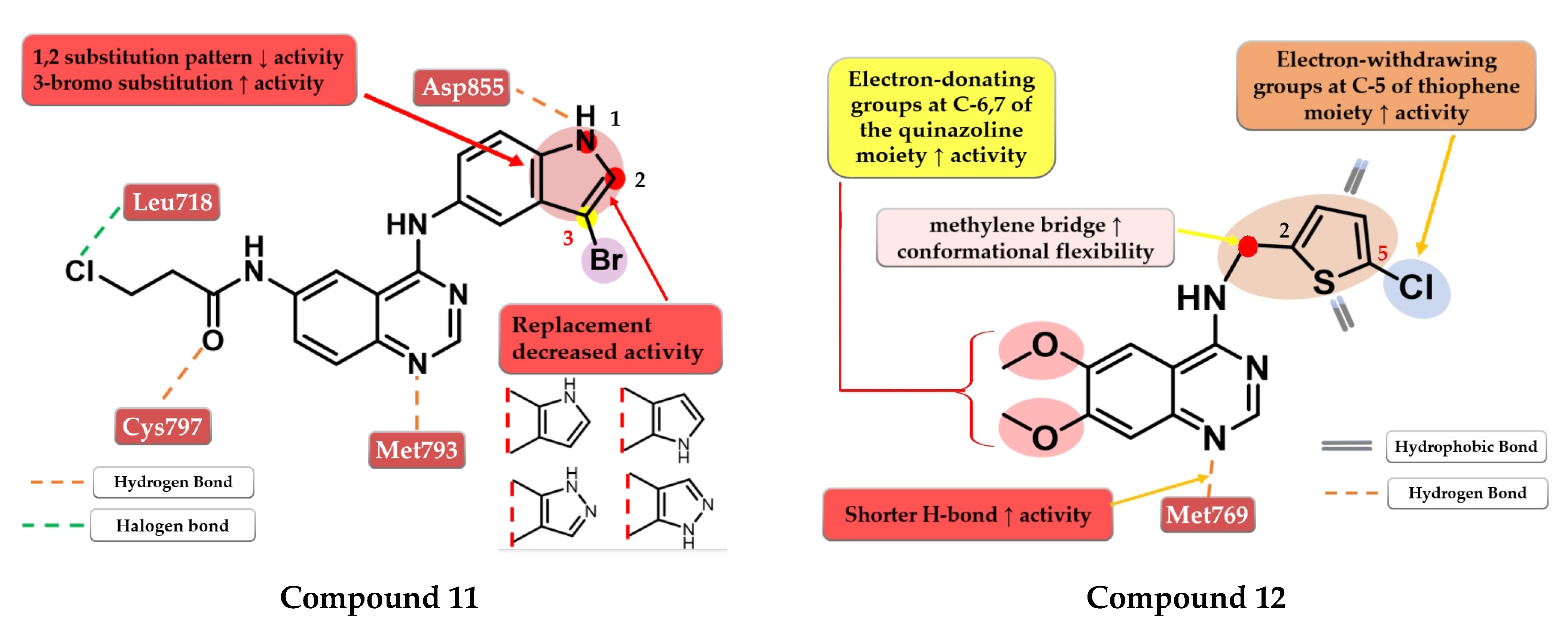
6.2. 6-Heteroaryl-thioacetamide-4-anilino-quinazoline Derivatives
6.3. 6-Acrylamide-4-amino-quinazoline Derivatives
6.4. Dihydro-6H-[1,4]Oxazino [3,2-g]Quinazoline Acrylamide Derivatives
6.5. N-(3-(Quinazolin-4-yl-amino)phenyl)acrylamide Derivatives
6.6. 2,5-Diazabicyclo [2.2.1]Heptane Linked 4-Anilino-quinazoline Acrylates
7. Third Generation of EGFR TKIs
8. Fourth Generation of EGFR TKIs
9. Novel Fourth-Generation Quinazoline EGFR TKIs (2017–Present)
9.1. 2-Aryl-4-amino-quinazoline Derivatives
9.2. 6-Methoxy-7-[(1-methylpiperidin-4-yl) methoxy]quinazolin-4-aniline Derivatives
10. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer Statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef] [PubMed]
- Lujambio, A.; Lowe, S.W. The Microcosmos of Cancer. Nature 2012, 482, 347–355. [Google Scholar] [CrossRef]
- Devita, V.T.; Rosenberg, S.A. Two Hundred Years of Cancer Research. N. Engl. J. Med. 2012, 366, 2207–2221. [Google Scholar] [CrossRef] [PubMed]
- Emamalipour, M.; Seidi, K.; Zununi Vahed, S.; Jahanban-Esfahlan, A.; Jaymand, M.; Majdi, H.; Amoozgar, Z.; Chitkushev, L.T.; Javaheri, T.; Jahanban-Esfahlan, R.; et al. Horizontal Gene Transfer: From Evolutionary Flexibility to Disease Progression. Front. Cell Dev. Biol. 2020, 8, 245. [Google Scholar] [CrossRef]
- Rahman, N. Realizing the Promise of Cancer Predisposition Genes. Nature 2014, 505, 302–308. [Google Scholar] [CrossRef] [PubMed]
- Irigaray, P.; Newby, J.A.; Clapp, R.; Hardell, L.; Howard, V.; Montagnier, L.; Epstein, S.; Belpomme, D. Lifestyle-Related Factors and Environmental Agents Causing Cancer: An Overview. Biomed. Pharmacother. 2007, 61, 640–658. [Google Scholar] [CrossRef]
- Ames, B.N.; Shigenaga, M.K.; Gold, L.S. DNA Lesions, Inducible DNA Repair, and Cell Division: Three Key Factors in Mutagenesis and Carcinogenesis. Environ. Health Perspect. 1993, 101, 35–44. [Google Scholar] [CrossRef]
- Basu, A.K. DNA Damage, Mutagenesis and Cancer. Int. J. Mol. Sci. 2018, 19, 970. [Google Scholar] [CrossRef]
- Stratton, M.R.; Campbell, P.J.; Futreal, P.A. The Cancer Genome. Nature 2009, 458, 719–724. [Google Scholar] [CrossRef]
- Russo, J.; Mailo, D.; Hu, Y.F.; Balogh, G.; Sheriff, F.; Russo, I.H.; Ingle, J.; Brodie, A.; Santen, R.; Colditz, G.; et al. Breast Differentiation and Its Implication in Cancer Prevention. Clin. Cancer Res. 2005, 11, 931s–936s. [Google Scholar] [CrossRef]
- Evan, G.I.; Vousden, K.H. Proliferation, Cell Cycle and Apoptosis in Cancer. Nature 2001, 411, 342–348. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. The Hallmarks of Cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef]
- Khalil, D.N.; Smith, E.L.; Brentjens, R.J.; Wolchok, J.D. The Future of Cancer Treatment: Immunomodulation, CARs and Combination Immunotherapy. Nat. Rev. Clin. Oncol. 2016, 13, 273–290. [Google Scholar] [CrossRef] [PubMed]
- Seton-Rogers, S. A Downside of Chemotherapy. Nat. Rev. Cancer 2012, 13, 5. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Sun, Y.; Liang, X.; Gu, X.; Ning, J.; Xu, Y.; Chen, S.; Pan, L. Emerging New Therapeutic Antibody Derivatives for Cancer Treatment. Signal Transduct. Target. Ther. 2022, 7, 39. [Google Scholar] [CrossRef]
- Cohen, P.; Cross, D.; Jänne, P.A. Kinase Drug Discovery 20 Years after Imatinib: Progress and Future Directions. Nat. Rev. Drug Discov. 2021, 20, 551–569. [Google Scholar] [CrossRef]
- Cheng, C.J.; Bahal, R.; Babar, I.A.; Pincus, Z.; Barrera, F.; Liu, C.; Svoronos, A.; Braddock, D.T.; Glazer, P.M.; Engelman, D.M.; et al. MicroRNA Silencing for Cancer Therapy Targeted to the Tumour Microenvironment. Nature 2014, 518, 107–110. [Google Scholar] [CrossRef]
- Fukuhara, H.; Ino, Y.; Todo, T. Oncolytic Virus Therapy: A New Era of Cancer Treatment at Dawn. Cancer Sci. 2016, 107, 1373–1379. [Google Scholar] [CrossRef]
- Troike, K.; Lathia, J.D. Optimising Gene Editing for Cancer Therapy. Nat. Cell Biol. 2020, 22, 259–261. [Google Scholar] [CrossRef]
- Kerru, N.; Gummidi, L.; Maddila, S.; Gangu, K.K.; Jonnalagadda, S.B. A Review on Recent Advances in Nitrogen-Containing Molecules and Their Biological Applications. Molecules 2020, 25, 1909. [Google Scholar] [CrossRef] [PubMed]
- Gomaa, H.A.M. A Comprehensive Review of Recent Advances in the Biological Activities of Quinazolines. Chem. Biol. Drug Des. 2022, 100, 639–655. [Google Scholar] [CrossRef]
- Haider, K.; Shafeeque, M.; Yahya, S.; Yar, M.S. A Comprehensive Review on Pyrazoline Based Heterocyclic Hybrids as Potent Anticancer Agents. Eur. J. Med. Chem. Rep. 2022, 5, 100042. [Google Scholar] [CrossRef]
- Sharma, S.V.; Bell, D.W.; Settleman, J.; Haber, D.A. Epidermal Growth Factor Receptor Mutations in Lung Cancer. Nat. Rev. Cancer 2007, 7, 169–181. [Google Scholar] [CrossRef]
- Roskoski, R. The ErbB/HER Family of Protein-Tyrosine Kinases and Cancer. Pharmacol. Res. 2014, 79, 34–74. [Google Scholar] [CrossRef]
- Jin, H.; Dan, H.G.; Rao, G.W. Research Progress in Quinazoline Derivatives as Multi-Target Tyrosine Kinase Inhibitors. Heterocycl. Commun. 2018, 24, 1–10. [Google Scholar] [CrossRef]
- Bhatia, P.; Sharma, V.; Alam, O.; Manaithiya, A.; Alam, P.; Kahksha; Alam, M.T.; Imran, M. Novel Quinazoline-Based EGFR Kinase Inhibitors: A Review Focussing on SAR and Molecular Docking Studies (2015–2019). Eur. J. Med. Chem. 2020, 204, 112640. [Google Scholar] [CrossRef]
- Reports Regarding the New Experimental Quinazoline Derivatives Series as EGFR Tirosin-Kinase Inhibitors. Available online: https://www.scopus.com (accessed on 24 March 2023).
- Tebbutt, N.; Pedersen, M.W.; Johns, T.G. Targeting the ERBB Family in Cancer: Couples Therapy. Nat. Rev. Cancer 2013, 13, 663–673. [Google Scholar] [CrossRef]
- Pennock, S.; Wang, Z. Stimulation of Cell Proliferation by Endosomal Epidermal Growth Factor Receptor as Revealed through Two Distinct Phases of Signaling. Mol. Cell. Biol. 2003, 23, 5803–5815. [Google Scholar] [CrossRef]
- Barberán, S.; Cebrià, F. The Role of the EGFR Signaling Pathway in Stem Cell Differentiation during Planarian Regeneration and Homeostasis. Semin. Cell Dev. Biol. 2019, 87, 45–57. [Google Scholar] [CrossRef]
- Chiasson-MacKenzie, C.; McClatchey, A.I. EGFR-Induced Cytoskeletal Changes Drive Complex Cell Behaviors: The Tip of the Iceberg. Sci. Signal. 2018, 11, eaas9473. [Google Scholar] [CrossRef] [PubMed]
- Kovacs, E.; Zorn, J.A.; Huang, Y.; Barros, T.; Kuriyan, J. A Structural Perspective on the Regulation of the Epidermal Growth Factor Receptor. Annu. Rev. Biochem. 2015, 84, 739–764. [Google Scholar] [CrossRef]
- Zhang, X.; Gureasko, J.; Shen, K.; Cole, P.A.; Kuriyan, J. An Allosteric Mechanism for Activation of the Kinase Domain of Epidermal Growth Factor Receptor. Cell 2006, 125, 1137–1149. [Google Scholar] [CrossRef] [PubMed]
- Burgess, A.W.; Cho, H.S.; Eigenbrot, C.; Ferguson, K.M.; Garrett, T.P.J.; Leahy, D.J.; Lemmon, M.A.; Sliwkowski, M.X.; Ward, C.W.; Yokoyama, S. An Open-and-Shut Case? Recent Insights into the Activation of EGF/ErbB Receptors. Mol. Cell 2003, 12, 541–552. [Google Scholar] [CrossRef]
- Sigismund, S.; Avanzato, D.; Lanzetti, L. Emerging Functions of the EGFR in Cancer. Mol. Oncol. 2018, 12, 3–20. [Google Scholar] [CrossRef] [PubMed]
- Pao, W.; Chmielecki, J. Rational, Biologically Based Treatment of EGFR-Mutant Non-Small-Cell Lung Cancer. Nat. Rev. Cancer 2010, 10, 760–774. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.C.; Vivanco, I.; Beroukhim, R.; Huang, J.H.Y.; Feng, W.L.; DeBiasi, R.M.; Yoshimoto, K.; King, J.C.; Nghiemphu, P.; Yuza, Y.; et al. Epidermal Growth Factor Receptor Activation in Glioblastoma through Novel Missense Mutations in the Extracellular Domain. PLoS Med. 2006, 3, e485. [Google Scholar] [CrossRef]
- Rogers, S.J.; Harrington, K.J.; Rhys-Evans, P.; O-Charoenrat, P.; Eccles, S.A. Biological Significance of C-ErbB Family Oncogenes in Head and Neck Cancer. Cancer Metastasis Rev. 2005, 24, 47–69. [Google Scholar] [CrossRef]
- Lo, H.W.; Hsu, S.C.; Hung, M.C. EGFR Signaling Pathway in Breast Cancers: From Traditional Signal Transduction to Direct Nuclear Translocalization. Breast Cancer Res. Treat. 2006, 95, 211–218. [Google Scholar] [CrossRef]
- Spano, J.P.; Lagorce, C.; Atlan, D.; Milano, G.; Domont, J.; Benamouzig, R.; Attar, A.; Benichou, J.; Martin, A.; Morere, J.F.; et al. Impact of EGFR Expression on Colorectal Cancer Patient Prognosis and Survival. Ann. Oncol. 2005, 16, 102–108. [Google Scholar] [CrossRef]
- Oliveira-Cunha, M.; Newman, W.G.; Siriwardena, A.K. Epidermal Growth Factor Receptor in Pancreatic Cancer. Cancers 2011, 3, 1513–1526. [Google Scholar] [CrossRef] [PubMed]
- Keller, S.; Schmidt, M.H.H. EGFR and EGFRvIII Promote Angiogenesis and Cell Invasion in Glioblastoma: Combination Therapies for an Effective Treatment. Int. J. Mol. Sci. 2017, 18, 1295. [Google Scholar] [CrossRef]
- Kedrin, D.; van Rheenen, J.; Hernandez, L.; Condeelis, J.; Segall, J.E. Cell Motility and Cytoskeletal Regulation in Invasion and Metastasis. J. Mammary Gland Biol. Neoplasia 2007, 12, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Shin, D.Y.; Na, I.I.; Kim, C.H.; Park, S.; Baek, H.; Yang, S.H. EGFR Mutation and Brain Metastasis in Pulmonary Adenocarcinomas. J. Thorac. Oncol. 2014, 9, 195–199. [Google Scholar] [CrossRef] [PubMed]
- Sordella, R.; Bell, D.W.; Haber, D.A.; Settleman, J. Gefitinib-Sensitizing EGFR Mutations in Lung Cancer Activate Anti-Apoptotic Pathways. Science 2004, 305, 1163–1167. [Google Scholar] [CrossRef] [PubMed]
- Sarris, E.G.; Saif, M.W.; Syrigos, K.N. The Biological Role of PI3K Pathway in Lung Cancer. Pharmaceuticals 2012, 5, 1236–1264. [Google Scholar] [CrossRef]
- Codony-Servat, C.; Codony-Servat, J.; Karachaliou, N.; Molina, M.A.; Chaib, I.; Ramirez, J.L.; Gil, M.d.l.L.; Solca, F.; Bivona, T.G.; Rosell, R. Activation of Signal Transducer and Activator of Transcription 3 (STAT3) Signaling in EGFR Mutant Non-Small-Cell Lung Cancer (NSCLC). Oncotarget 2017, 8, 47316. [Google Scholar] [CrossRef]
- Wee, P.; Wang, Z. Epidermal Growth Factor Receptor Cell Proliferation Signaling Pathways. Cancers 2017, 9, 52. [Google Scholar] [CrossRef]
- Yun, C.-H.; Boggon, T.J.; Li, Y.; Woo, M.S.; Greulich, H.; Meyerson, M.; Eck, M.J. Structures of Lung Cancer-Derived EGFR Mutants and Inhibitor Complexes: Mechanism of Activation and Insights into Differential Inhibitor Sensitivity. Cancer Cell 2007, 11, 217–227. [Google Scholar] [CrossRef]
- Troiani, T.; Napolitano, S.; Della Corte, C.M.; Martini, G.; Martinelli, E.; Morgillo, F.; Ciardiello, F. Therapeutic Value of EGFR Inhibition in CRC and NSCLC: 15 Years of Clinical Evidence. ESMO Open 2016, 1, e000088. [Google Scholar] [CrossRef] [PubMed]
- Gan, H.K.; Cvrljevic, A.N.; Johns, T.G. The Epidermal Growth Factor Receptor Variant III (EGFRvIII): Where Wild Things Are Altered. FEBS J. 2013, 280, 5350–5370. [Google Scholar] [CrossRef]
- Martinelli, E.; Ciardiello, D.; Martini, G.; Troiani, T.; Cardone, C.; Vitiello, P.P.; Normanno, N.; Rachiglio, A.M.; Maiello, E.; Latiano, T.; et al. Implementing Anti-Epidermal Growth Factor Receptor (EGFR) Therapy in Metastatic Colorectal Cancer: Challenges and Future Perspectives. Ann. Oncol. 2020, 31, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.F.; Liu, Z.C.; Xie, B.F.; Li, Z.M.; Feng, G.K.; Yang, D.; Zeng, Y.X. EGFR Tyrosine Kinase Inhibitor AG1478 Inhibits Cell Proliferation and Arrests Cell Cycle in Nasopharyngeal Carcinoma Cells. Cancer Lett. 2001, 169, 27–32. [Google Scholar] [CrossRef]
- Gonzales, A.J.; Fry, D.W. G1 Cell Cycle Arrest Due to the Inhibition of ErbB Family Receptor Tyrosine Kinases Does Not Require the Retinoblastoma Protein. Exp. Cell Res. 2005, 303, 56–67. [Google Scholar] [CrossRef] [PubMed]
- Ahn, S.H.; Jeong, E.H.; Lee, T.G.; Kim, S.Y.; Kim, H.R.; Kim, C.H. Gefitinib Induces Cytoplasmic Translocation of the CDK Inhibitor P27 and Its Binding to a Cleaved Intermediate of Caspase 8 in Non-Small Cell Lung Cancer Cells. Cell. Oncol. 2014, 37, 377–386. [Google Scholar] [CrossRef]
- Gong, Y.; Somwar, R.; Politi, K.; Balak, M.; Chmielecki, J.; Jiang, X.; Pao, W. Induction of BIM Is Essential for Apoptosis Triggered by EGFR Kinase Inhibitors in Mutant EGFR-Dependent Lung Adenocarcinomas. PLoS Med. 2007, 4, e294. [Google Scholar] [CrossRef]
- Costa, D.B.; Halmos, B.; Kumar, A.; Schumer, S.T.; Huberman, M.S.; Boggon, T.J.; Tenen, D.G.; Kobayashi, S. BIM Mediates EGFR Tyrosine Kinase Inhibitor-Induced Apoptosis in Lung Cancers with Oncogenic EGFR Mutations. PLoS Med. 2007, 4, e315. [Google Scholar] [CrossRef]
- Santos, E.d.S.; Nogueira, K.A.B.; Fernandes, L.C.C.; Martins, J.R.P.; Reis, A.V.F.; Neto, J.d.B.V.; Júnior, I.J.d.S.; Pessoa, C.; Petrilli, R.; Eloy, J.O. EGFR Targeting for Cancer Therapy: Pharmacology and Immunoconjugates with Drugs and Nanoparticles. Int. J. Pharm. 2021, 592, 120082. [Google Scholar] [CrossRef]
- Wu, S.; Deng, C.; Zhang, C.; Xiong, J.; Gu, X.; Wang, Z.; Tu, J.; Xie, J. Preparation of a Novel EGFR Specific Immunotoxin and Its Efficacy of Anti-Colorectal Cancer In Vitro and In Vivo. Clin. Transl. Oncol. 2021, 23, 1549–1560. [Google Scholar] [CrossRef]
- Bauman, J.E.; Duvvuri, U.; Thomas, S.; Gooding, W.E.; Clump, D.A.; Karlovits, B.; Wehbe, A.; Miller, F.R.; Kim, S.; Sen, M.; et al. Phase 1 Study of EGFR-Antisense DNA, Cetuximab, and Radiotherapy in Head and Neck Cancer with Preclinical Correlatives. Cancer 2018, 124, 3881–3889. [Google Scholar] [CrossRef] [PubMed]
- Malkki, H. Glioblastoma Vaccine Therapy Disappointment in Phase III Trial. Nat. Rev. Neurol. 2016, 12, 190. [Google Scholar] [CrossRef]
- Du, Y.; Chen, Y.; Wang, Y.; Chen, J.; Lu, X.; Zhang, L.; Li, Y.; Wang, Z.; Ye, G.; Zhang, G. HJM-561, a Potent, Selective, and Orally Bioavailable EGFR PROTAC That Overcomes Osimertinib-Resistant EGFR Triple Mutations. Mol. Cancer Ther. 2022, 21, 1060–1066. [Google Scholar] [CrossRef]
- Mok, T.S.; Wu, Y.-L.; Thongprasert, S.; Yang, C.-H.; Chu, D.-T.; Saijo, N.; Sunpaweravong, P.; Han, B.; Margono, B.; Ichinose, Y.; et al. Gefitinib or Carboplatin–Paclitaxel in Pulmonary Adenocarcinoma. N. Engl. J. Med. 2009, 361, 947–957. [Google Scholar] [CrossRef] [PubMed]
- Li, A.R.; Chitale, D.; Riely, G.J.; Pao, W.; Miller, V.A.; Zakowski, M.F.; Rusch, V.; Kris, M.G.; Ladanyi, M. Clinical Testing Experience and Relationship to EGFR Gene Copy Number and Immunohistochemical Expression. J. Mol. Diagn. 2008, 10, 242–248. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.L.; Chu, D.T.; Han, B.; Liu, X.; Zhang, L.; Zhou, C.; Liao, M.; Mok, T.; Jiang, H.; Duffield, E.; et al. Phase III, Randomized, Open-Label, First-Line Study in Asia of Gefitinib versus Carboplatin/Paclitaxel in Clinically Selected Patients with Advanced Non-Small-Cell Lung Cancer: Evaluation of Patients Recruited from Mainland China. Asia. Pac. J. Clin. Oncol. 2012, 8, 232–243. [Google Scholar] [CrossRef]
- Food and Drug Administration (FDA) Label—IRESSA® (Gefitinib) 250mg Tablets for Oral Use. Available online: www.fda.gov/medwatch (accessed on 15 February 2023).
- Patil, V.M.; Noronha, V.; Joshi, A.; Choughule, A.B.; Bhattacharjee, A.; Kumar, R.; Goud, S.; More, S.; Ramaswamy, A.; Karpe, A.; et al. Phase III Study of Gefitinib or Pemetrexed with Carboplatin in EGFR-Mutated Advanced Lung Adenocarcinoma. ESMO Open 2017, 2, eooo25. [Google Scholar] [CrossRef]
- Park, K.; Tan, E.H.; O’Byrne, K.; Zhang, L.; Boyer, M.; Mok, T.; Hirsh, V.; Yang, J.C.H.; Lee, K.H.; Lu, S.; et al. Afatinib versus Gefitinib as First-Line Treatment of Patients with EGFR Mutation-Positive Non-Small-Cell Lung Cancer (LUX-Lung 7): A Phase 2B, Open-Label, Randomised Controlled Trial. Lancet Oncol. 2016, 17, 577–589. [Google Scholar] [CrossRef]
- Food and Drug Administration (FDA) Label—TARCEVA® (Erlotinib) 25 Mg, 100 Mg and 150 Mg Tablets. Available online: www.fda.gov/medwatch (accessed on 15 February 2023).
- Moore, M.J.; Goldstein, D.; Hamm, J.; Figer, A.; Hecht, J.R.; Gallinger, S.; Au, H.J.; Murawa, P.; Walde, D.; Wolff, R.A.; et al. Erlotinib plus Gemcitabine Compared with Gemcitabine Alone in Patients with Advanced Pancreatic Cancer: A Phase III Trial of the National Cancer Institute of Canada Clinical Trials Group. J. Clin. Oncol. 2007, 25, 1960–1966. [Google Scholar] [CrossRef]
- Shepherd, F.A.; Pereira, J.; Ciuleanu, T.E.; Tan, E.H.; Hirsh, V.; Thongprasert, S.; Bezjak, A.; Tu, D.; Santabárbara, P.; Seymour, L. A Randomized Placebo-Controlled Trial of Erlotinib in Patients with Advanced Non-Small Cell Lung Cancer (NSCLC) Following Failure of 1st Line or 2nd Line Chemotherapy. A National Cancer Institute of Canada Clinical Trials Group (NCIC CTG) Trial. J. Clin. Oncol. 2004, 22, 7022. [Google Scholar] [CrossRef]
- Shi, Y.; Wang, L.; Han, B.; Li, W.; Yu, P.; Liu, Y.; Ding, C.; Song, X.; Yong, M.Z.; Ren, X.; et al. First-Line Icotinib versus Cisplatine/Pemetrexed plus Pemetrexed Maintenance Therapy in Lung Adenocarcinoma Patients with Sensitizing EGFR Mutation (CONVINCE). J. Clin. Oncol. 2016, 34, 9041. [Google Scholar] [CrossRef]
- Shi, Y.; Zhang, L.; Liu, X.; Zhou, C.; Zhang, S.; Wang, D.; Li, Q.; Qin, S.; Hu, C.; Zhang, Y.; et al. Icotinib versus Gefitinib in Previously Treated Advanced Non-Small-Cell Lung Cancer (ICOGEN): A Randomised, Double-Blind Phase 3 Non-Inferiority Trial. Lancet Oncol. 2013, 14, 953–961. [Google Scholar] [CrossRef] [PubMed]
- Thongprasert, S.; Duffield, E.; Saijo, N.; Wu, Y.L.; Yang, J.C.H.; Chu, D.T.; Liao, M.; Chen, Y.M.; Kuo, H.P.; Negoro, S.; et al. Health-Related Quality-of-Life in a Randomized Phase III First-Line Study of Gefitinib Versus Carboplatin/Paclitaxel in Clinically Selected Patients from Asia with Advanced NSCLC (IPASS). J. Thorac. Oncol. 2011, 6, 1872–1880. [Google Scholar] [CrossRef] [PubMed]
- Rangachari, D.; To, C.; Shpilsky, J.E.; VanderLaan, P.A.; Kobayashi, S.S.; Mushajiang, M.; Lau, C.J.; Paweletz, C.P.; Oxnard, G.R.; Jänne, P.A.; et al. EGFR-Mutated Lung Cancers Resistant to Osimertinib through EGFR C797S Respond to First-Generation Reversible EGFR Inhibitors but Eventually Acquire EGFR T790M/C797S in Preclinical Models and Clinical Samples. J. Thorac. Oncol. 2019, 14, 1995–2002. [Google Scholar] [CrossRef]
- Pao, W.; Miller, V.A.; Politi, K.A.; Riely, G.J.; Somwar, R.; Zakowski, M.F.; Kris, M.G.; Varmus, H. Acquired Resistance of Lung Adenocarcinomas to Gefitinib or Erlotinib Is Associated with a Second Mutation in the EGFR Kinase Domain. PLoS Med. 2005, 2, 0225–0235. [Google Scholar] [CrossRef]
- Oxnard, G.R.; Arcila, M.E.; Sima, C.S.; Riely, G.J.; Chmielecki, J.; Kris, M.G.; Pao, W.; Ladanyi, M.; Miller, V.A. Acquired Resistance to EGFR Tyrosine Kinase Inhibitors in EGFR-Mutant Lung Cancer: Distinct Natural History of Patients with Tumors Harboring the T790M Mutation. Clin. Cancer Res. 2011, 17, 1616–1622. [Google Scholar] [CrossRef]
- Ogino, A.; Kitao, H.; Hirano, S.; Uchida, A.; Ishiai, M.; Kozuki, T.; Takigawa, N.; Takata, M.; Kiura, K.; Tanimoto, M. Emergence of Epidermal Growth Factor Receptor T790M Mutation during Chronic Exposure to Gefitinib in a Non–Small Cell Lung Cancer Cell Line. Cancer Res. 2007, 67, 7807–7814. [Google Scholar] [CrossRef]
- Chiu, C.H.; Yang, C.T.; Shih, J.Y.; Huang, M.S.; Su, W.C.; Lai, R.S.; Wang, C.C.; Hsiao, S.H.; Lin, Y.C.; Ho, C.L.; et al. Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitor Treatment Response in Advanced Lung Adenocarcinomas with G719X/L861Q/S768I Mutations. J. Thorac. Oncol. 2015, 10, 793–799. [Google Scholar] [CrossRef]
- Kancha, R.K.; Von Bubnoff, N.; Peschel, C.; Duyster, J. Functional Analysis of Epidermal Growth Factor Receptor (EGFR) Mutations and Potential Implications for EGFR Targeted Therapy. Clin. Cancer Res. 2009, 15, 460–467. [Google Scholar] [CrossRef]
- Lei, L.; Wang, W.; Zhu, Y.; Li, J.; Fang, Y.; Wang, H.; Zhuang, W.; Zhang, Y.; Wang, L.; Fang, M.; et al. Potential Mechanism of Primary Resistance to Icotinib in Patients with Advanced Non–Small Cell Lung Cancer Harboring Uncommon Mutant Epidermal Growth Factor Receptor: A Multi-Center Study. Cancer Sci. 2020, 111, 679–686. [Google Scholar] [CrossRef]
- Vyse, S.; Huang, P.H. Targeting EGFR Exon 20 Insertion Mutations in Non-Small Cell Lung Cancer. Signal Transduct. Target. Ther. 2019, 4, 51. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.W.; Kim, H.P.; Im, S.A.; Kang, S.; Hur, H.S.; Yoon, Y.K.; Oh, D.Y.; Kim, J.H.; Lee, D.S.; Kim, T.Y.; et al. The Growth Inhibitory Effect of Lapatinib, a Dual Inhibitor of EGFR and HER2 Tyrosine Kinase, in Gastric Cancer Cell Lines. Cancer Lett. 2008, 272, 296–306. [Google Scholar] [CrossRef]
- Food and Drug Administration (FDA) Label—TYKERB® (Lapatinib) 250 Mg Tablets. Available online: www.fda.gov/medwatch (accessed on 15 February 2023).
- Morabito, A.; Piccirillo, M.C.; Falasconi, F.; De Feo, G.; Del Giudice, A.; Bryce, J.; Di Maio, M.; De Maio, E.; Normanno, N.; Perrone, F. Vandetanib (ZD6474), a Dual Inhibitor of Vascular Endothelial Growth Factor Receptor (VEGFR) and Epidermal Growth Factor Receptor (EGFR) Tyrosine Kinases: Current Status and Future Directions. Oncologist 2009, 14, 378–390. [Google Scholar] [CrossRef] [PubMed]
- Food and Drug Administration (FDA) Label—CAPRELSA® (Vandetanib) 100 Mg and 300 Mg Tablets for Oral Use. Available online: www.fda.gov/medwatch (accessed on 15 February 2023).
- Zhang, D.; Pal, A.; Bornmann, W.G.; Yamasaki, F.; Esteva, F.J.; Hortobagyi, G.N.; Bartholomeusz, C.; Ueno, N.T. Activity of Lapatinib Is Independent of EGFR Expression Level in HER2-Overexpressing Breast Cancer Cells. Mol. Cancer Ther. 2008, 7, 1846–1850. [Google Scholar] [CrossRef]
- Ahn, M.J.; Kim, D.W.; Cho, B.C.; Kim, S.W.; Lee, J.S.; Ahn, J.S.; Kim, T.M.; Lin, C.C.; Kim, H.R.; John, T.; et al. Activity and Safety of AZD3759 in EGFR-Mutant Non-Small-Cell Lung Cancer with CNS Metastases (BLOOM): A Phase 1, Open-Label, Dose-Escalation and Dose-Expansion Study. Lancet Respir. Med. 2017, 5, 891–902. [Google Scholar] [CrossRef] [PubMed]
- Milik, S.N.; Lasheen, D.S.; Serya, R.A.T.; Abouzid, K.A.M. How to Train Your Inhibitor: Design Strategies to Overcome Resistance to Epidermal Growth Factor Receptor Inhibitors. Eur. J. Med. Chem. 2017, 142, 131–151. [Google Scholar] [CrossRef]
- Zuccotto, F.; Ardini, E.; Casale, E.; Angiolini, M. Through the “Gatekeeper Door”: Exploiting the Active Kinase Conformation. J. Med. Chem. 2010, 53, 2681–2694. [Google Scholar] [CrossRef]
- Aertgeerts, K.; Skene, R.; Yano, J.; Sang, B.-C.; Zou, H.; Snell, G.; Jennings, A.; Iwamoto, K.; Habuka, N.; Hirokawa, A.; et al. Structural Analysis of the Mechanism of Inhibition and Allosteric Activation of the Kinase Domain of HER2 Protein. J. Biol. Chem. 2011, 286, 18756–18765. [Google Scholar] [CrossRef]
- Tong, M.; Seeliger, M.A. Targeting Conformational Plasticity of Protein Kinases. ACS Chem. Biol. 2015, 10, 190–200. [Google Scholar] [CrossRef]
- Vivanco, I.; Ian Robins, H.; Rohle, D.; Campos, C.; Grommes, C.; Nghiemphu, P.L.; Kubek, S.; Oldrini, B.; Chheda, M.G.; Yannuzzi, N.; et al. Differential Sensitivity of Glioma- versus Lung Cancer-Specific EGFR Mutations to EGFR Kinase Inhibitors. Cancer Discov. 2012, 2, 458–471. [Google Scholar] [CrossRef]
- Park, J.H.; Liu, Y.; Lemmon, M.A.; Radhakrishnan, R. Erlotinib Binds Both Inactive and Active Conformations of the EGFR Tyrosine Kinase Domain. Biochem. J. 2012, 448, 423. [Google Scholar] [CrossRef] [PubMed]
- Ward, W.H.J.; Cook, P.N.; Slater, A.M.; Davies, D.H.; Holdgate, G.A.; Green, L.R. Epidermal Growth Factor Receptor Tyrosine Kinase: Investigation of Catalytic Mechanism, Structure-Based Searching and Discovery of a Potent Inhibitor. Biochem. Pharmacol. 1994, 48, 659–666. [Google Scholar] [CrossRef]
- Verma, N.; Rai, A.K.; Kaushik, V.; Brünnert, D.; Chahar, K.R.; Pandey, J.; Goyal, P. Identification of Gefitinib Off-Targets Using a Structure-Based Systems Biology Approach; Their Validation with Reverse Docking and Retrospective Data Mining. Sci. Rep. 2016, 6, 33949. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Li, M.; Qu, Y.; Tang, W.; Zheng, Y.; Lian, J.; Ji, M.; Xu, L. Design and Synthesis of Novel Gefitinib Analogues with Improved Anti-Tumor Activity. Bioorg. Med. Chem. 2010, 18, 3812–3822. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Sun, M.; Wu, X.; Chen, J.; Wang, P.; Zong, X.; Ji, M. Design and Synthesis of Novel 4-Benzothiazole Amino Quinazolines Dasatinib Derivatives as Potential Anti-Tumor Agents. Eur. J. Med. Chem. 2013, 63, 702–712. [Google Scholar] [CrossRef]
- Wood, E.R.; Truesdale, A.T.; McDonald, O.B.; Yuan, D.; Hassell, A.; Dickerson, S.H.; Ellis, B.; Pennisi, C.; Horne, E.; Lackey, K.; et al. A Unique Structure for Epidermal Growth Factor Receptor Bound to GW572016 (Lapatinib). Cancer Res. 2004, 64, 6652–6659. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, Y.; Liu, J.; Wang, W.; Li, X.; Zhao, L.; Wang, W.; Li, B. Novel 4-Arylaminoquinazoline Derivatives with (E)-Propen-1-Yl Moiety as Potent EGFR Inhibitors with Enhanced Antiproliferative Activities against Tumor Cells. Eur. J. Med. Chem. 2017, 138, 689–697. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, L.; Xu, H.; Li, X.; Zhao, L.; Wang, W.; Li, B.; Zhang, X. 6,7-Dimorpholinoalkoxy Quinazoline Derivatives as Potent EGFR Inhibitors with Enhanced Antiproliferative Activities against Tumor Cells. Eur. J. Med. Chem. 2018, 147, 77–89. [Google Scholar] [CrossRef]
- Tu, Y.; Wang, C.; Xu, S.; Lan, Z.; Li, W.; Han, J.; Zhou, Y.; Zheng, P.; Zhu, W. Design, Synthesis, and Docking Studies of Quinazoline Analogues Bearing Aryl Semicarbazone Scaffolds as Potent EGFR Inhibitors. Bioorg. Med. Chem. 2017, 25, 3148–3157. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Xu, S.; Peng, L.; Zhang, B.; Zhang, H.; Hu, Y.; Zheng, P.; Zhu, W. Design, Synthesis and Biological Evaluation of Novel 4-Anlinoquinazoline Derivatives as EGFR Inhibitors with the Potential to Inhibit the Gefitinib-Resistant Nonsmall Cell Lung Cancers. J. Enzyme Inhib. Med. Chem. 2019, 34, 204–218. [Google Scholar] [CrossRef]
- Solca, F.; Dahl, G.; Zoephel, A.; Bader, G.; Sanderson, M.; Klein, C.; Kraemer, O.; Himmelsbach, F.; Haaksma, E.; Adolf, G.R. Target Binding Properties and Cellular Activity of Afatinib (BIBW 2992), an Irreversible ErbB Family Blocker. J. Pharmacol. Exp. Ther. 2012, 343, 342–350. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.Q.; Gong, F.H.; Ye, J.Q.; Zhang, C.; Yue, X.H.; Li, C.G.; Xu, Y.G.; Sun, L.P. Design and Discovery of 4-Anilinoquinazoline-Urea Derivatives as Dual TK Inhibitors of EGFR and VEGFR-2. Eur. J. Med. Chem. 2017, 125, 245–254. [Google Scholar] [CrossRef]
- Sun, S.; Zhang, J.; Wang, N.; Kong, X.; Fu, F.; Wang, H.; Yao, J. Design and Discovery of Quinazoline- and Thiourea-Containing Sorafenib Analogs as EGFR and VEGFR-2 Dual TK Inhibitors. Molecules 2017, 23, 24. [Google Scholar] [CrossRef]
- Hamed, M.M.; Darwish, S.S.; Herrmann, J.; Abadi, A.H.; Engel, M. First Bispecific Inhibitors of the Epidermal Growth Factor Receptor Kinase and the NF-ΚB Activity as Novel Anticancer Agents. J. Med. Chem. 2017, 60, 2853–2868. [Google Scholar] [CrossRef] [PubMed]
- Gan, W.; Wang, C.; Pan, Q.; Li, Y.; Guo, Y.; Fan, D.; Peng, Y.; Rao, Z.; Xu, S.; Zheng, P.; et al. Discovery of Novel 4-Arylamino-Quinazoline Derivatives as EGFRL858R/T790M Inhibitors with the Potential to Inhibit the Non-Small Cell Lung Cancers. Bioorg. Chem. 2022, 127, 106011. [Google Scholar] [CrossRef]
- Hou, W.; Ren, Y.; Zhang, Z.; Sun, H.; Ma, Y.; Yan, B. Novel Quinazoline Derivatives Bearing Various 6-Benzamide Moieties as Highly Selective and Potent EGFR Inhibitors. Bioorg. Med. Chem. 2018, 26, 1740–1750. [Google Scholar] [CrossRef]
- Zhang, B.; Xu, Z.; Liu, Q.; Xia, S.; Liu, Z.; Liao, Z.; Gou, S. Design, Synthesis and Biological Evaluation of Cinnamamide-Quinazoline Derivatives as Potential EGFR Inhibitors to Reverse T790M Mutation. Bioorg. Chem. 2021, 117, 105432. [Google Scholar] [CrossRef]
- Yun, C.H.; Mengwasser, K.E.; Toms, A.V.; Woo, M.S.; Greulich, H.; Wong, K.K.; Meyerson, M.; Eck, M.J. The T790M Mutation in EGFR Kinase Causes Drug Resistance by Increasing the Affinity for ATP. Proc. Natl. Acad. Sci. USA 2008, 105, 2070. [Google Scholar] [CrossRef]
- Tang, Q.; Peng, T.; Hu, J.; Zhang, T.; Chen, P.; Chen, D.; Wang, Y.; Chen, L.; Tong, L.; Chen, Y.; et al. Discovery of N-(3-Bromo-1H-Indol-5-Yl)-Quinazolin-4-Amine as an Effective Molecular Skeleton to Develop Reversible/Irreversible Pan-HER Inhibitors. Eur. J. Med. Chem. 2022, 233, 114262. [Google Scholar] [CrossRef] [PubMed]
- Zou, M.; Jin, B.; Liu, Y.; Chen, H.; Zhang, Z.; Zhang, C.; Zhao, Z.; Zheng, L. Synthesis and Biological Evaluation of Some Novel Thiophene-Bearing Quinazoline Derivatives as EGFR Inhibitors. Lett. Drug Des. Discov. 2018, 16, 102–110. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, Y.; Liu, J.; Chen, L.; Zhao, L.; Li, B.; Wang, W. Synthesis and in Vitro Biological Evaluation of Novel Quinazoline Derivatives. Bioorg. Med. Chem. Lett. 2017, 27, 1584–1587. [Google Scholar] [CrossRef]
- Ding, C.; Chen, S.; Zhang, C.; Hu, G.; Zhang, W.; Li, L.; Chen, Y.Z.; Tan, C.; Jiang, Y. Synthesis and Investigation of Novel 6-(1,2,3-Triazol-4-Yl)-4-Aminoquinazolin Derivatives Possessing Hydroxamic Acid Moiety for Cancer Therapy. Bioorg. Med. Chem. 2017, 25, 27–37. [Google Scholar] [CrossRef]
- Wei, H.; Duan, Y.; Gou, W.; Cui, J.; Ning, H.; Li, D.; Qin, Y.; Liu, Q.; Li, Y. Design, Synthesis and Biological Evaluation of Novel 4-Anilinoquinazoline Derivatives as Hypoxia-Selective EGFR and VEGFR-2 Dual Inhibitors. Eur. J. Med. Chem. 2019, 181, 111564. [Google Scholar] [CrossRef]
- Cheng, W.; Wang, S.; Yang, Z.; Tian, X.; Hu, Y. Design, Synthesis, and Biological Study of 4-[(2-Nitroimidazole-1H-Alkyloxyl)Aniline]-Quinazolines as EGFR Inhibitors Exerting Cytotoxicities Both under Normoxia and Hypoxia. Drug Des. Devel. Ther. 2019, 13, 3079–3089. [Google Scholar] [CrossRef] [PubMed]
- Elkamhawy, A.; Son, S.; Lee, H.Y.; El-Maghrabey, M.H.; El Hamd, M.A.; Alshammari, S.O.; Abdelhameed, A.A.; Alshammari, Q.A.; Abdeen, A.; Ibrahim, S.F.; et al. Design, Synthesis, Biological Evaluation, and Molecular Dynamics Studies of Novel Lapatinib Derivatives. Pharmaceuticals 2022, 16, 43. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Yang, L.; Liu, P.; Yang, L.; Chen, L.; Hu, L.; Jiang, M. Design, Synthesis and Biological Evaluation of 2,3-Dihydro-[1,4]Dioxino[2,3-f]Quinazoline Derivatives as EGFR Inhibitors. Bioorg. Chem. 2021, 110, 104755. [Google Scholar] [CrossRef] [PubMed]
- Stamos, J.; Sliwkowski, M.X.; Eigenbrot, C. Structure of the Epidermal Growth Factor Receptor Kinase Domain Alone and in Complex with a 4-Anilinoquinazoline Inhibitor. J. Biol. Chem. 2002, 277, 46265–46272. [Google Scholar] [CrossRef]
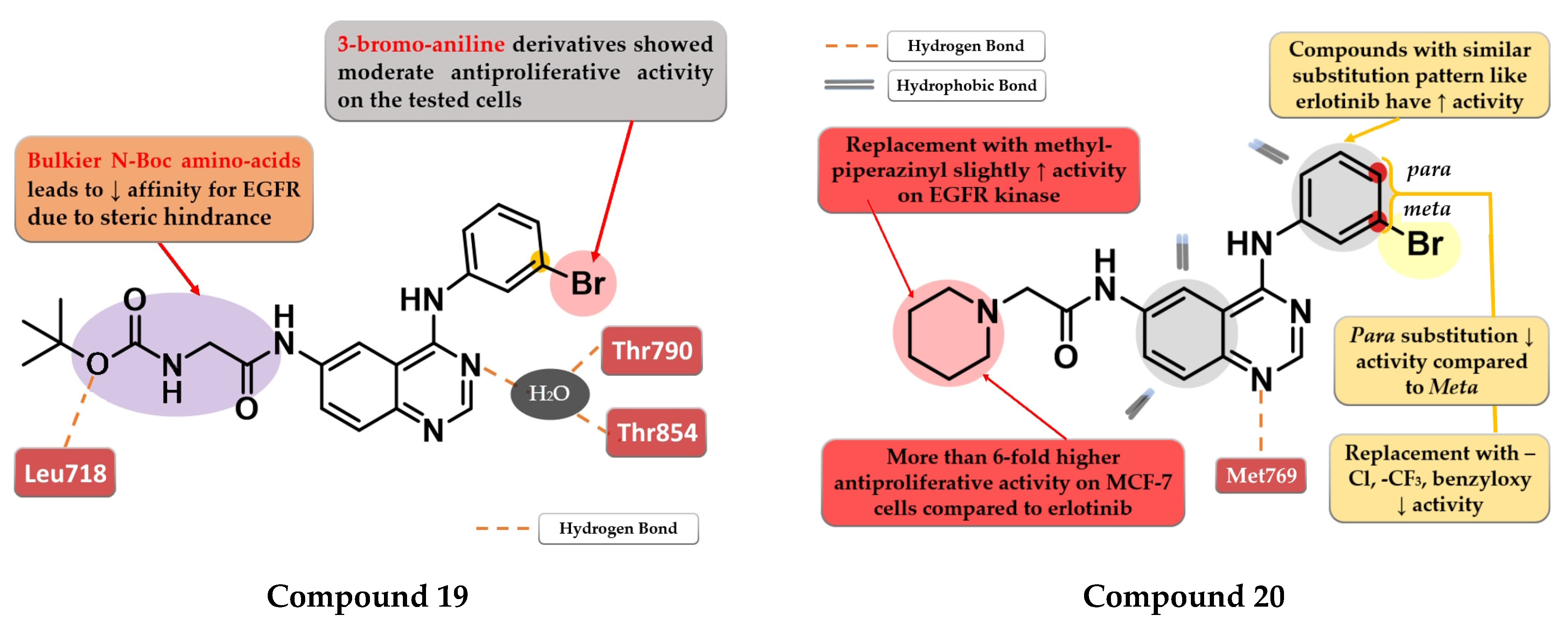
- Zheng, Y.G.; Su, J.; Gao, C.Y.; Jiang, P.; An, L.; Xue, Y.S.; Gao, J.; Liu, Y. Design, Synthesis, and Biological Evaluation of Novel 4-Anilinoquinazoline Derivatives Bearing Amino Acid Moiety as Potential EGFR Kinase Inhibitors. Eur. J. Med. Chem. 2017, 130, 393–405. [Google Scholar] [CrossRef]
- Ismail, R.S.M.; Abou-Seri, S.M.; Eldehna, W.M.; Ismail, N.S.M.; Elgazwi, S.M.; Ghabbour, H.A.; Ahmed, M.S.; Halaweish, F.T.; Abou El Ella, D.A. Novel Series of 6-(2-Substitutedacetamido)-4-Anilinoquinazolines as EGFR-ERK Signal Transduction Inhibitors in MCF-7 Breast Cancer Cells. Eur. J. Med. Chem. 2018, 155, 782–796. [Google Scholar] [CrossRef] [PubMed]
- Farouk, A.K.B.A.W.; Abdelrasheed Allam, H.; Rashwan, E.; George, R.F.; Abbas, S.E.S. Design and Synthesis of Some New 6-Bromo-2-(Pyridin-3-Yl)-4-Substituted Quinazolines as Multi Tyrosine Kinase Inhibitors. Bioorg. Chem. 2022, 128, 10710. [Google Scholar] [CrossRef] [PubMed]
- Mphahlele, M.J.; Mmonwa, M.M.; Aro, A.; McGaw, L.J.; Choong, Y.S. Synthesis, Biological Evaluation and Molecular Docking of Novel Indole-Aminoquinazoline Hybrids for Anticancer Properties. Int. J. Mol. Sci. 2018, 19, 2232. [Google Scholar] [CrossRef]
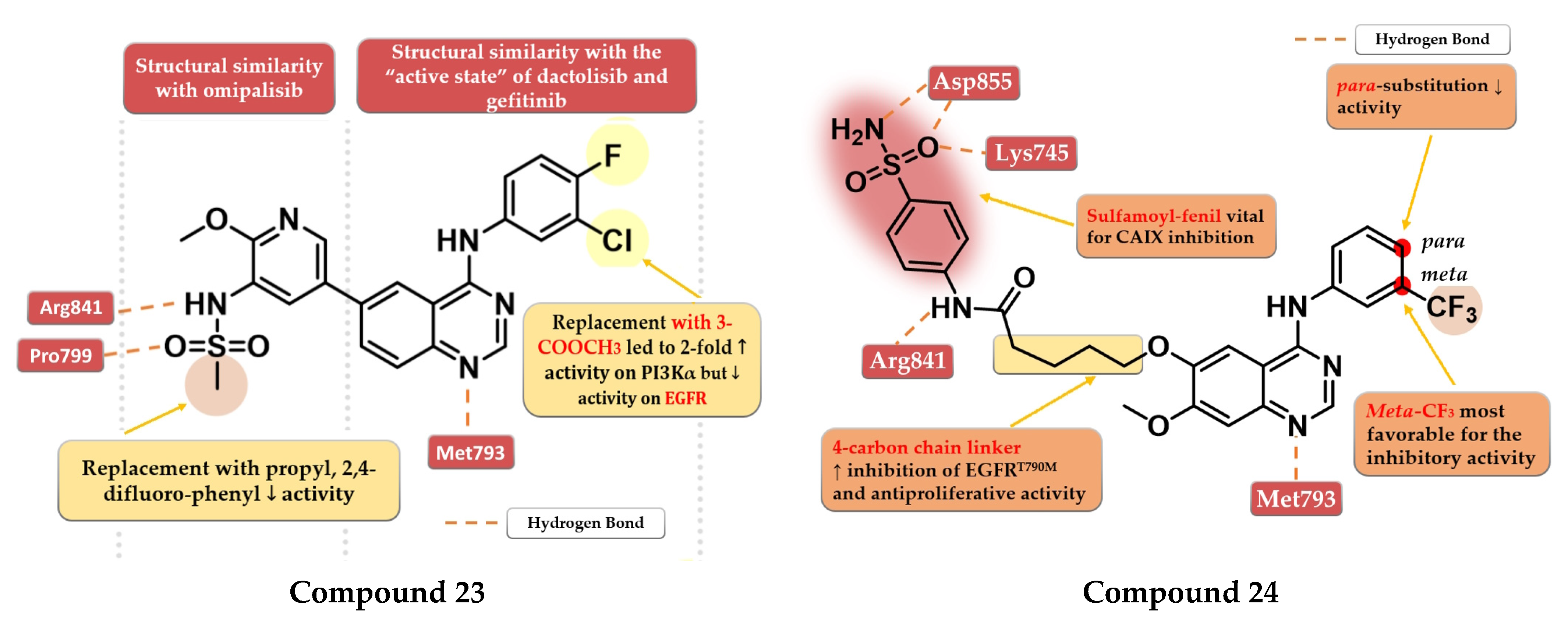
- Ding, H.W.; Deng, C.L.; Li, D.D.; Liu, D.D.; Chai, S.M.; Wang, W.; Zhang, Y.; Chen, K.; Li, X.; Wang, J.; et al. Design, Synthesis and Biological Evaluation of Novel 4-Aminoquinazolines as Dual Target Inhibitors of EGFR-PI3Kα. Eur. J. Med. Chem. 2018, 146, 460–470. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Liu, Z.; Xia, S.; Liu, Q.; Gou, S. Design, Synthesis and Biological Evaluation of Sulfamoylphenyl-Quinazoline Derivatives as Potential EGFR/CAIX Dual Inhibitors. Eur. J. Med. Chem. 2021, 216, 113300. [Google Scholar] [CrossRef] [PubMed]
- Yosaatmadja, Y.; Squire, C.J. 1.85 Angstrom Structure of EGFR Kinase Domain with Gefitinib. Available online: https://www.rcsb.org/structure/4WKQ (accessed on 22 March 2023).
- Chang, J.; Ren, H.; Zhao, M.; Chong, Y.; Zhao, W.; He, Y.; Zhao, Y.; Zhang, H.; Qi, C. Development of a Series of Novel 4-Anlinoquinazoline Derivatives Possessing Quinazoline Skeleton: Design, Synthesis, EGFR Kinase Inhibitory Efficacy, and Evaluation of Anticancer Activities in Vitro. Eur. J. Med. Chem. 2017, 138, 669–688. [Google Scholar] [CrossRef]
- Wang, L.; Li, P.; Li, B.; Wang, Y.; Li, J.; Song, L. Design, Synthesis, and Antitumor Activity of Novel Quinazoline Derivatives. Molecules 2017, 22, 1624. [Google Scholar] [CrossRef]
- Ju, Y.; Wu, J.; Yuan, X.; Zhao, L.; Zhang, G.; Li, C.; Qiao, R. Design and Evaluation of Potent EGFR Inhibitors through the Incorporation of Macrocyclic Polyamine Moieties into the 4-Anilinoquinazoline Scaffold. J. Med. Chem. 2018, 61, 11372–11383. [Google Scholar] [CrossRef]
- Amrhein, J.A.; Beyett, T.S.; Feng, W.W.; Krämer, A.; Weckesser, J.; Schaeffner, I.K.; Rana, J.K.; Jänne, P.A.; Eck, M.J.; Knapp, S.; et al. Macrocyclization of Quinazoline-Based EGFR Inhibitors Leads to Exclusive Mutant Selectivity for EGFR L858R and Del19. J. Med. Chem. 2022, 65, 15679–15697. [Google Scholar] [CrossRef]
- Yamahana, H.; Kunieda, Y.; Tominaga, M.; Yamada, H.; Uto, Y. Development of a Novel Acetyl Glucose-Modified Gefitinib Derivative to Enhance the Radiosensitizing Effect. Bioorg. Med. Chem. 2021, 29, 115889. [Google Scholar] [CrossRef]
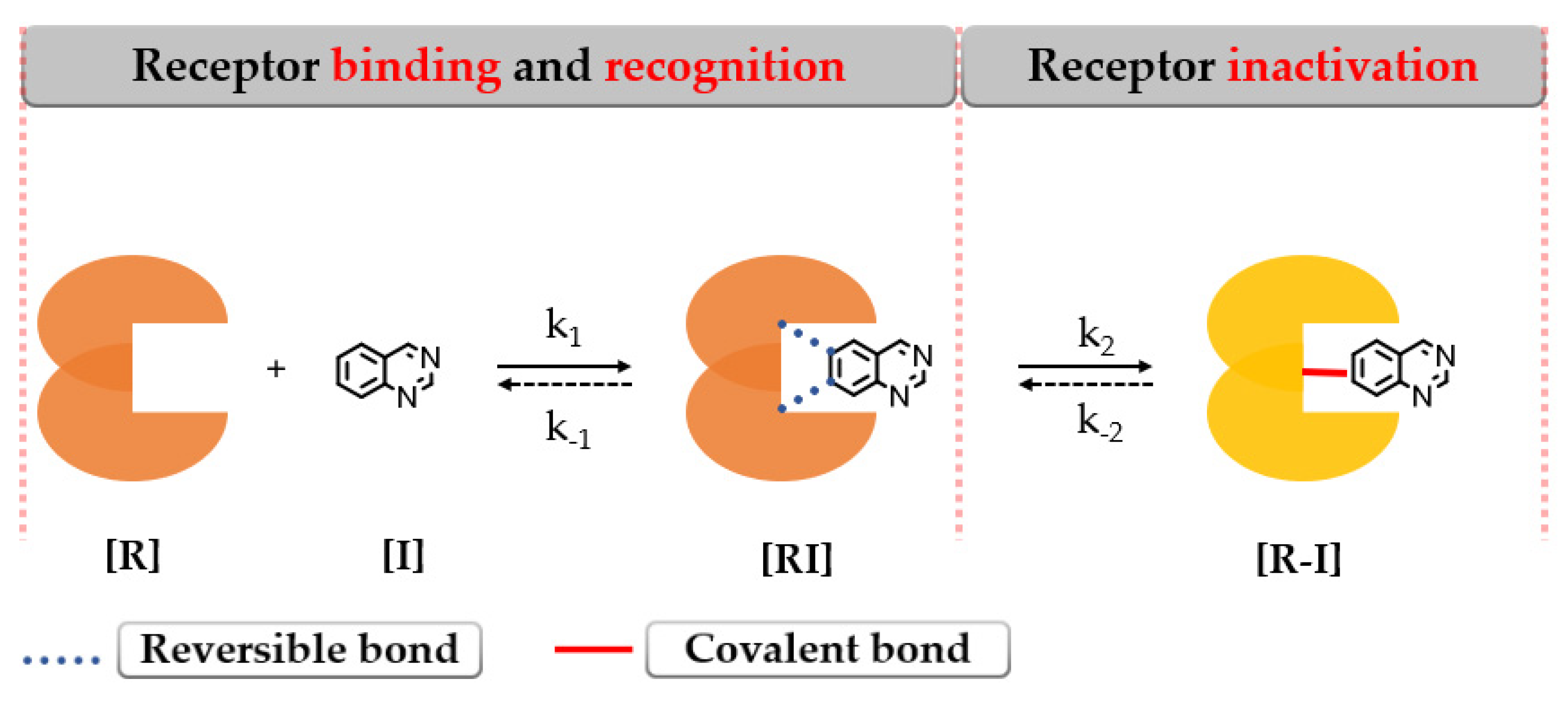
- Hossam, M.; Lasheen, D.S.; Abouzid, K.A.M. Covalent EGFR Inhibitors: Binding Mechanisms, Synthetic Approaches, and Clinical Profiles. Arch. Pharm. 2016, 349, 573–593. [Google Scholar] [CrossRef] [PubMed]
- Singh, J.; Dobrusin, E.M.; Fry, D.W.; Haske, T.; Whitty, A.; McNamara, D.J. Structure-Based Design of a Potent, Selective, and Irreversible Inhibitor of the Catalytic Domain of the ErbB Receptor Subfamily of Protein Tyrosine Kinases. J. Med. Chem. 1997, 40, 1130–1135. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Liu, Q.; Bliven, S.; Xie, L.; Bourne, P.E. Determining Cysteines Available for Covalent Inhibition Across the Human Kinome. J. Med. Chem. 2017, 60, 2879–2889. [Google Scholar] [CrossRef] [PubMed]
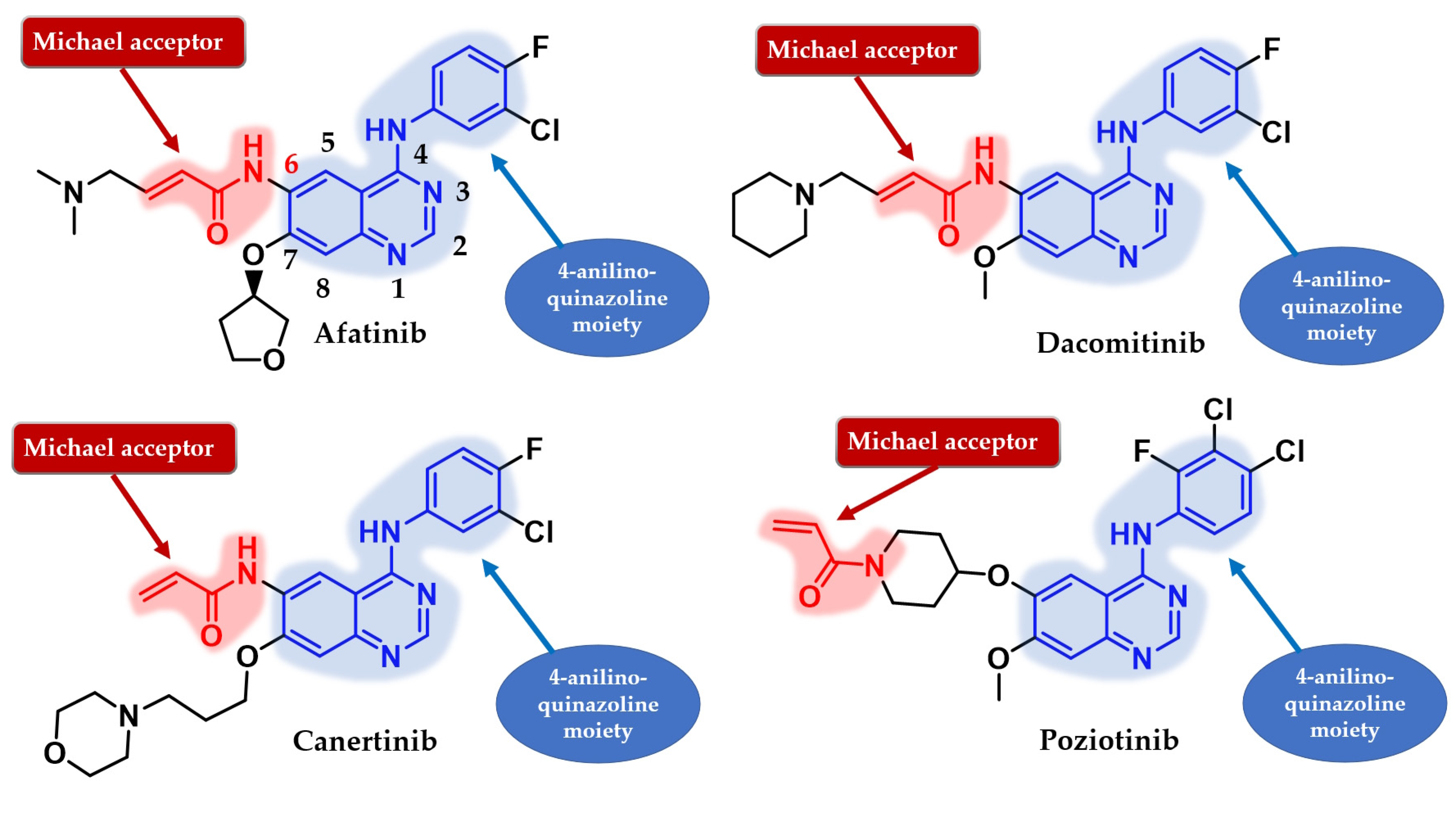
- Ou, S.H.I. Second-Generation Irreversible Epidermal Growth Factor Receptor (EGFR) Tyrosine Kinase Inhibitors (TKIs): A Better Mousetrap? A Review of the Clinical Evidence. Crit. Rev. Oncol. Hematol. 2012, 83, 407–421. [Google Scholar] [CrossRef]
- Baillie, T.A. Targeted Covalent Inhibitors for Drug Design. Angew. Chem. Int. Ed. 2016, 55, 13408–13421. [Google Scholar] [CrossRef]
- Lonsdale, R.; Burgess, J.; Colclough, N.; Davies, N.L.; Lenz, E.M.; Orton, A.L.; Ward, R.A. Expanding the Armory: Predicting and Tuning Covalent Warhead Reactivity. J. Chem. Inf. Model. 2017, 57, 3124–3137. [Google Scholar] [CrossRef]
- Tuley, A.; Fast, W. The Taxonomy of Covalent Inhibitors. Biochemistry 2018, 57, 3326–3337. [Google Scholar] [CrossRef]
- Food and Drug Administration (FDA) Label—GILOTRIFTM (Afatinib): 40 Mg, 30 Mg, and 20 Mg Tablets, for Oral Use. Available online: www.fda.gov/medwatch (accessed on 18 February 2023).
- Food and Drug Administration (FDA) Label—VIZIMPRO® (Dacomitinib) 15 Mg, 30 Mg, and 45 Mg Tablets, for Oral Use. Available online: www.fda.gov/medwatch (accessed on 18 February 2023).
- Shen, Y.C.; Tseng, G.C.; Tu, C.Y.; Chen, W.C.; Liao, W.C.; Chen, W.C.; Li, C.H.; Chen, H.J.; Hsia, T.C. Comparing the Effects of Afatinib with Gefitinib or Erlotinib in Patients with Advanced-Stage Lung Adenocarcinoma Harboring Non-Classical Epidermal Growth Factor Receptor Mutations. Lung Cancer 2017, 110, 56–62. [Google Scholar] [CrossRef]
- Hsu, P.C.; Lee, S.H.; Chiu, L.C.; Lee, C.S.; Wu, C.E.; Kuo, S.C.H.; Ju, J.S.; Huang, A.C.C.; Li, S.H.; Ko, H.W.; et al. Afatinib in Untreated Stage IIIB/IV Lung Adenocarcinoma with Major Uncommon Epidermal Growth Factor Receptor (EGFR) Mutations (G719X/L861Q/S768I): A Multicenter Observational Study in Taiwan. Target. Oncol. 2023, 18, 195–207. [Google Scholar] [CrossRef]
- Kosaka, T.; Tanizaki, J.; Paranal, R.M.; Endoh, H.; Lydon, C.; Capelletti, M.; Repellin, C.E.; Choi, J.; Ogino, A.; Calles, A.; et al. Response Heterogeneity of EGFR and HER2 Exon 20 Insertions to Covalent EGFR and HER2 Inhibitors. Cancer Res. 2017, 77, 2712–2721. [Google Scholar] [CrossRef]
- van Veggel, B.; de Langen, A.J.; Hashemi, S.M.S.; Monkhorst, K.; Heideman, D.A.M.; Thunnissen, E.; Smit, E.F. Afatinib and Cetuximab in Four Patients with EGFR Exon 20 Insertion-Positive Advanced NSCLC. J. Thorac. Oncol. 2018, 13, 1222–1226. [Google Scholar] [CrossRef] [PubMed]
- Metro, G.; Crinò, L. The LUX-Lung Clinical Trial Program of Afatinib for Non-Small-Cell Lung Cancer. Expert Rev. Anticancer Ther. 2014, 11, 673–682. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.L.; Sequist, L.V.; Tan, E.H.; Geater, S.L.; Orlov, S.; Zhang, L.; Lee, K.H.; Tsai, C.M.; Kato, T.; Barrios, C.H.; et al. Afatinib as First-Line Treatment of Older Patients with EGFR Mutation-Positive Non-Small-Cell Lung Cancer: Subgroup Analyses of the LUX-Lung 3, LUX-Lung 6, and LUX-Lung 7 Trials. Clin. Lung Cancer 2018, 19, e465–e479. [Google Scholar] [CrossRef] [PubMed]
- Tagliamento, M.; Genova, C.; Rijavec, E.; Rossi, G.; Biello, F.; Dal Bello, M.G.; Alama, A.; Coco, S.; Boccardo, S.; Grossi, F. Afatinib and Erlotinib in the Treatment of Squamous-Cell Lung Cancer. Expert Opin. Pharmacother. 2018, 19, 2055–2062. [Google Scholar] [CrossRef]
- Ohashi, K.; Ninomiya, K.; Yoshioka, H.; Bessho, A.; Shibayama, T.; Aoe, K.; Ishikawa, N.; Kozuki, T.; Kawai, H.; Kuyama, S.; et al. Impact of HER2 Expression on EGFR-TKI Treatment Outcomes in Lung Tumors Harboring EGFR Mutations: A HER2-CS Study Subset Analysis. Lung Cancer 2020, 150, 83–89. [Google Scholar] [CrossRef]
- Nagasaka, M.; Singh, V.; Baca, Y.; Sukari, A.; Kim, C.; Mamdani, H.; Spira, A.I.; Uprety, D.; Bepler, G.; Kim, E.S.; et al. The Effects of HER2 Alterations in EGFR Mutant Non-Small Cell Lung Cancer. Clin. Lung Cancer 2022, 23, 52–59. [Google Scholar] [CrossRef]
- Wind, S.; Schnell, D.; Ebner, T.; Freiwald, M.; Stopfer, P. Clinical Pharmacokinetics and Pharmacodynamics of Afatinib. Clin. Pharmacokinet. 2017, 56, 235–250. [Google Scholar] [CrossRef]
- Ding, P.N.; Lord, S.J.; Gebski, V.; Links, M.; Bray, V.; Gralla, R.J.; Yang, J.C.H.; Lee, C.K. Risk of Treatment-Related Toxicities from EGFR Tyrosine Kinase Inhibitors: A Meta-Analysis of Clinical Trials of Gefitinib, Erlotinib, and Afatinib in Advanced EGFR-Mutated Non–Small Cell Lung Cancer. J. Thorac. Oncol. 2017, 12, 633–643. [Google Scholar] [CrossRef]
- Han, W.; Du, Y. Recent Development of the Second and Third Generation Irreversible Epidermal Growth Factor Receptor Inhibitors. Chem. Biodivers. 2017, 14, e1600393. [Google Scholar] [CrossRef]
- Sequist, L.V.; Yang, J.C.H.; Yamamoto, N.; O’Byrne, K.; Hirsh, V.; Mok, T.; Geater, S.L.; Orlov, S.; Tsai, C.M.; Boyer, M.; et al. Phase III Study of Afatinib or Cisplatin plus Pemetrexed in Patients with Metastatic Lung Adenocarcinoma with EGFR Mutations. J. Clin. Oncol. 2013, 31, 3327–3334. [Google Scholar] [CrossRef]
- Yang, J.C.H.; Wu, Y.L.; Schuler, M.; Sebastian, M.; Popat, S.; Yamamoto, N.; Zhou, C.; Hu, C.P.; O’Byrne, K.; Feng, J.; et al. Afatinib versus Cisplatin-Based Chemotherapy for EGFR Mutation-Positive Lung Adenocarcinoma (LUX-Lung 3 and LUX-Lung 6): Analysis of Overall Survival Data from Two Randomised, Phase 3 Trials. Lancet Oncol. 2015, 16, 141–151. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.L.; Zhou, C.; Hu, C.P.; Feng, J.; Lu, S.; Huang, Y.; Li, W.; Hou, M.; Shi, J.H.; Lee, K.Y.; et al. Afatinib versus Cisplatin plus Gemcitabine for First-Line Treatment of Asian Patients with Advanced Non-Small-Cell Lung Cancer Harbouring EGFR Mutations (LUX-Lung 6): An Open-Label, Randomised Phase 3 Trial. Lancet Oncol. 2014, 15, 213–222. [Google Scholar] [CrossRef] [PubMed]
- Miyahisa, I.; Sameshima, T.; Hixon, M.S. Rapid Determination of the Specificity Constant of Irreversible Inhibitors (Kinact/KI) by Means of an Endpoint Competition Assay. Angew. Chem. 2015, 127, 14305–14308. [Google Scholar] [CrossRef]
- Schwartz, P.A.; Kuzmic, P.; Solowiej, J.; Bergqvist, S.; Bolanos, B.; Almaden, C.; Nagata, A.; Ryan, K.; Feng, J.; Dalvie, D.; et al. Covalent EGFR Inhibitor Analysis Reveals Importance of Reversible Interactions to Potency and Mechanisms of Drug Resistance. Proc. Natl. Acad. Sci. USA 2014, 111, 173–178. [Google Scholar] [CrossRef] [PubMed]
- Gajiwala, K.S.; Feng, J.; Ferre, R.; Ryan, K.; Brodsky, O.; Weinrich, S.; Kath, J.C.; Stewart, A. Insights into the Aberrant Activity of Mutant EGFR Kinase Domain and Drug Recognition. Structure 2013, 21, 209–219. [Google Scholar] [CrossRef]
- Lategahn, J.; Tumbrink, H.L.; Schultz-Fademrecht, C.; Heimsoeth, A.; Werr, L.; Niggenaber, J.; Keul, M.; Parmaksiz, F.; Baumann, M.; Menninger, S.; et al. Insight into Targeting Exon20 Insertion Mutations of the Epidermal Growth Factor Receptor with Wild Type-Sparing Inhibitors. J. Med. Chem. 2022, 65, 6643–6655. [Google Scholar] [CrossRef]
- De Cesco, S.; Kurian, J.; Dufresne, C.; Mittermaier, A.K.; Moitessier, N. Covalent Inhibitors Design and Discovery. Eur. J. Med. Chem. 2017, 138, 96–114. [Google Scholar] [CrossRef]
- Wissner, A.; Fraser, H.L.; Ingalls, C.L.; Dushin, R.G.; Floyd, M.B.; Cheung, K.; Nittoli, T.; Ravi, M.R.; Tan, X.; Loganzo, F. Dual Irreversible Kinase Inhibitors: Quinazoline-Based Inhibitors Incorporating Two Independent Reactive Centers with Each Targeting Different Cysteine Residues in the Kinase Domains of EGFR and VEGFR-2. Bioorg. Med. Chem. 2007, 15, 3635–3648. [Google Scholar] [CrossRef]
- Blair, J.A.; Rauh, D.; Kung, C.; Yun, C.H.; Fan, Q.W.; Rode, H.; Zhang, C.; Eck, M.J.; Weiss, W.A.; Shokat, K.M. Structure-Guided Development of Affinity Probes for Tyrosine Kinases Using Chemical Genetics. Nat. Chem. Biol. 2007, 3, 229–238. [Google Scholar] [CrossRef]
- Martin, J.S.; MacKenzie, C.J.; Fletcher, D.; Gilbert, I.H. Characterising Covalent Warhead Reactivity. Bioorg. Med. Chem. 2019, 27, 2066–2074. [Google Scholar] [CrossRef]
- Flanagan, M.E.; Abramite, J.A.; Anderson, D.P.; Aulabaugh, A.; Dahal, U.P.; Gilbert, A.M.; Li, C.; Montgomery, J.; Oppenheimer, S.R.; Ryder, T.; et al. Chemical and Computational Methods for the Characterization of Covalent Reactive Groups for the Prospective Design of Irreversible Inhibitors. J. Med. Chem. 2014, 57, 10072–10079. [Google Scholar] [CrossRef] [PubMed]
- Ábrányi-Balogh, P.; Petri, L.; Imre, T.; Szijj, P.; Scarpino, A.; Hrast, M.; Mitrović, A.; Fonovič, U.P.; Németh, K.; Barreteau, H.; et al. A Road Map for Prioritizing Warheads for Cysteine Targeting Covalent Inhibitors. Eur. J. Med. Chem. 2018, 160, 94–107. [Google Scholar] [CrossRef] [PubMed]
- Abdeldayem, A.; Raouf, Y.S.; Constantinescu, S.N.; Moriggl, R.; Gunning, P.T. Advances in Covalent Kinase Inhibitors. Chem. Soc. Rev. 2020, 49, 2617–2687. [Google Scholar] [CrossRef] [PubMed]
- Tsou, H.R.; Mamuya, N.; Johnson, B.D.; Reich, M.F.; Gruber, B.C.; Ye, F.; Nilakantan, R.; Shen, R.; Discafani, C.; DeBlanc, R.; et al. 6-Substituted-4-(3-Bromophenylamino) Quinazolines as Putative Irreversible Inhibitors of the Epidermal Growth Factor Receptor (EGFR) and Human Epidermal Growth Factor Receptor (HER-2) Tyrosine Kinases with Enhanced Antitumor Activity. J. Med. Chem. 2001, 44, 2719–2734. [Google Scholar] [CrossRef] [PubMed]
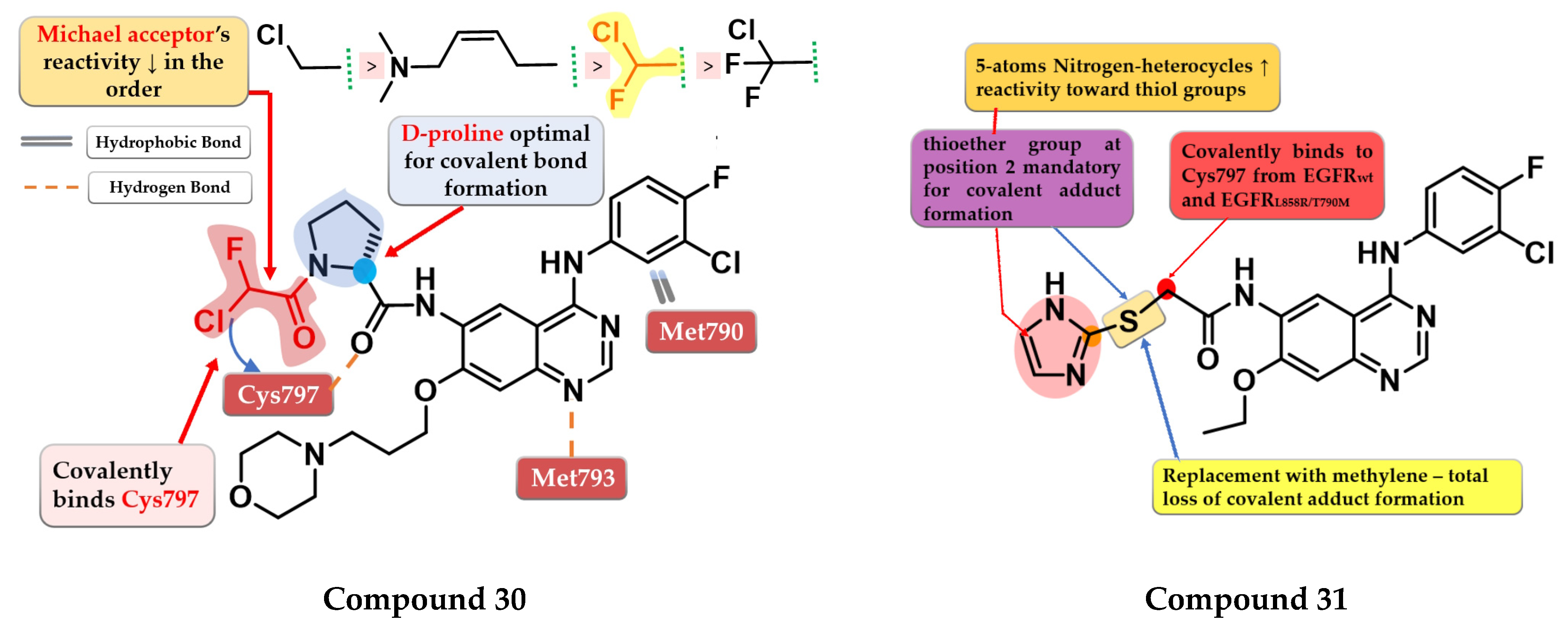
- Shindo, N.; Fuchida, H.; Sato, M.; Watari, K.; Shibata, T.; Kuwata, K.; Miura, C.; Okamoto, K.; Hatsuyama, Y.; Tokunaga, K.; et al. Selective and Reversible Modification of Kinase Cysteines with Chlorofluoroacetamides. Nat. Chem. Biol. 2019, 15, 250–258. [Google Scholar] [CrossRef]
- Castelli, R.; Bozza, N.; Cavazzoni, A.; Bonelli, M.; Vacondio, F.; Ferlenghi, F.; Callegari, D.; Silva, C.; Rivara, S.; Lodola, A.; et al. Balancing Reactivity and Antitumor Activity: Heteroarylthioacetamide Derivatives as Potent and Time-Dependent Inhibitors of EGFR. Eur. J. Med. Chem. 2019, 162, 507–524. [Google Scholar] [CrossRef]
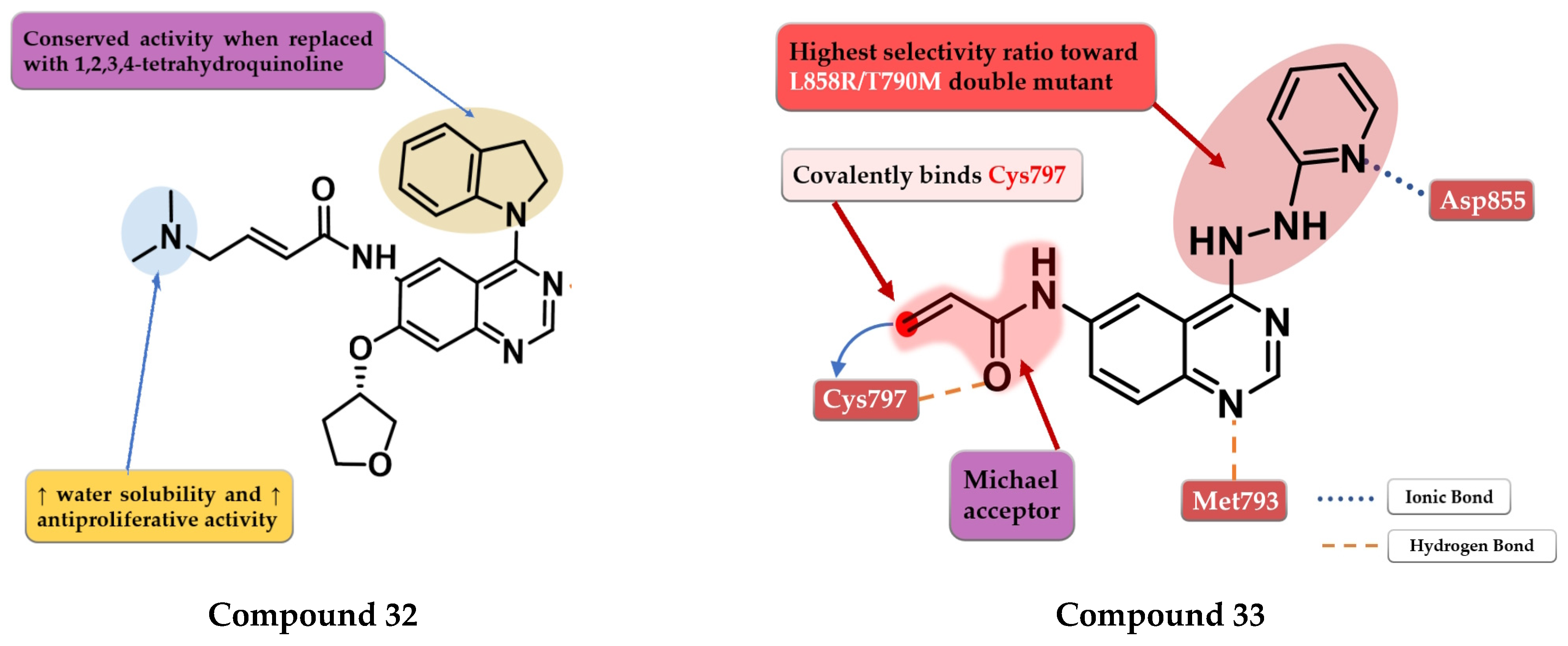
- OuYang, Y.; Zou, W.; Peng, L.; Yang, Z.; Tang, Q.; Chen, M.; Jia, S.; Zhang, H.; Lan, Z.; Zheng, P.; et al. Design, Synthesis, Antiproliferative Activity and Docking Studies of Quinazoline Derivatives Bearing 2,3-Dihydro-Indole or 1,2,3,4-Tetrahydroquinoline as Potential EGFR Inhibitors. Eur. J. Med. Chem. 2018, 154, 29–43. [Google Scholar] [CrossRef]
- Pawara, R.; Ahmad, I.; Nayak, D.; Wagh, S.; Wadkar, A.; Ansari, A.; Belamkar, S.; Surana, S.; Nath Kundu, C.; Patil, C.; et al. Novel, Selective Acrylamide Linked Quinazolines for the Treatment of Double Mutant EGFR-L858R/T790M Non-Small-Cell Lung Cancer (NSCLC). Bioorg. Chem. 2021, 115, 105234. [Google Scholar] [CrossRef]
- Zhou, W.; Ercan, D.; Chen, L.; Yun, C.H.; Li, D.; Capelletti, M.; Cortot, A.B.; Chirieac, L.; Iacob, R.E.; Padera, R.; et al. Novel Mutant-Selective EGFR Kinase Inhibitors against EGFR T790M. Nature 2009, 462, 1070–1074. [Google Scholar] [CrossRef]
- Zhao, L.; Fan, T.; Shi, Z.; Ding, C.; Zhang, C.; Yuan, Z.; Sun, Q.; Tan, C.; Chu, B.; Jiang, Y. Design, Synthesis and Evaluation of Novel ErbB/HDAC Multitargeted Inhibitors with Selectivity in EGFRT790M Mutant Cell Lines. Eur. J. Med. Chem. 2021, 213, 113173. [Google Scholar] [CrossRef]
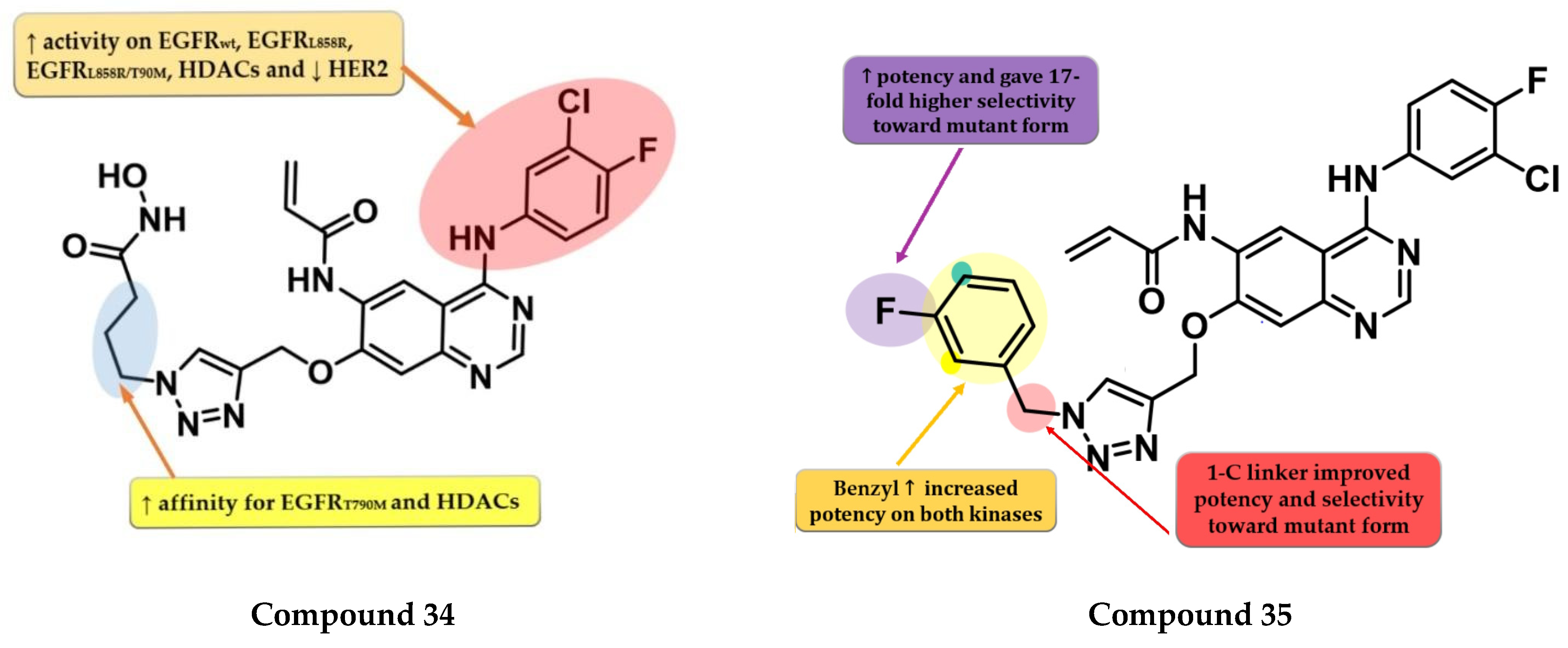
- Song, J.; Jang, S.; Lee, J.W.; Jung, D.; Lee, S.; Min, K.H. Click Chemistry for Improvement in Selectivity of Quinazoline-Based Kinase Inhibitors for Mutant Epidermal Growth Factor Receptors. Bioorg. Med. Chem. Lett. 2019, 29, 477–480. [Google Scholar] [CrossRef]
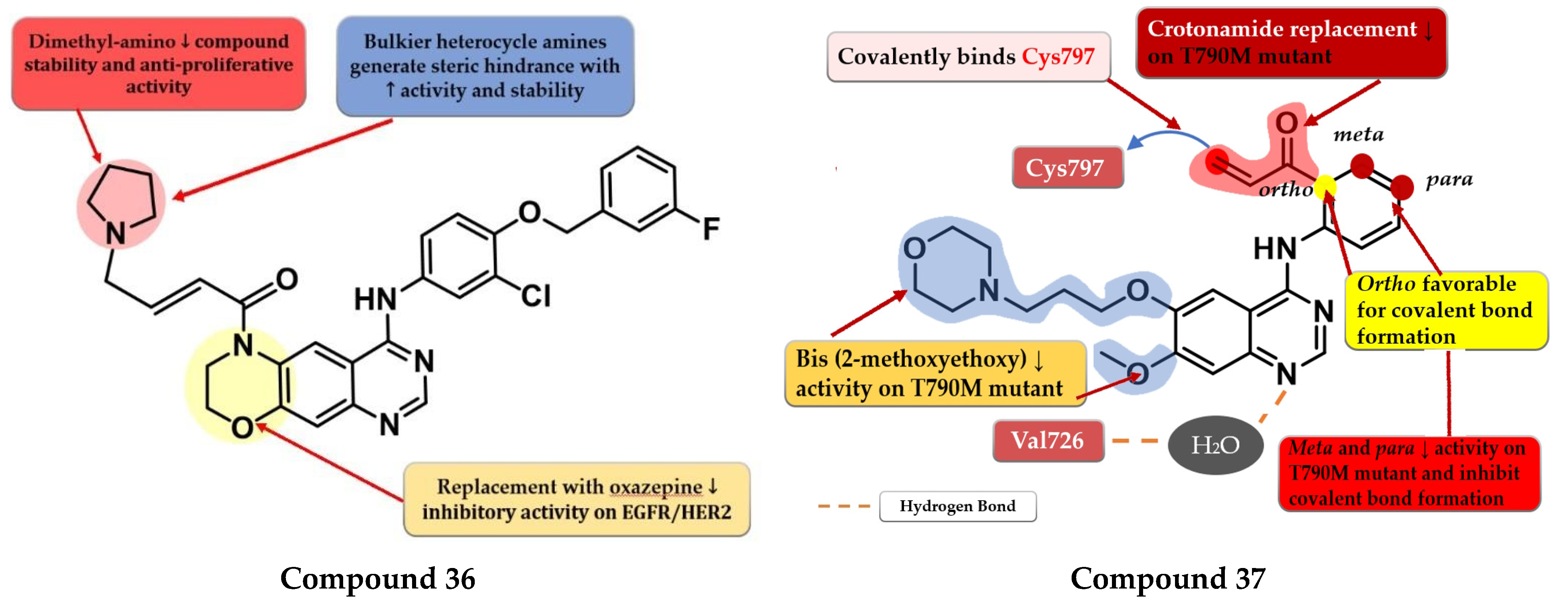
- Sun, M.; Jia, J.; Sun, H.; Wang, F. Design and Synthesis of a Novel Class EGFR/HER2 Dual Inhibitors Containing Tricyclic Oxazine Fused Quinazolines Scaffold. Bioorg. Med. Chem. Lett. 2020, 30, 127060. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Wang, L.; Feng, M.; Yi, Y.; Zhang, W.; Liu, W.; Li, L.; Liu, Z.; Li, Y.; Ma, X. New Acrylamide-Substituted Quinazoline Derivatives with Enhanced Potency for the Treatment of EGFR T790M-Mutant Non-Small-Cell Lung Cancers. Bioorg. Chem. 2018, 77, 593–599. [Google Scholar] [CrossRef] [PubMed]
- Jiao, X.; Zhang, Q.; Zhang, Y.; Shao, J.; Ding, L.; Tang, C.; Feng, B. Synthesis and Biological Evaluation of New Series of Quinazoline Derivatives as EGFR/HER2 Dual-Target Inhibitors. Bioorg. Med. Chem. Lett. 2022, 67, 128703. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.S.; Kumarakulasinghe, N.B.; Huang, Y.Q.; Ang, Y.L.E.; Choo, J.R.E.; Goh, B.C.; Soo, R.A. Third Generation EGFR TKIs: Current Data and Future Directions. Mol. Cancer 2018, 17, 29. [Google Scholar] [CrossRef] [PubMed]
- Patel, H.; Pawara, R.; Ansari, A.; Surana, S. Recent Updates on Third Generation EGFR Inhibitors and Emergence of Fourth Generation EGFR Inhibitors to Combat C797S Resistance. Eur. J. Med. Chem. 2017, 142, 32–47. [Google Scholar] [CrossRef] [PubMed]
- Murtuza, A.; Bulbul, A.; Shen, J.P.; Keshavarzian, P.; Woodward, B.D.; Lopez-Diaz, F.J.; Lippman, S.M.; Husain, H. Novel Third-Generation EGFR Tyrosine Kinase Inhibitors and Strategies to Overcome Therapeutic Resistance in Lung Cancer. Cancer Res. 2019, 79, 689–698. [Google Scholar] [CrossRef]
- Remon, J.; Steuer, C.E.; Ramalingam, S.S.; Felip, E. Osimertinib and Other Third-Generation EGFR TKI in EGFR-Mutant NSCLC Patients. Ann. Oncol. 2018, 29, i20–i27. [Google Scholar] [CrossRef]
- Jänne, P.A.; Yang, J.C.-H.; Kim, D.-W.; Planchard, D.; Ohe, Y.; Ramalingam, S.S.; Ahn, M.-J.; Kim, S.-W.; Su, W.-C.; Horn, L.; et al. AZD9291 in EGFR Inhibitor–Resistant Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2015, 372, 1689–1699. [Google Scholar] [CrossRef]
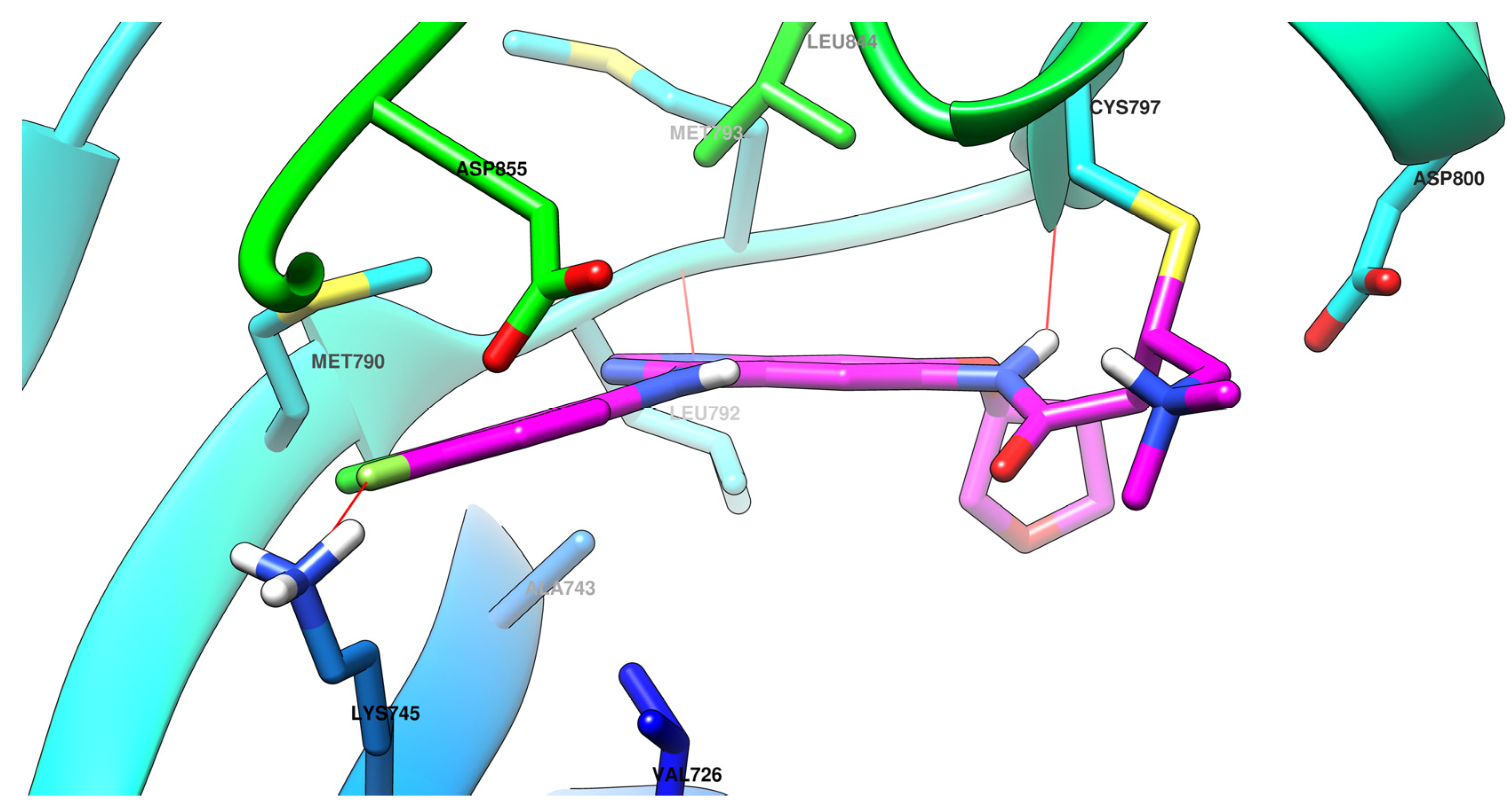
- Ward, R.A.; Anderton, M.J.; Ashton, S.; Bethel, P.A.; Box, M.; Butterworth, S.; Colclough, N.; Chorley, C.G.; Chuaqui, C.; Cross, D.A.E.; et al. Structure- and Reactivity-Based Development of Covalent Inhibitors of the Activating and Gatekeeper Mutant Forms of the Epidermal Growth Factor Receptor (EGFR). J. Med. Chem. 2013, 56, 7025–7048. [Google Scholar] [CrossRef]
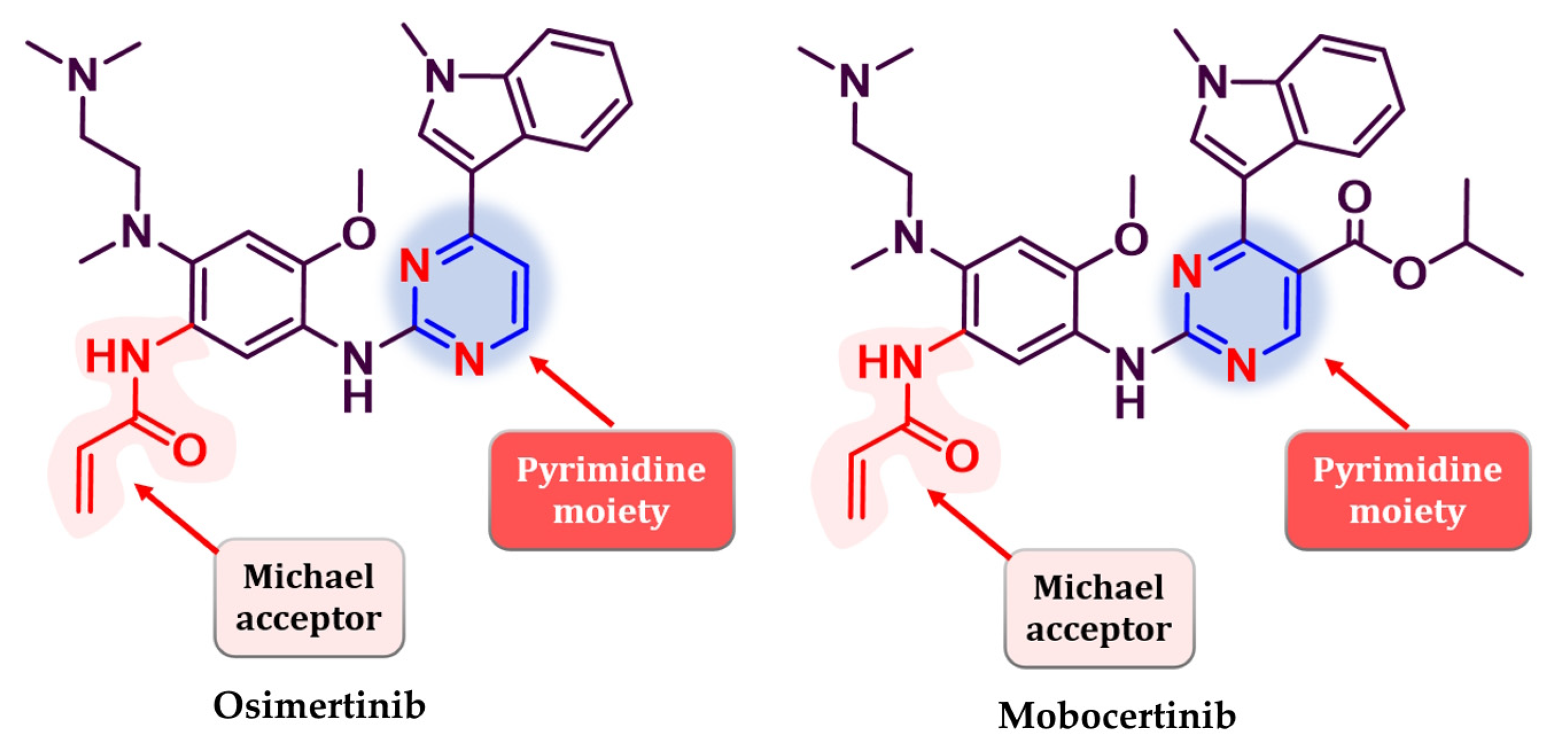
- Yver, A. Osimertinib (AZD9291)-a Sciencedriven, Collaborative Approach to Rapid Drug Design and Development. Ann. Oncol. 2016, 27, 1165–1170. [Google Scholar] [CrossRef] [PubMed]
- Food and Drug Administration (FDA) Label—TAGRISSO® (Osimertinib) 80 Mg and 40 Mg Tablets, for Oral Use. Available online: www.fda.gov/medwatch (accessed on 21 February 2023).
- Soria, J.-C.; Ohe, Y.; Vansteenkiste, J.; Reungwetwattana, T.; Chewaskulyong, B.; Lee, K.H.; Dechaphunkul, A.; Imamura, F.; Nogami, N.; Kurata, T.; et al. Osimertinib in Untreated EGFR -Mutated Advanced Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2018, 378, 113–125. [Google Scholar] [CrossRef] [PubMed]
- Ramalingam, S.S.; Zhou, C.; Kim, T.M.; Kim, S.-W.; Yang, J.C.-H.; Riely, G.J.; Mekhail, T.; Nguyen, D.; García Campelo, M.R.; Felip, E.; et al. Mobocertinib (TAK-788) in EGFR Exon 20 Insertion (Ex20ins)+ Metastatic NSCLC (MNSCLC): Additional Results from Platinum-Pretreated Patients (Pts) and EXCLAIM Cohort of Phase 1/2 Study. J. Clin. Oncol. 2021, 39, 9014. [Google Scholar] [CrossRef]
- Gonzalvez, F.; Vincent, S.; Baker, T.E.; Gould, A.E.; Li, S.; Wardwell, S.D.; Nadworny, S.; Ning, Y.; Zhang, S.; Huang, W.S.; et al. Mobocertinib (Tak-788): A Targeted Inhibitor of Egfr Exon 20 Insertion Mutants in Non–Small Cell Lung Cancer. Cancer Discov. 2021, 11, 1672–1687. [Google Scholar] [CrossRef]
- Food and Drug Administration (FDA) Label—EXKIVITY® (Mobocertinib) 40 Mg Capsules, for Oral Use. Available online: www.fda.gov/medwatch (accessed on 7 March 2023).
- Zhang, S.S.; Zhu, V.W. Spotlight on Mobocertinib (TAK-788) in NSCLC with EGFR Exon 20 Insertion Mutations. Lung Cancer Targets Ther. 2021, 12, 61–65. [Google Scholar] [CrossRef]
- Starrett, J.H.; Guernet, A.A.; Cuomo, M.E.; Poels, K.E.; van Alderwerelt van Rosenburgh, I.K.; Nagelberg, A.; Farnsworth, D.; Price, K.S.; Khan, H.; Ashtekar, K.D.; et al. Drug Sensitivity and Allele Specificity of First-Line Osimertinib Resistance EGFR Mutations. Cancer Res. 2020, 80, 2017–2030. [Google Scholar] [CrossRef]
- Thress, K.S.; Paweletz, C.P.; Felip, E.; Cho, B.C.; Stetson, D.; Dougherty, B.; Lai, Z.; Markovets, A.; Vivancos, A.; Kuang, Y.; et al. Acquired EGFR C797S Mutation Mediates Resistance to AZD9291 in Non-Small Cell Lung Cancer Harboring EGFR T790M. Nat. Med. 2015, 21, 560–562. [Google Scholar] [CrossRef] [PubMed]
- Ercan, D.; Choi, H.G.; Yun, C.H.; Capelletti, M.; Xie, T.; Eck, M.J.; Gray, N.S.; Jänne, P.A. EGFR Mutations and Resistance to Irreversible Pyrimidine-Based EGFR Inhibitors. Clin. Cancer Res. 2015, 21, 3913–3923. [Google Scholar] [CrossRef] [PubMed]
- Chic, N.; Mayo-de-las-Casas, C.; Reguart, N. Successful Treatment with Gefitinib in Advanced Non–Small Cell Lung Cancer after Acquired Resistance to Osimertinib. J. Thorac. Oncol. 2017, 12, e78–e80. [Google Scholar] [CrossRef]
- Arulananda, S.; Do, H.; Musafer, A.; Mitchell, P.; Dobrovic, A.; John, T. Combination Osimertinib and Gefitinib in C797S and T790M EGFR-Mutated Non–Small Cell Lung Cancer. J. Thorac. Oncol. 2017, 12, 1728–1732. [Google Scholar] [CrossRef]
- Sullivan, I.; Planchard, D. Osimertinib in the Treatment of Patients with Epidermal Growth Factor Receptor T790M Mutation-Positive Metastatic Non-Small Cell Lung Cancer: Clinical Trial Evidence and Experience. Ther. Adv. Respir. Dis. 2016, 10, 549–565. [Google Scholar] [CrossRef] [PubMed]
- Scalvini, L.; Castelli, R.; La Monica, S.; Tiseo, M.; Alfieri, R. Fighting Tertiary Mutations in EGFR-Driven Lung-Cancers: Current Advances and Future Perspectives in Medicinal Chemistry. Biochem. Pharmacol. 2021, 190, 114643. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Xu, B.; Wang, J.; Gao, Y.; He, X.; Xie, T.; Ye, X.Y. Recent Advances of Novel Fourth Generation EGFR Inhibitors in Overcoming C797S Mutation of Lung Cancer Therapy. Eur. J. Med. Chem. 2023, 245, 114900. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Yao, M.Y.; Zhu, S.J.; Chen, J.Y.; Yun, C.H. Crystal Structure of EGFR T790M/C797S/V948R in Complex with EAI045. Biochem. Biophys. Res. Commun. 2018, 502, 332–337. [Google Scholar] [CrossRef]
- Jia, Y.; Yun, C.H.; Park, E.; Ercan, D.; Manuia, M.; Juarez, J.; Xu, C.; Rhee, K.; Chen, T.; Zhang, H.; et al. Overcoming EGFR(T790M) and EGFR(C797S) Resistance with Mutant-Selective Allosteric Inhibitors. Nature 2016, 534, 129–132. [Google Scholar] [CrossRef]
- Li, Q.; Zhang, T.; Li, S.; Tong, L.; Li, J.; Su, Z.; Feng, F.; Sun, D.; Tong, Y.; Wang, X.; et al. Discovery of Potent and Noncovalent Reversible EGFR Kinase Inhibitors of EGFRL858R/T790M/C797S. ACS Med. Chem. Lett. 2019, 10, 869–873. [Google Scholar] [CrossRef]
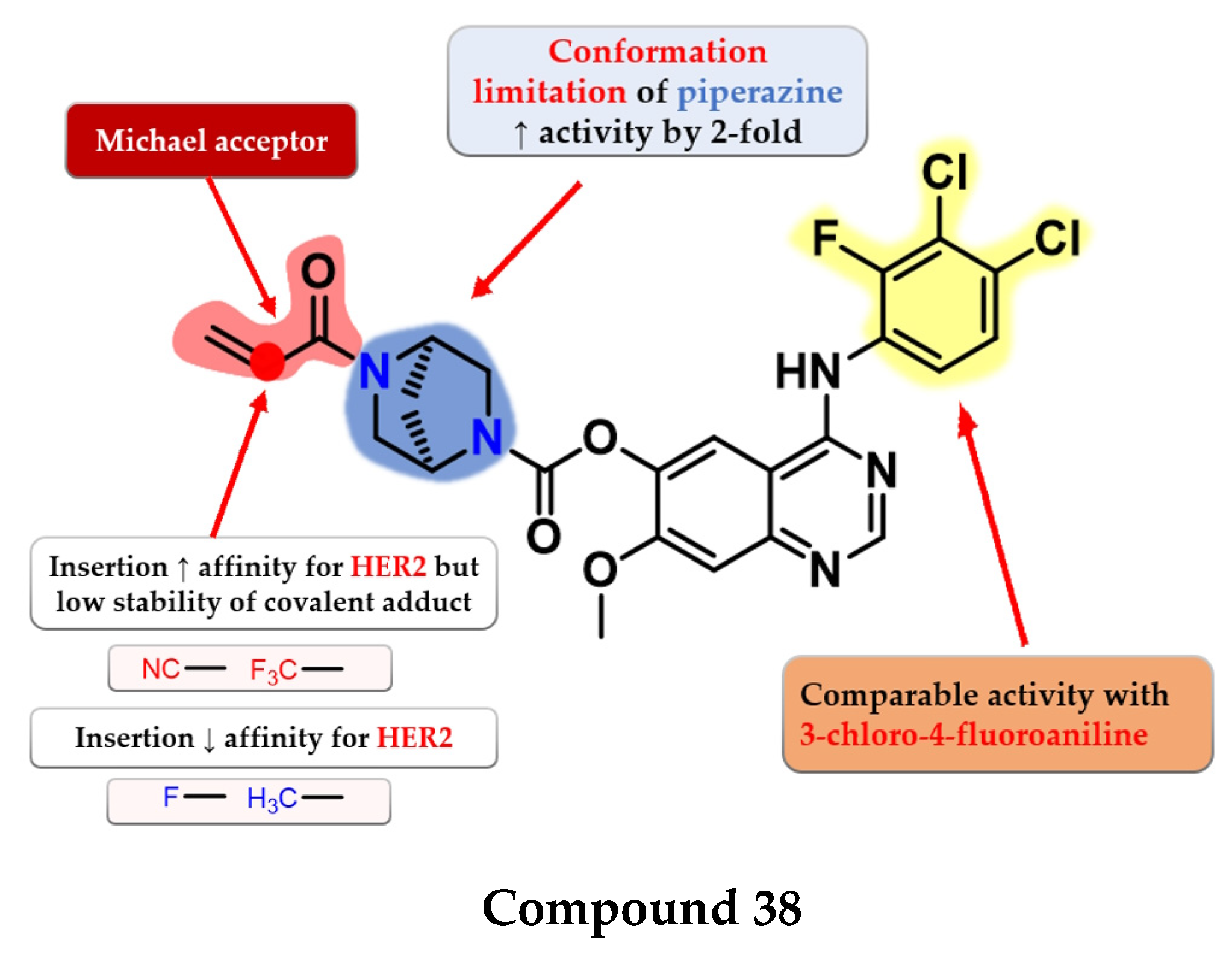
- Zhou, Z.; He, J.; Yang, F.; Pan, Q.; Yang, Z.; Zheng, P.; Xu, S.; Zhu, W. Design, Synthesis and Evaluation of Anti-Proliferative Activity of 2-Aryl-4-Aminoquinazoline Derivatives as EGFR Inhibitors. Bioorg. Chem. 2021, 112, 104848. [Google Scholar] [CrossRef]
- Park, H.; Jung, H.Y.; Mah, S.; Hong, S. Discovery of EGF Receptor Inhibitors That Are Selective for the D746-750/T790M/C797S Mutant through Structure-Based de Novo Design. Angew. Chem. Int. Ed. Engl. 2017, 56, 7634–7638. [Google Scholar] [CrossRef]
- Sogabe, S.; Kawakita, Y.; Igaki, S.; Iwata, H.; Miki, H.; Cary, D.R.; Takagi, T.; Takagi, S.; Ohta, Y.; Ishikawa, T. Structure-Based Approach for the Discovery of Pyrrolo[3,2-d]Pyrimidine-Based EGFR T790M/L858R Mutant Inhibitors. ACS Med. Chem. Lett. 2012, 4, 201–205. [Google Scholar] [CrossRef]
- Kong, L.L.; Ma, R.; Yao, M.Y.; Yan, X.E.; Zhu, S.J.; Zhao, P.; Yun, C.H. Structural Pharmacological Studies on EGFR T790M/C797S. Biochem. Biophys. Res. Commun. 2017, 488, 266–272. [Google Scholar] [CrossRef]
- Dou, D.; Wang, J.; Qiao, Y.; Wumaier, G.; Sha, W.; Li, W.; Mei, W.; Yang, T.; Zhang, C.; He, H.; et al. Discovery and Optimization of 4-Anilinoquinazoline Derivatives Spanning ATP Binding Site and Allosteric Site as Effective EGFR-C797S Inhibitors. Eur. J. Med. Chem. 2022, 244, 114856. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Șandor, A.; Ionuț, I.; Marc, G.; Oniga, I.; Eniu, D.; Oniga, O. Structure–Activity Relationship Studies Based on Quinazoline Derivatives as EGFR Kinase Inhibitors (2017–Present). Pharmaceuticals 2023, 16, 534. https://doi.org/10.3390/ph16040534
Șandor A, Ionuț I, Marc G, Oniga I, Eniu D, Oniga O. Structure–Activity Relationship Studies Based on Quinazoline Derivatives as EGFR Kinase Inhibitors (2017–Present). Pharmaceuticals. 2023; 16(4):534. https://doi.org/10.3390/ph16040534
Chicago/Turabian StyleȘandor, Alexandru, Ioana Ionuț, Gabriel Marc, Ilioara Oniga, Dan Eniu, and Ovidiu Oniga. 2023. "Structure–Activity Relationship Studies Based on Quinazoline Derivatives as EGFR Kinase Inhibitors (2017–Present)" Pharmaceuticals 16, no. 4: 534. https://doi.org/10.3390/ph16040534
APA StyleȘandor, A., Ionuț, I., Marc, G., Oniga, I., Eniu, D., & Oniga, O. (2023). Structure–Activity Relationship Studies Based on Quinazoline Derivatives as EGFR Kinase Inhibitors (2017–Present). Pharmaceuticals, 16(4), 534. https://doi.org/10.3390/ph16040534