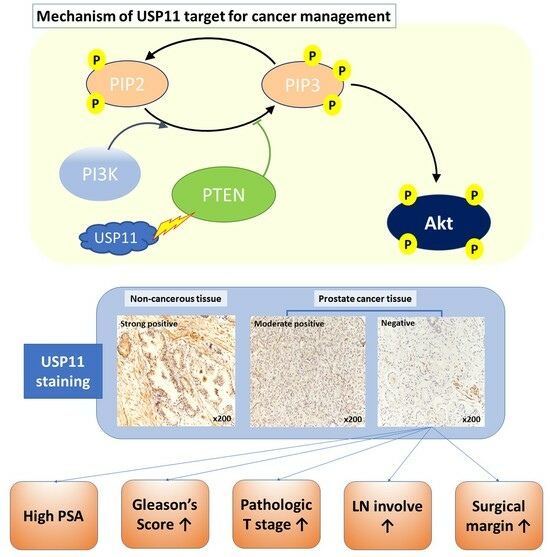
The Correlations between the Intensity of Histopathological Ubiquitin-Specific Protease 11 Staining and Progression of Prostate Cancer
Abstract
:1. Introduction
2. Results
2.1. Clinicopathologic Properties of Patients with Prostate Cancer
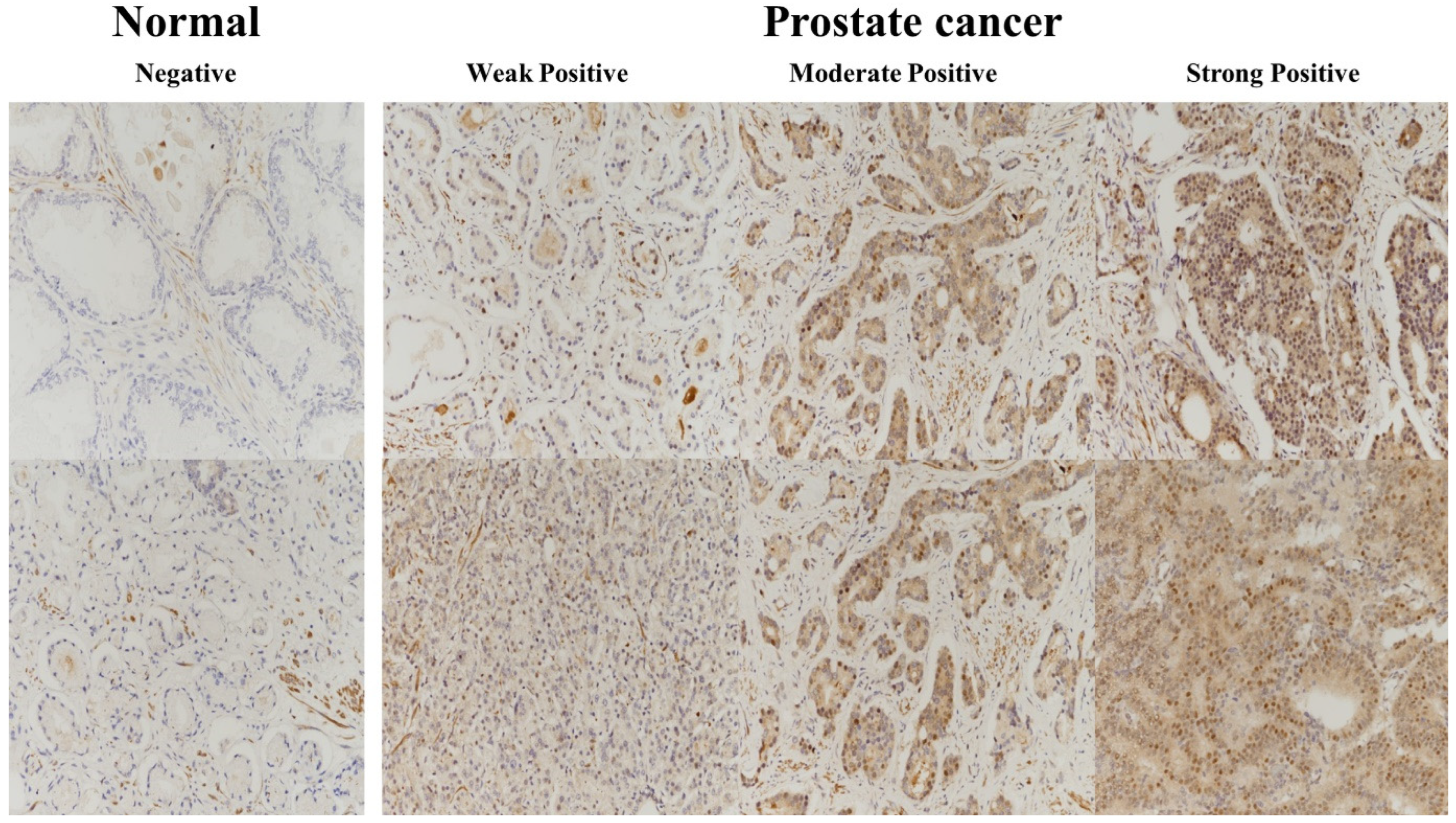
2.2. USP11 Expression
2.3. Association of USP11 Expression with Clinicopathologic Parameters through a Cox Proportional Hazard Model Test
2.3.1. Univariable Analysis
2.3.2. Multivariable Analysis
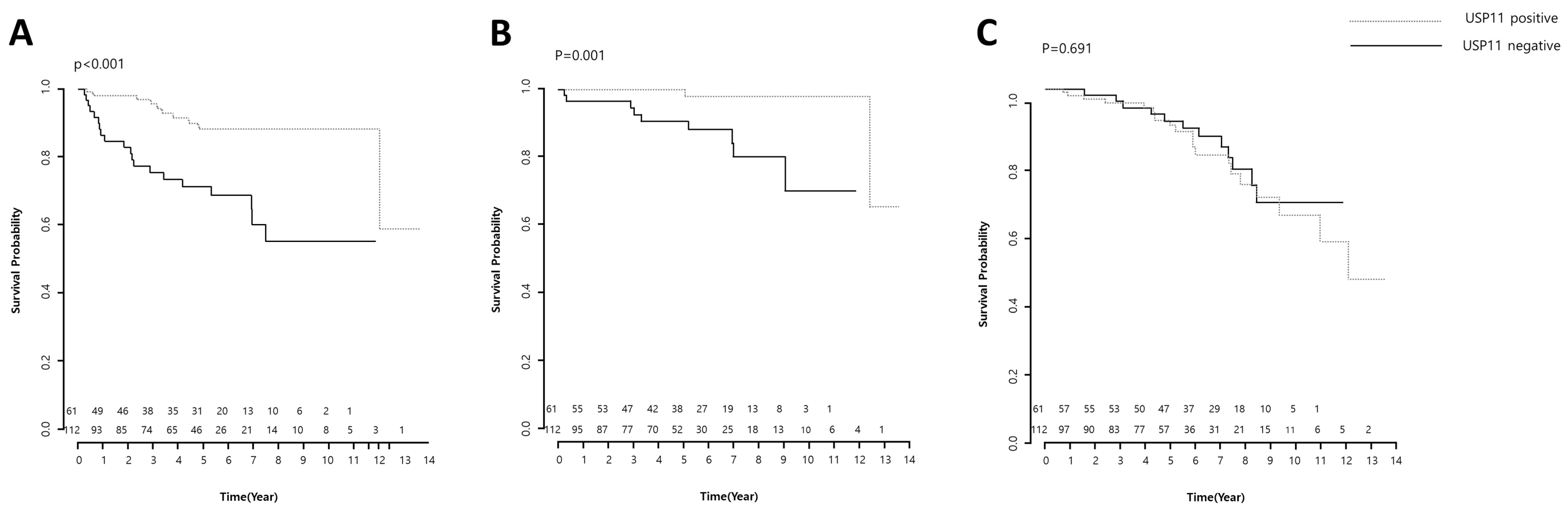
2.3.3. Survival Analysis
3. Discussion
4. Materials and Methods
4.1. Patients and Specimen Preparation
4.2. Construction of Tissue Microarrays
4.3. Purchase of Additional Tissues for Microarrays
4.4. Immunohistochemical Staining of Tissue Microarrays
4.5. Definition of Survival Analysis
4.6. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Maehama, T.; Dixon, J.E. The tumor suppressor, pten/mmac1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J. Biol. Chem. 1998, 273, 13375–13378. [Google Scholar] [CrossRef]
- Stambolic, V.; Suzuki, A.; de la Pompa, J.L.; Brothers, G.M.; Mirtsos, C.; Sasaki, T.; Ruland, J.; Penninger, J.M.; Siderovski, D.P.; Mak, T.W. Negative regulation of pkb/akt-dependent cell survival by the tumor suppressor pten. Cell 1998, 95, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Song, M.S.; Salmena, L.; Pandolfi, P.P. The functions and regulation of the pten tumour suppressor. Nat. Rev. Mol. Cell Biol. 2012, 13, 283–296. [Google Scholar] [CrossRef] [PubMed]
- Chung, J.S.; Morgan, T.M.; Hong, S.K. Clinical implications of genomic evaluations for prostate cancer risk stratification, screening, and treatment: A narrative review. Prostate Int. 2020, 8, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Ke, J.Y.; Dai, C.J.; Wu, W.L.; Gao, J.H.; Xia, A.J.; Liu, G.P.; Lv, K.S.; Wu, C.L. Usp11 regulates p53 stability by deubiquitinating p53. J. Zhejiang Univ. Sci. B 2014, 15, 1032–1038. [Google Scholar] [CrossRef]
- Sun, W.; Tan, X.; Shi, Y.; Xu, G.; Mao, R.; Gu, X.; Fan, Y.; Yu, Y.; Burlingame, S.; Zhang, H.; et al. Usp11 negatively regulates tnfalpha-induced nf-kappab activation by targeting on ikappabalpha. Cell Signal 2010, 22, 386–394. [Google Scholar] [CrossRef]
- Wu, H.C.; Lin, Y.C.; Liu, C.H.; Chung, H.C.; Wang, Y.T.; Lin, Y.W.; Ma, H.I.; Tu, P.H.; Lawler, S.E.; Chen, R.H. Usp11 regulates pml stability to control notch-induced malignancy in brain tumours. Nat. Commun. 2014, 5, 3214. [Google Scholar] [CrossRef]
- Hatano, K.; Nonomura, N. Genomic profiling of prostate cancer: An updated review. World J. Men’s Health 2022, 40, 368–379. [Google Scholar] [CrossRef]
- Wang, X.; Trotman, L.C.; Koppie, T.; Alimonti, A.; Chen, Z.; Gao, Z.; Wang, J.; Erdjument-Bromage, H.; Tempst, P.; Cordon-Cardo, C.; et al. Nedd4-1 is a proto-oncogenic ubiquitin ligase for pten. Cell 2007, 128, 129–139. [Google Scholar] [CrossRef]
- Maddika, S.; Kavela, S.; Rani, N.; Palicharla, V.R.; Pokorny, J.L.; Sarkaria, J.N.; Chen, J. Wwp2 is an e3 ubiquitin ligase for pten. Nat. Cell Biol. 2011, 13, 728–733. [Google Scholar] [CrossRef]
- Ahmed, S.F.; Deb, S.; Paul, I.; Chatterjee, A.; Mandal, T.; Chatterjee, U.; Ghosh, M.K. The chaperone-assisted e3 ligase c terminus of hsc70-interacting protein (chip) targets pten for proteasomal degradation. J. Biol. Chem. 2012, 287, 15996–16006. [Google Scholar] [CrossRef] [PubMed]
- Van Themsche, C.; Leblanc, V.; Parent, S.; Asselin, E. X-linked inhibitor of apoptosis protein (xiap) regulates pten ubiquitination, content, and compartmentalization. J. Biol. Chem. 2009, 284, 20462–20466. [Google Scholar] [CrossRef] [PubMed]
- Song, M.S.; Salmena, L.; Carracedo, A.; Egia, A.; Lo-Coco, F.; Teruya-Feldstein, J.; Pandolfi, P.P. The deubiquitinylation and localization of pten are regulated by a hausp-pml network. Nature 2008, 455, 813–817. [Google Scholar] [CrossRef] [PubMed]
- Sacco, J.J.; Yau, T.Y.; Darling, S.; Patel, V.; Liu, H.; Urbé, S.; Clague, M.J.; Coulson, J.M. The deubiquitylase ataxin-3 restricts pten transcription in lung cancer cells. Oncogene 2014, 33, 4265–4272. [Google Scholar] [CrossRef]
- Yuan, L.; Lv, Y.; Li, H.; Gao, H.; Song, S.; Zhang, Y.; Xing, G.; Kong, X.; Wang, L.; Li, Y.; et al. Deubiquitylase otud3 regulates pten stability and suppresses tumorigenesis. Nat. Cell Biol. 2015, 17, 1169–1181. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, P.; Wei, Y.; Piao, H.L.; Wang, W.; Maddika, S.; Wang, M.; Chen, D.; Sun, Y.; Hung, M.C.; et al. Deubiquitylation and stabilization of pten by usp13. Nat. Cell Biol. 2013, 15, 1486–1494. [Google Scholar] [CrossRef]
- Swanson, D.A.; Freund, C.L.; Ploder, L.; McInnes, R.R.; Valle, D. A ubiquitin c-terminal hydrolase gene on the proximal short arm of the x chromosome: Implications for x-linked retinal disorders. Hum. Mol. Genet. 1996, 5, 533–538. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cbioportal. Sci. Signal 2013, 6, pl1. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cbio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Győrffy, B. Discovery and ranking of the most robust prognostic biomarkers in serous ovarian cancer. Geroscience 2023, 45, 1889–1898. [Google Scholar] [CrossRef]
- Yang-Feng, T.L.; Li, S.; Han, H.; Schwartz, P.E. Frequent loss of heterozygosity on chromosomes xp and 13q in human ovarian cancer. Int. J. Cancer 1992, 52, 575–580. [Google Scholar] [CrossRef]
- Spatz, A.; Borg, C.; Feunteun, J. X-chromosome genetics and human cancer. Nat. Rev. Cancer 2004, 4, 617–629. [Google Scholar] [CrossRef]
- Stephenson, A.J.; Smith, A.; Kattan, M.W.; Satagopan, J.; Reuter, V.E.; Scardino, P.T.; Gerald, W.L. Integration of gene expression profiling and clinical variables to predict prostate carcinoma recurrence after radical prostatectomy. Cancer 2005, 104, 290–298. [Google Scholar] [CrossRef] [PubMed]
- Taylor, B.S.; Schultz, N.; Hieronymus, H.; Gopalan, A.; Xiao, Y.; Carver, B.S.; Arora, V.K.; Kaushik, P.; Cerami, E.; Reva, B.; et al. Integrative genomic profiling of human prostate cancer. Cancer Cell 2010, 18, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.Z.; Graham, K.; Glubrecht, D.D.; Germain, D.R.; Mackey, J.R.; Godbout, R. Association of fabp5 expression with poor survival in triple-negative breast cancer: Implication for retinoic acid therapy. Am. J. Pathol. 2011, 178, 997–1008. [Google Scholar] [CrossRef] [PubMed]
- Györffy, B.; Schäfer, R. Meta-analysis of gene expression profiles related to relapse-free survival in 1079 breast cancer patients. Breast Cancer Res. Treat. 2009, 118, 433–441. [Google Scholar] [CrossRef]
- Park, M.K.; Yao, Y.; Xia, W.; Setijono, S.R.; Kim, J.H.; Vila, I.K.; Chiu, H.H.; Wu, Y.; Billalabeitia, E.G.; Lee, M.G.; et al. Pten self-regulates through usp11 via the pi3k-foxo pathway to stabilize tumor suppression. Nat. Commun. 2019, 10, 636. [Google Scholar] [CrossRef]

| Prognostic Factors | n (%) |
|---|---|
| Age | 66 (61, 70) |
| PSA | |
| ≤10 | 141 (53.82%) |
| 10–20 | 69 (26.34%) |
| >20 | 52 (19.85%) |
| Gleason score | |
| ≤6 | 41 (15.65%) |
| 7–8 | 85 (32.44%) |
| 8–10 | 52 (19.85%) |
| Pathological stage | |
| ≤T2 | 112 (42.75%) |
| ≥T3 | 150 (57.25%) |
| Seminal vesicle invasion | |
| Negative | 146 (84.88%) |
| Positive | 26 (15.12%) |
| Lymph node involvement | |
| Negative | 165 (96.49%) |
| Positive | 6 (3.51%) |
| Surgical margin * | |
| Negative | 65 (55.23%) |
| Positive | 77 (44.77%) |
| Expression of USP11 # | 115 (44.75%) |
| 142 (55.25%) |
| UPS11 | Adjacent Non-Neoplastic Tissues | Prostate Cancer | Total | p-Value |
|---|---|---|---|---|
| 0 | 5 | 135 | 140 | <0.05 |
| 1 | 15 | 131 | 146 |
| USP11 | p-Value | ||
|---|---|---|---|
| 0 (n = 115) | 1 (n = 142) | ||
| Age | 66 (62, 70) | 66 (61, 70) | 0.614 |
| PSA | <0.001 | ||
| ≤10 | 44 | 92 | |
| 10–20 | 37 | 29 | |
| >20 | 31 | 19 | |
| Gleason score | <0.001 | ||
| ≤6 | 0 | 41 | |
| 7–8 | 4 | 80 | |
| 8–10 | 108 | 19 | |
| Pathological stage | <0.001 | ||
| ≤T2 | 28 | 84 | |
| ≥T3 | 84 | 56 | |
| Seminal vesicle invasion | <0.001 | ||
| Negative | 43 | 103 | |
| Positive | 17 | 9 | |
| Lymph node involvement | 0.1834 | ||
| Negative | 55 | 110 | |
| Positive | 4 | 2 | |
| Surgical margin * | <0.05 | ||
| Negative | 26 | 69 | |
| Positive | 34 | 43 | |
| USP11 | n | Event | Mean Survival Time (Estimated ± SE) | Log-Rank Test |
|---|---|---|---|---|
| Biochemical recurrence-free survival | ||||
| 0 | 61 | 20 | 8.118 ± 0.666 | 0.000 |
| 1 | 112 | 10 | 11.991 ± 0.558 | |
| Clinical recurrence-free survival | ||||
| 0 | 61 | 9 | 10.111 ± 0.527 | 0.001 |
| 1 | 112 | 2 | 13.023 ± 0.350 | |
| Overall survival | ||||
| 0 | 61 | 12 | 10.014 ± 0.460 | 0.691 |
| 1 | 112 | 20 | 10.487 ± 0.552 | |
| Univariable Analysis | Multivariable Analysis | |||
|---|---|---|---|---|
| HR (95% CI) | p-Value | HR (95% CI) | p-Value | |
| Age | 1.023 (0.962–1.087) | 0.477 | ||
| USP11 grade | ||||
| 0 | Reference | |||
| 1 | 0.253 (0.115–0.556) | 0.001 | 0.413 (0.180–0.951) | 0.0338 |
| PSA | ||||
| ≤10 | Reference | |||
| 10–20 | 1.648 (0.673–4.036) | 0.274 | ||
| >20 | 2.364 (1.009–5.539 | 0.048 | ||
| Gleason score | ||||
| ≤6 | Reference | |||
| 7–8 | 1.545 (0.311–7.669) | 0.595 | ||
| 8–10 | 4.939 (1.158–21.070) | 0.031 | ||
| Pathological stage | ||||
| ≤T2 | Reference | |||
| ≥T3 | 3.516 (1.672–7.392) | 0.001 | ||
| Seminal vesicle invasion | ||||
| Negative | Reference | |||
| Positive | 4.444 (2.104–9.389) | <0.001 | 2.186 (0.963–4.965) | 0.062 |
| Lymph node involvement | ||||
| Negative | Reference | |||
| Positive | 5.863 (2.225–15.446) | <0.001 | 3.342 (1.209–9.236) | 0.020 |
| Surgical margin | ||||
| Negative | Reference | |||
| Positive | 5.861 (2.392–14.361) | <0.001 | 3.769 (1.454–9.772) | 0.006 |
| Univariable Analysis | Multivariable Analysis | |||
|---|---|---|---|---|
| HR (95% CI) | p-Value | HR (95% CI) | p-Value | |
| Age | 1.095 (0.983–1.218) | 0.099 | ||
| USP11 grade | ||||
| 0 | Reference | |||
| 1 | 0.075 (0.010–0.593) | 0.014 | 0.241 (0.026–2.272) | 0.214 |
| PSA | ||||
| ≤10 | Reference | |||
| 10–20 | 1.632 (0.435–6.118) | 0.468 | 1.325 (0.200–8.780) | 0.770 |
| >20 | 0.968 (0.186–5.039) | 0.969 | 0.198 (0.023–1.716) | 0.142 |
| Pathological stage | ||||
| ≤T2 | Reference | |||
| ≥T3 | 3.008 (0.877–10.311) | 0.080 | ||
| Seminal vesicle invasion | ||||
| Negative | Reference | |||
| Positive | 3.910 (1.083–14.121) | 0.037 | 5.297 (0.835–33.606) | 0.077 |
| Lymph node involvement | ||||
| Negative | Reference | |||
| Positive | 13.802 (3.870–49.211) | <0.001 | 36.850 (3.131–433.667) | 0.004 |
| Surgical margin | ||||
| Negative | Reference | |||
| Positive | 3.341 (0.082–12.654) | 0.076 | ||
| Univariable Analysis | Multivariable Analysis | |||
|---|---|---|---|---|
| HR (95% CI) | p-Value | HR (95% CI) | p-Value | |
| Age | 1.028 (0.968–1.091) | 0.371 | ||
| USP11 grade | ||||
| 0 | Reference | |||
| 1 | 1.159 (0.559–2.400) | 0.692 | 1.377 (0.651–2.991) | 0.403 |
| PSA | ||||
| ≤10 | Reference | |||
| 10–20 | 1.186 (0.509–2.765) | 0.692 | ||
| >20 | 1.408 (0.605–3.273) | 0.427 | ||
| Gleason score | ||||
| ≤6 | Reference | |||
| 7–8 | 0.945 (0.354–2.523) | 0.909 | ||
| 8–10 | 0.834 (0.318–2.186) | 0.712 | ||
| Pathological stage | ||||
| ≤T2 | Reference | |||
| ≥T3 | 1.911 (0.946–3.859) | 0.071 | ||
| Seminal vesicle invasion | ||||
| Negative | Reference | |||
| Positive | 2.441 (1.137–5.242) | 0.022 | 2.631 (1.203–5.755) | 0.015 |
| Lymph node involvement | ||||
| Negative | Reference | |||
| Positive | 1.751 (0.530–5.791) | 0.358 | ||
| Surgical margin | ||||
| Negative | Reference | |||
| Positive | 1.349 (0.664–2.740) | 0.408 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.H.; Yang, H.J.; Lee, K.W.; Park, J.J.; Lee, C.-H.; Jeon, Y.S.; Kim, J.H.; Park, S.; Song, S.J.; Lee, J.-H.; et al. The Correlations between the Intensity of Histopathological Ubiquitin-Specific Protease 11 Staining and Progression of Prostate Cancer. Pharmaceuticals 2023, 16, 1703. https://doi.org/10.3390/ph16121703
Kim JH, Yang HJ, Lee KW, Park JJ, Lee C-H, Jeon YS, Kim JH, Park S, Song SJ, Lee J-H, et al. The Correlations between the Intensity of Histopathological Ubiquitin-Specific Protease 11 Staining and Progression of Prostate Cancer. Pharmaceuticals. 2023; 16(12):1703. https://doi.org/10.3390/ph16121703
Chicago/Turabian StyleKim, Jae Heon, Hee Jo Yang, Kwang Woo Lee, Jae Joon Park, Chang-Ho Lee, Youn Soo Jeon, Jae Ho Kim, Suyeon Park, Su Jung Song, Ji-Hye Lee, and et al. 2023. "The Correlations between the Intensity of Histopathological Ubiquitin-Specific Protease 11 Staining and Progression of Prostate Cancer" Pharmaceuticals 16, no. 12: 1703. https://doi.org/10.3390/ph16121703
APA StyleKim, J. H., Yang, H. J., Lee, K. W., Park, J. J., Lee, C.-H., Jeon, Y. S., Kim, J. H., Park, S., Song, S. J., Lee, J.-H., Moon, A., Kim, Y. H., & Song, Y. S. (2023). The Correlations between the Intensity of Histopathological Ubiquitin-Specific Protease 11 Staining and Progression of Prostate Cancer. Pharmaceuticals, 16(12), 1703. https://doi.org/10.3390/ph16121703