Discovery of a Multifunctional Octapeptide from Lingzhi with Antioxidant and Tyrosinase Inhibitory Activity
Abstract
:1. Introduction
2. Results
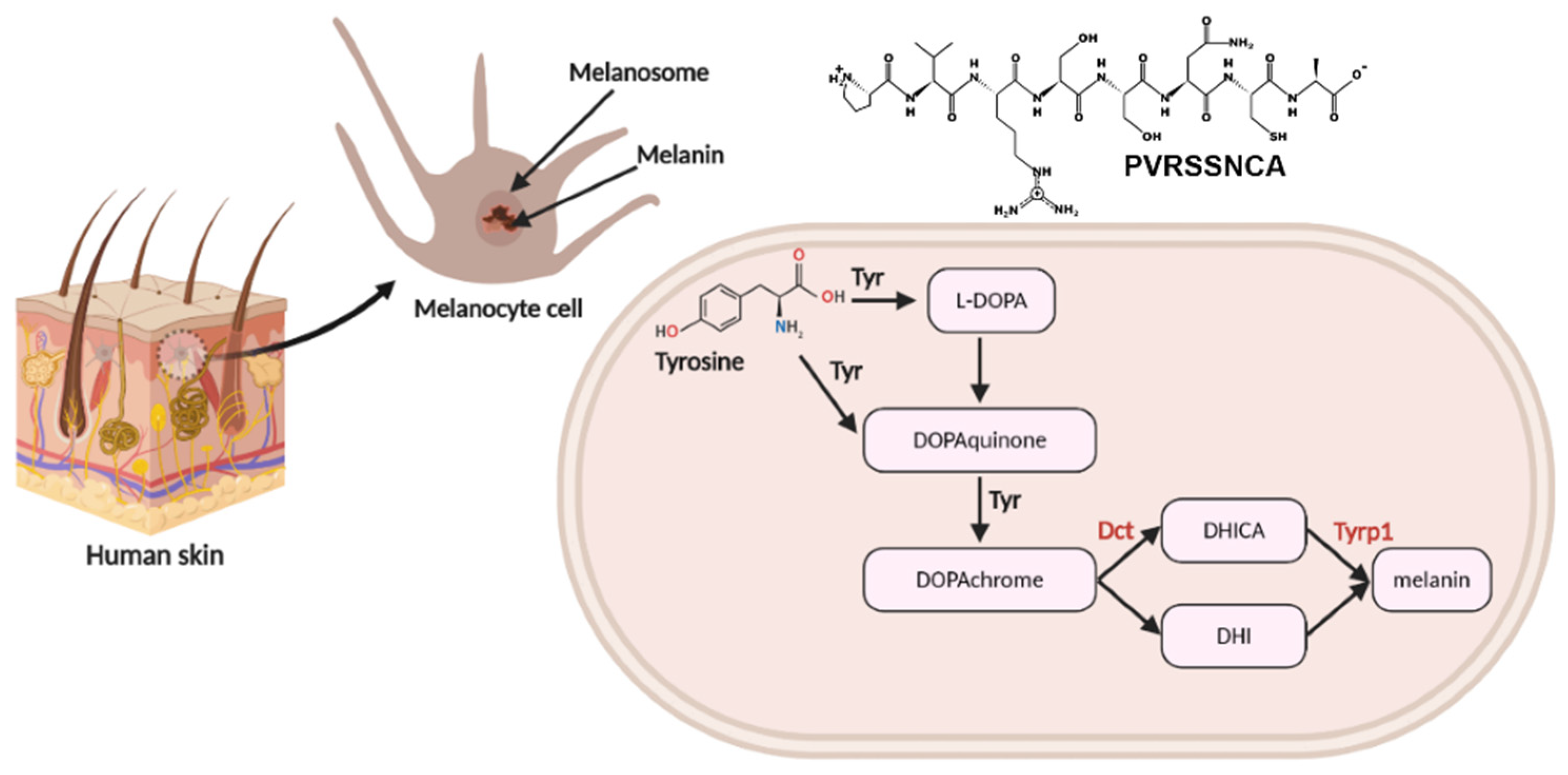
2.1. Isolation and Identification of Multifunctional Octapeptide Derived from Lingzhi Protein Hydrolysate
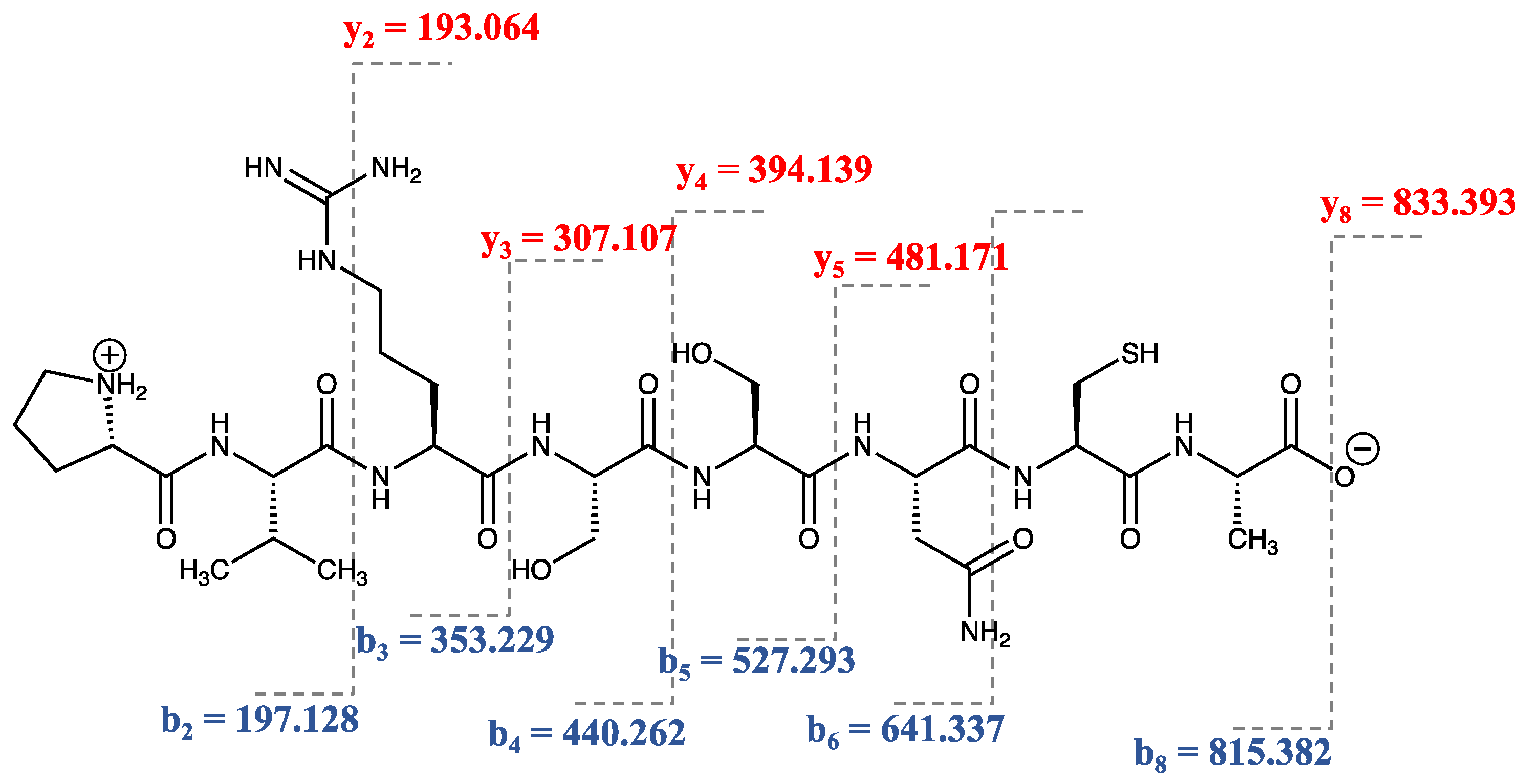
2.2. Peptide Synthesis by SPPS Approach
2.3. In Vitro Radical Scavenging Assay of the Octapeptide by DPPH, ABTS, and FRAP Assays
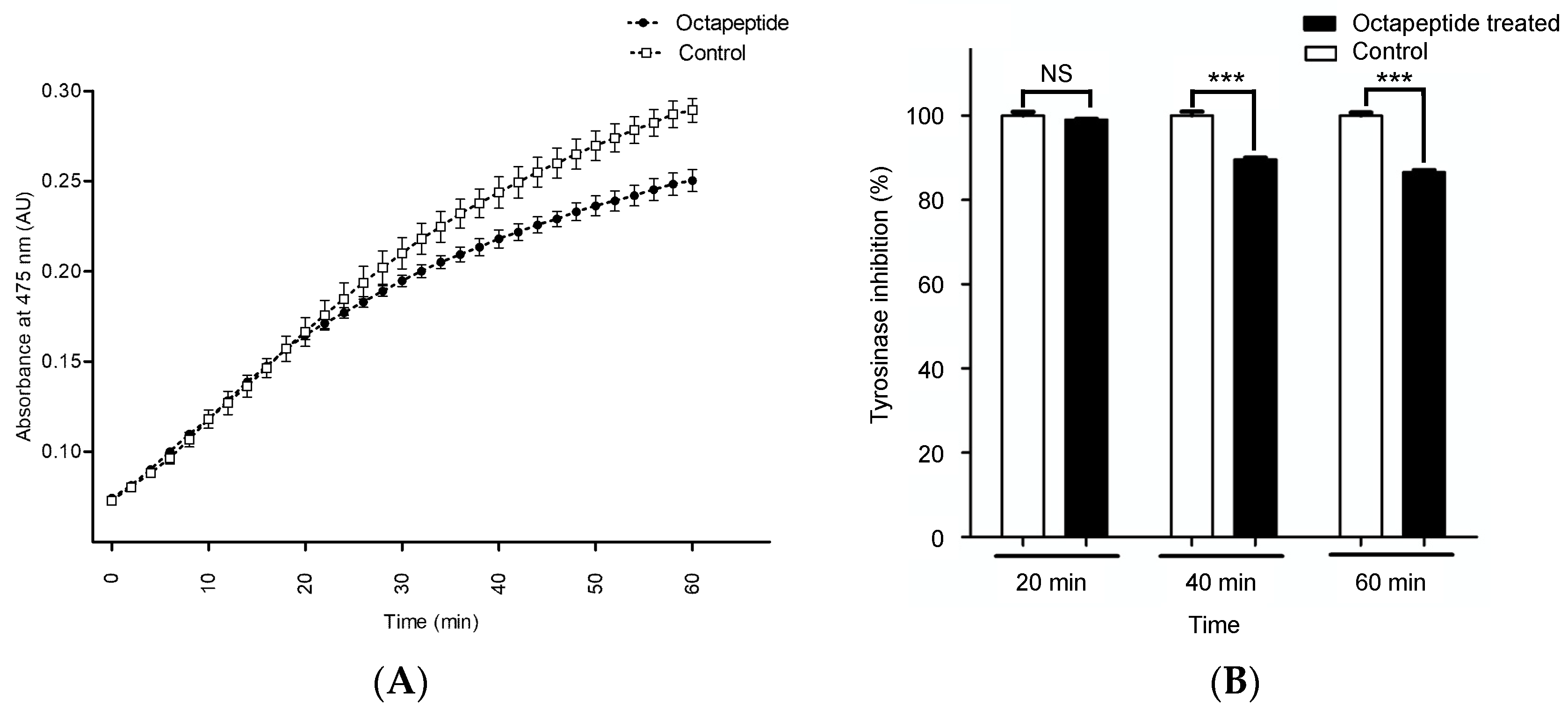
2.4. In Vitro Tyrosinase Inhibition Assay
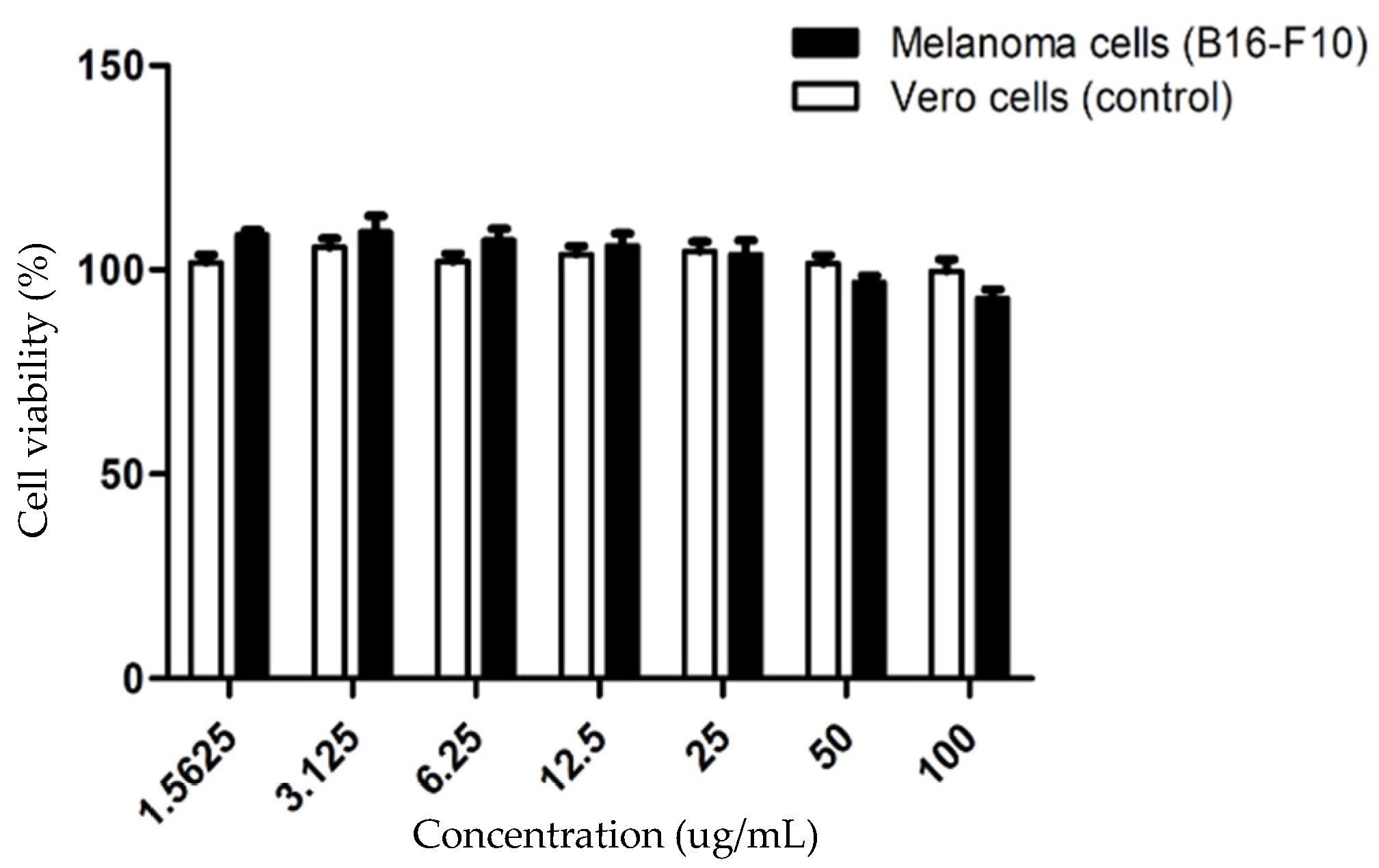
2.5. Cytotoxicity Assessment and Treatment Condition
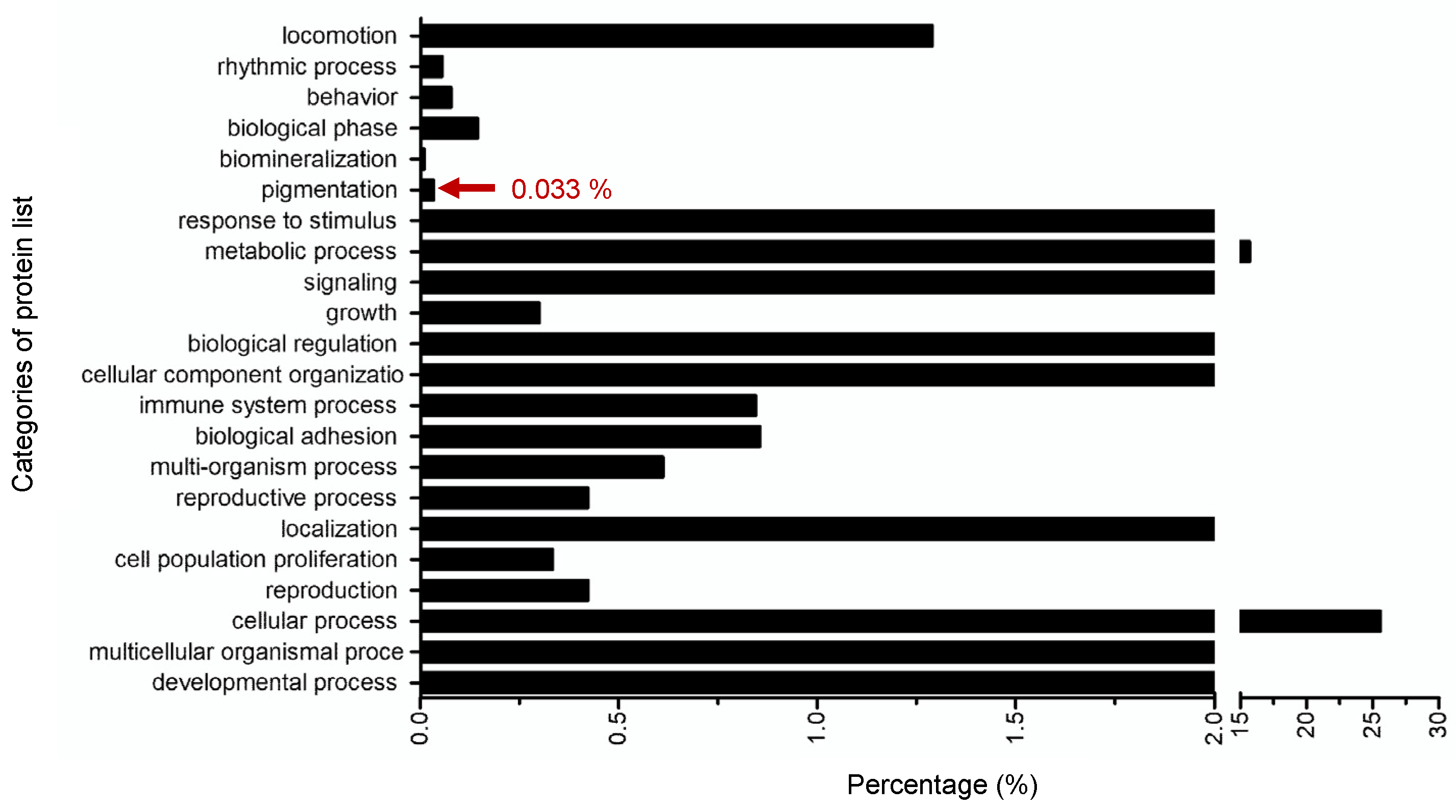
2.6. Targeted-Proteomics Analysis of the Octapeptide in Melanoma Cells
3. Discussion
4. Materials and Methods
4.1. Purification and Identification of Multifunctional Octapeptide Derived from Lingzhi Protein Hydrolysate
4.2. Peptide Synthesis by Solid Phase Peptide Synthesis (SPPS) Approach
4.3. In Vitro Radical Scavenging Assay of the Octapeptide by DPPH, ABTS and Ferric Reducing Antioxidant Power (FRAP) Assay and Tyrosinase Activity Inhibition Assay
4.4. Tyrosinase Inhibition Assay
4.5. Cell Culture and Treatment Condition
4.6. Determination of the Peptide Concentration in Culture Medium Using LC-MS/MS
4.7. Targeted-Proteome Quantitation and Data Analysis
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Huang, S.Y.; Huang, G.J.; Wu, H.C.; Kao, M.C.; Huang, W.C. Ganoderma tsugae Inhibits the SREBP-1/AR Axis Leading to Suppression of Cell Growth and Activation of Apoptosis in Prostate Cancer Cells. Molecules 2018, 23, 2539. [Google Scholar] [CrossRef] [Green Version]
- Winska, K.; Maczka, W.; Gabryelska, K.; Grabarczyk, M. Mushrooms of the Genus Ganoderma Used to Treat Diabetes and Insulin Resistance. Molecules 2019, 24, 4075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krobthong, S.; Choowongkomon, K.; Suphakun, P.; Kuaprasert, B.; Samutrtai, P.; Yingchutrakul, Y. The anti-oxidative effect of Lingzhi protein hydrolysates on lipopolysaccharide-stimulated A549 cells. Food Biosci. 2021, 41, 101093. [Google Scholar] [CrossRef]
- Habtemariam, S. Modulation of Reactive Oxygen Species in Health and Disease. Antioxidants 2019, 8, 513. [Google Scholar] [CrossRef] [Green Version]
- Yu, N.; Huang, Y.; Jiang, Y.; Zou, L.; Liu, X.; Liu, S.; Chen, F.; Luo, J.; Zhu, Y. Ganoderma lucidum Triterpenoids (GLTs) Reduce Neuronal Apoptosis via Inhibition of ROCK Signal Pathway in APP/PS1 Transgenic Alzheimer’s Disease Mice. Oxid. Med. Cell Longev. 2020, 2020, 9894037. [Google Scholar] [CrossRef]
- de Oliveira Pateis, V.; Bracht, L.; Dos Santos Castro, L.; Bueno Franco Salla, G.; Comar, J.F.; Valderrama Parizotto, A.; Peralta, R.M.; Bracht, A. The food additive BHA modifies energy metabolism in the perfused rat liver. Toxicol. Lett. 2018, 299, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Zaidi, K.U.; Ali, S.A.; Ali, A.S. Effect of Purified Mushroom Tyrosinase on Melanin Content and Melanogenic Protein Expression. Biotechnol. Res. Int. 2016, 2016, 9706214. [Google Scholar] [CrossRef] [Green Version]
- Asadzadeh, A.; Sirous, H.; Pourfarzam, M.; Yaghmaei, P.; Afshin, F. In vitro and in silico studies of the inhibitory effects of some novel kojic acid derivatives on tyrosinase enzyme. Iran J. Basic Med. Sci. 2016, 19, 132–144. [Google Scholar]
- Garcia-Jimenez, A.; Teruel-Puche, J.A.; Berna, J.; Rodriguez-Lopez, J.N.; Tudela, J.; Garcia-Canovas, F. Action of tyrosinase on alpha and beta-arbutin: A kinetic study. PLoS ONE 2017, 12, e0177330. [Google Scholar] [CrossRef] [Green Version]
- Oiso, N.; Tatebayashi, M.; Hoshiyama, Y.; Kawada, A. Allergic contact dermatitis caused by arbutin and dipotassium glycyrrhizate in skin-lightening products. Contact. Dermat. 2017, 77, 51–53. [Google Scholar] [CrossRef]
- Krobthong, S.; Yingchutrakul, Y.; Samutrtai, P.; Choowongkomon, K. The C-terminally shortened analogs of a hexapeptide derived from Lingzhi hydrolysate with enhanced tyrosinase-inhibitory activity. Arch. Pharm. (Weinheim) 2021, 354, e2100204. [Google Scholar] [CrossRef]
- Babini, E.; Tagliazucchi, D.; Martini, S.; Più, L.D.; Gianotti, A. LC-ESI-QTOF-MS identification of novel antioxidant peptides obtained by enzymatic and microbial hydrolysis of vegetable proteins. Food Chem. 2017, 228, 186–196. [Google Scholar] [CrossRef]
- Chakrabarti, S.; Guha, S.; Majumder, K. Food-Derived Bioactive Peptides in Human Health: Challenges and Opportunities. Nutrients 2018, 10, 1738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, G.-l.; He, W.; Wu, Y.-g.; Chen, J.; Hu, X.-w.; Yu, J. Purification of Angiotensin-I-Converting Enzyme Inhibitory Peptides Derived from Camellia oleifera Abel Seed Meal Hydrolysate. J. Food Qual. 2019, 2019, 7364213. [Google Scholar] [CrossRef] [Green Version]
- Krobthong, S.; Yingchutrakul, Y. Identification and enhancement of antioxidant P1-peptide isolated from Ganoderma lucidum hydrolysate. Food Biotechnol. 2020, 34, 338–351. [Google Scholar] [CrossRef]
- Chen, W.C.; Tseng, T.S.; Hsiao, N.W.; Lin, Y.L.; Wen, Z.H.; Tsai, C.C.; Lee, Y.C.; Lin, H.H.; Tsai, K.C. Discovery of highly potent tyrosinase inhibitor, T1, with significant anti-melanogenesis ability by zebrafish in vivo assay and computational molecular modeling. Sci. Rep. 2015, 5, 7995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, P.Y.; You, Y.J.; Liu, Y.J.; Hou, C.W.; Wu, C.S.; Wen, K.C.; Lin, C.Y.; Chiang, H.M. Sesamol Inhibited Melanogenesis by Regulating Melanin-Related Signal Transduction in B16F10 Cells. Int. J. Mol. Sci. 2018, 19, 1108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, H.C.; Chiu, S.H.; Chang, T.M. Inhibitory effect of [6]-gingerol on melanogenesis in B16F10 melanoma cells and a possible mechanism of action. Biosci. Biotechnol. Biochem. 2011, 75, 1067–1072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ujan, R.; Saeed, A.; Ashraf, S.; Channar, P.A.; Abbas, Q.; Rind, M.A.; Hassan, M.; Raza, H.; Seo, S.Y.; El-Seedi, H.R. Synthesis, computational studies and enzyme inhibitory kinetics of benzothiazole-linked thioureas as mushroom tyrosinase inhibitors. J. Biomol. Struct. Dyn. 2021, 39, 7035–7043. [Google Scholar] [CrossRef]
- Lai, X.; Wichers, H.J.; Soler-Lopez, M.; Dijkstra, B.W. Structure of Human Tyrosinase Related Protein 1 Reveals a Binuclear Zinc Active Site Important for Melanogenesis. Angew. Chem. Int. Ed. Engl. 2017, 56, 9812–9815. [Google Scholar] [CrossRef]
- Pigat, S.; Connolly, A.; Cushen, M.; Cullen, M.; O’Mahony, C. A probabilistic intake model to estimate the impact of reformulation by the food industry among Irish consumers. Int. J. Food Sci. Nutr. 2018, 69, 938–945. [Google Scholar] [CrossRef]
- Krobthong, S.; Yingchutrakul, Y.; Visessanguan, W.; Mahatnirunkul, T.; Samutrtai, P.; Chaichana, C.; Papan, P.; Choowongkomon, K. Study of the Lipolysis Effect of Nanoliposome-Encapsulated Ganoderma lucidum Protein Hydrolysates on Adipocyte Cells Using Proteomics Approach. Foods 2021, 10, 2157. [Google Scholar] [CrossRef] [PubMed]
- Oh, Y.; Ahn, C.B.; Nam, K.H.; Kim, Y.K.; Yoon, N.Y.; Je, J.Y. Amino Acid Composition, Antioxidant, and Cytoprotective Effect of Blue Mussel (Mytilus edulis) Hydrolysate through the Inhibition of Caspase-3 Activation in Oxidative Stress-Mediated Endothelial Cell Injury. Mar. Drugs 2019, 17, 135. [Google Scholar] [CrossRef] [Green Version]
- Ranathunga, S.; Rajapakse, N.; Kim, S.-K. Purification and characterization of antioxidative peptide derived from muscle of conger eel (Conger myriaster). Eur. Food Res. Technol. 2005, 222, 310–315. [Google Scholar] [CrossRef]
- Dolinska, M.B.; Young, K.L., 2nd; Kassouf, C.; Dimitriadis, E.K.; Wingfield, P.T.; Sergeev, Y.V. Protein Stability and Functional Characterization of Intra-Melanosomal Domain of Human Recombinant Tyrosinase-Related Protein 1. Int. J. Mol. Sci. 2020, 21, 331. [Google Scholar] [CrossRef] [Green Version]
- Kuhn, M.; von Mering, C.; Campillos, M.; Jensen, L.J.; Bork, P. STITCH: Interaction networks of chemicals and proteins. Nucleic Acids Res. 2008, 36, D684–D688. [Google Scholar] [CrossRef] [PubMed]
- Saternus, R.; Pilz, S.; Graber, S.; Kleber, M.; Marz, W.; Vogt, T.; Reichrath, J. A closer look at evolution: Variants (SNPs) of genes involved in skin pigmentation, including EXOC2, TYR, TYRP1, and DCT, are associated with 25(OH)D serum concentration. Endocrinology 2015, 156, 39–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirosaki, K.; Yamashita, T.; Wada, I.; Jin, H.Y.; Jimbow, K. Tyrosinase and tyrosinase-related protein 1 require Rab7 for their intracellular transport. J. Investig. Dermatol. 2002, 119, 475–480. [Google Scholar] [CrossRef]
- Guyonneau, L.; Murisier, F.; Rossier, A.; Moulin, A.; Beermann, F. Melanocytes and pigmentation are affected in dopachrome tautomerase knockout mice. Mol. Cell Biol. 2004, 24, 3396–3403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coin, I.; Beyermann, M.; Bienert, M. Solid-phase peptide synthesis: From standard procedures to the synthesis of difficult sequences. Nat. Protoc. 2007, 2, 3247–3256. [Google Scholar] [CrossRef] [PubMed]
- Brenton, A.G.; Godfrey, A.R. Accurate mass measurement: Terminology and treatment of data. J. Am. Soc. Mass. Spectrom. 2010, 21, 1821–1835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benzie, I.F.; Strain, J.J. The ferric reducing ability of plasma (FRAP) as a measure of “antioxidant power”: The FRAP assay. Anal. Biochem. 1996, 239, 70–76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riss, T.L.; Moravec, R.A.; Niles, A.L.; Duellman, S.; Benink, H.A.; Worzella, T.J.; Minor, L. Cell Viability Assays. In Assay Guidance Manual; Sittampalam, G.S., Grossman, A., Brimacombe, K., Arkin, M., Auld, D., Austin, C.P., Baell, J., Bejcek, B., Caaveiro, J.M.M., Chung, T.D.Y., et al., Eds.; Eli Lilly & Company and the National Center for Advancing Translational Sciences: Bethesda, MD, USA, 2004; pp. 355–386. [Google Scholar]
- Zhang, Y.; Liu, L.; Ren, L. Liquid chromatography-tandem mass spectrometry (LC-MS/MS) determination of cantharidin in biological specimens and application to postmortem interval estimation in cantharidin poisoning. Sci. Rep. 2020, 10, 10438. [Google Scholar] [CrossRef] [PubMed]
- Johansson, C.; Samskog, J.; Sundstrom, L.; Wadensten, H.; Bjorkesten, L.; Flensburg, J. Differential expression analysis of Escherichia coli proteins using a novel software for relative quantitation of LC-MS/MS data. Proteomics 2006, 6, 4475–4485. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Huang, X.; Muruganujan, A.; Tang, H.; Mills, C.; Kang, D.; Thomas, P.D. PANTHER version 11: Expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017, 45, D183–D189. [Google Scholar] [CrossRef] [Green Version]
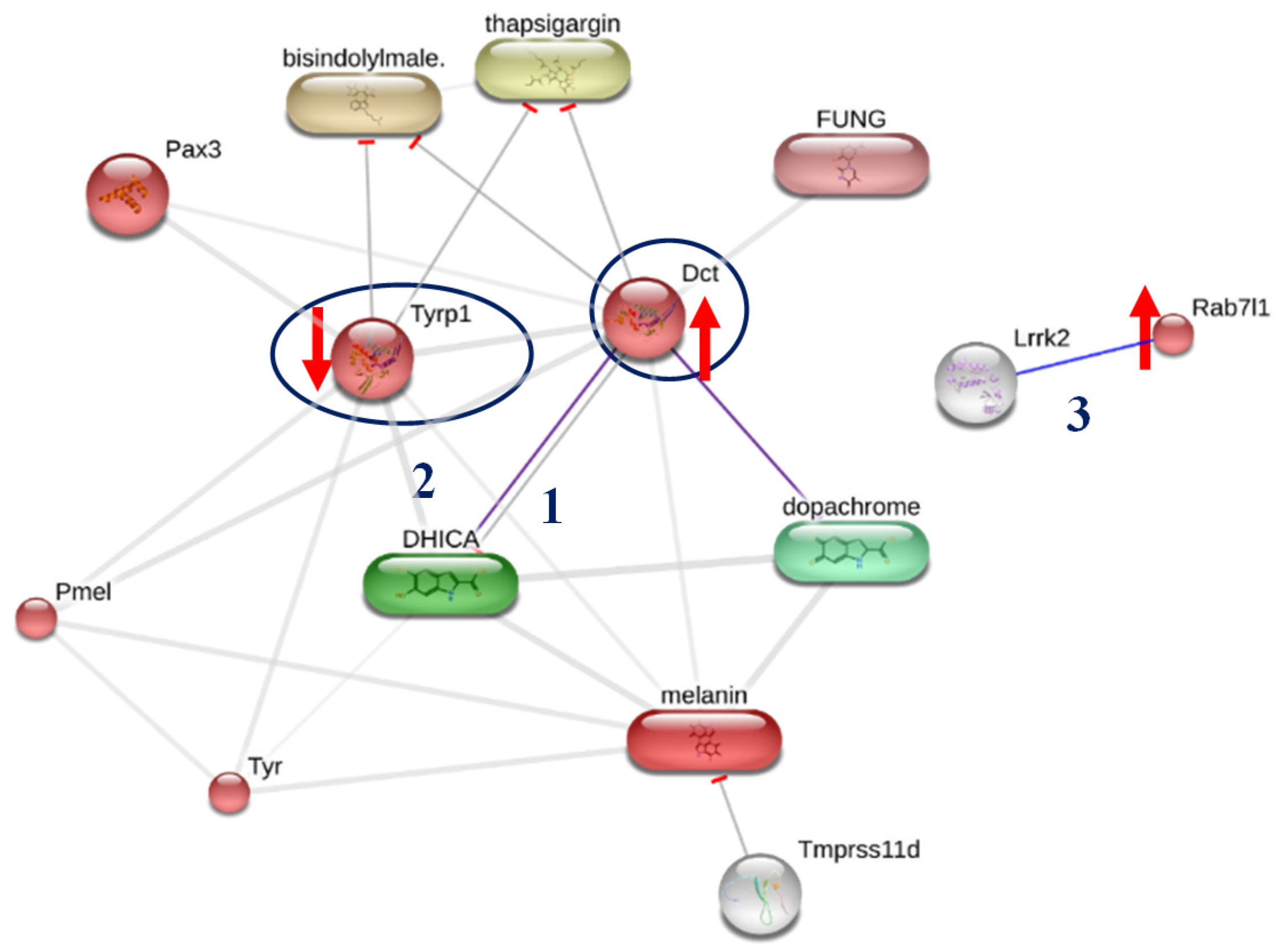
| Protein ID | Protein Name | PANTHER Family/Subfamily | PANTHER Protein Class | Relative Expression (Fold) |
|---|---|---|---|---|
| Q91YQ1 | Ras-related protein Rab-7L1; Rab29 | PTHR24073:SF315 | - | 2.0646 |
| P29812 | L-dopachrome tautomerase; Dct | PTHR11474:SF4 | oxygenase | 2.0460 |
| P07147 | 5,6-dihydroxyindole-2-carboxylic acid oxidase; Tyrp1 | PTHR11474:SF3 | oxygenase | 0.5034 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yingchutrakul, Y.; Krobthong, S.; Choowongkomon, K.; Papan, P.; Samutrtai, P.; Mahatnirunkul, T.; Chomtong, T.; Srimongkolpithak, N.; Jaroenchuensiri, T.; Aonbangkhen, C. Discovery of a Multifunctional Octapeptide from Lingzhi with Antioxidant and Tyrosinase Inhibitory Activity. Pharmaceuticals 2022, 15, 684. https://doi.org/10.3390/ph15060684
Yingchutrakul Y, Krobthong S, Choowongkomon K, Papan P, Samutrtai P, Mahatnirunkul T, Chomtong T, Srimongkolpithak N, Jaroenchuensiri T, Aonbangkhen C. Discovery of a Multifunctional Octapeptide from Lingzhi with Antioxidant and Tyrosinase Inhibitory Activity. Pharmaceuticals. 2022; 15(6):684. https://doi.org/10.3390/ph15060684
Chicago/Turabian StyleYingchutrakul, Yodying, Sucheewin Krobthong, Kiattawee Choowongkomon, Phakorn Papan, Pawitrabhorn Samutrtai, Thanisorn Mahatnirunkul, Thitikorn Chomtong, Nitipol Srimongkolpithak, Theeranuch Jaroenchuensiri, and Chanat Aonbangkhen. 2022. "Discovery of a Multifunctional Octapeptide from Lingzhi with Antioxidant and Tyrosinase Inhibitory Activity" Pharmaceuticals 15, no. 6: 684. https://doi.org/10.3390/ph15060684
APA StyleYingchutrakul, Y., Krobthong, S., Choowongkomon, K., Papan, P., Samutrtai, P., Mahatnirunkul, T., Chomtong, T., Srimongkolpithak, N., Jaroenchuensiri, T., & Aonbangkhen, C. (2022). Discovery of a Multifunctional Octapeptide from Lingzhi with Antioxidant and Tyrosinase Inhibitory Activity. Pharmaceuticals, 15(6), 684. https://doi.org/10.3390/ph15060684






