Structure-Activity Relationship Investigations of Novel Constrained Chimeric Peptidomimetics of SOCS3 Protein Targeting JAK2
Abstract
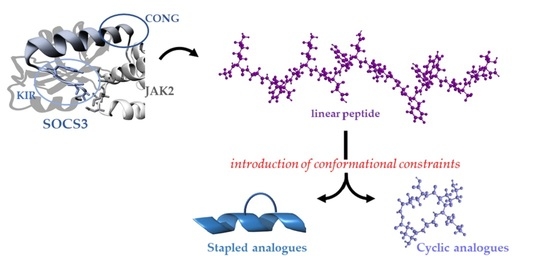
:1. Introduction
2. Materials and Methods
2.1. Peptide Synthesis
2.2. Shake Flask Procedure for Determination of Log P
2.3. Circular Dichroism (CD) Spectroscopy
2.4. NMR Studies
2.5. NMR Structure Calculations and Analysis
2.6. Serum Stability
2.7. Microscale Thermophoresis
3. Results
3.1. Design of Constrained KIRCONG Chim Mimetics
3.2. Conformational Studies of Constrained KIRCONG Analogues
3.2.1. Circular Dichroism
3.2.2. NMR Studies
3.3. Serum Stability
3.4. MST Investigations
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yoshimura, A.; Ito, M.; Mise-Omata, S.; Ando, M. SOCS: Negative regulators of cytokine signaling for immune tolerance. Int. Immunol. 2021, 33, 711–716. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Li, J.; Fu, M.; Zhao, X.; Wang, W. The JAK/STAT signaling pathway: From bench to clinic. Signal Transduct Target Ther. 2021, 6, 402. [Google Scholar] [CrossRef] [PubMed]
- Galic, S.; Sachithanandan, N.; Kay, T.W.; Steinberg, G.R. Suppressor of cytokine signalling (SOCS) proteins as guardians of inflammatory responses critical for regulating insulin sensitivity. Biochem. J. 2014, 461, 177–188. [Google Scholar] [CrossRef] [PubMed]
- Yoshimura, A.; Yasukawa, H. JAK’s SOCS: A mechanism of inhibition. Immunity 2012, 36, 157–159. [Google Scholar] [CrossRef] [Green Version]
- Croker, B.A.; Kiu, H.; Nicholson, S.E. SOCS regulation of the JAK/STAT signalling pathway. Semin. Cell Dev. Biol. 2008, 19, 414–422. [Google Scholar] [CrossRef] [Green Version]
- Liau, N.P.D.; Laktyushin, A.; Lucet, I.S.; Murphy, J.M.; Yao, S.; Whitlock, E.; Callaghan, K.; Nicola, N.A.; Kershaw, N.J.; Babon, J.J. The molecular basis of JAK/STAT inhibition by SOCS1. Nat. Commun. 2018, 9, 1558. [Google Scholar] [CrossRef]
- Kershaw, N.J.; Murphy, J.M.; Liau, N.P.; Varghese, L.N.; Laktyushin, A.; Whitlock, E.L.; Lucet, I.S.; Nicola, N.A.; Babon, J.J. SOCS3 binds specific receptor-JAK complexes to control cytokine signaling by direct kinase inhibition. Nat. Struct. Mol. Biol. 2013, 20, 469–476. [Google Scholar] [CrossRef]
- Kishore, R.; Verma, S.K. Roles of STATs signaling in cardiovascular diseases. JAKSTAT 2012, 1, 118–124. [Google Scholar] [CrossRef] [Green Version]
- Banerjee, S.; Biehl, A.; Gadina, M.; Hasni, S.; Schwartz, D.M. JAK-STAT Signaling as a Target for Inflammatory and Autoimmune Diseases: Current and Future Prospects. Drugs 2017, 77, 521–546. [Google Scholar] [CrossRef]
- Blumer, T.; Coto-Llerena, M.; Duong, F.H.T.; Heim, M.H. SOCS1 is an inducible negative regulator of interferon lambda (IFN-lambda)-induced gene expression in vivo. J. Biol. Chem. 2017, 292, 17928–17938. [Google Scholar] [CrossRef] [Green Version]
- Gao, A.H.; Hu, Y.R.; Zhu, W.P. IFN-gamma inhibits ovarian cancer progression via SOCS1/JAK/STAT signaling pathway. Clin. Transl. Oncol. 2022, 24, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Davey, G.M.; Starr, R.; Cornish, A.L.; Burghardt, J.T.; Alexander, W.S.; Carbone, F.R.; Surh, C.D.; Heath, W.R. SOCS-1 regulates IL-15-driven homeostatic proliferation of antigen-naive CD8 T cells, limiting their autoimmune potential. J. Exp. Med. 2005, 202, 1099–1108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eyles, J.L.; Metcalf, D.; Grusby, M.J.; Hilton, D.J.; Starr, R. Negative regulation of interleukin-12 signaling by suppressor of cytokine signaling-1. J. Biol. Chem. 2002, 277, 43735–43740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamiya, T.; Kashiwagi, I.; Takahashi, R.; Yasukawa, H.; Yoshimura, A. Suppressors of Cytokine Signaling (SOCS) Proteins and JAK/STAT Pathways Regulation of T-Cell Inflammation by SOCS1 and SOCS3. Arter. Throm. Vas. 2011, 31, 980–985. [Google Scholar] [CrossRef] [Green Version]
- Yoshimura, A.; Suzuki, M.; Sakaguchi, R.; Hanada, T.; Yasukawa, H. SOCS, Inflammation, and Autoimmunity. Front. Immunol. 2012, 3, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Madonna, S.; Scarponi, C.; De Pita, O.; Albanesi, C. Suppressor of cytokine signaling 1 inhibits IFN-gamma inflammatory signaling in human keratinocytes by sustaining ERK1/2 activation. FASEB J. 2008, 22, 3287–3297. [Google Scholar] [CrossRef] [PubMed]
- Jung, S.H.; Kim, S.M.; Lee, C.E. Mechanism of suppressors of cytokine signaling 1 inhibition of epithelial-mesenchymal transition signaling through ROS regulation in colon cancer cells: Suppression of Src leading to thioredoxin up-regulation. Oncotarget 2016, 7, 62559–62571. [Google Scholar] [CrossRef] [Green Version]
- Schuett, J.; Kreutz, J.; Grote, K.; Vlacil, A.K.; Schuett, H.; Oberoi, R.; Schmid, A.; Witten, A.; Stoll, M.; Schieffer, B.; et al. Suppressor of Cytokine Signaling 1 is Involved in Gene Regulation Which Controls the Survival of Ly6C(low) Monocytes in Mice. Cell. Physiol. Biochem. 2019, 52, 336–353. [Google Scholar] [CrossRef]
- Miklossy, G.; Hilliard, T.S.; Turkson, J. Therapeutic modulators of STAT signalling for human diseases. Nat. Rev. Drug Discov. 2013, 12, 611–629. [Google Scholar] [CrossRef] [Green Version]
- Torella, D.; Curcio, A.; Gasparri, C.; Galuppo, V.; De Serio, D.; Surace, F.C.; Cavaliere, A.L.; Leone, A.; Coppola, C.; Ellison, G.M.; et al. Fludarabine prevents smooth muscle proliferation in vitro and neointimal hyperplasia in vivo through specific inhibition of STAT-1 activation. Am. J. Physiol. Heart Circ. Physiol. 2007, 292, H2935–H2943. [Google Scholar] [CrossRef]
- Du, Y.; Yang, F.; Wang, Q.; Xu, N.; Xie, Y.; Chen, S.; Qin, T.; Peng, D. Influenza a virus antagonizes type I and type II interferon responses via SOCS1-dependent ubiquitination and degradation of JAK1. Virol. J. 2020, 17, 74. [Google Scholar] [CrossRef] [PubMed]
- Seong, R.K.; Lee, J.K.; Shin, O.S. Zika Virus-Induction of the Suppressor of Cytokine Signaling 1/3 Contributes to the Modulation of Viral Replication. Pathogens 2020, 9, 163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, M.G.M.; Ghosh, A.; Variya, B.; Santharam, M.A.; Kandhi, R.; Ramanathan, S.; Ilangumaran, S. Hepatocyte growth control by SOCS1 and SOCS3. Cytokine 2019, 121, 154733. [Google Scholar] [CrossRef] [PubMed]
- Chu, Q.; Shen, D.; He, L.; Wang, H.; Liu, C.; Zhang, W. Prognostic significance of SOCS3 and its biological function in colorectal cancer. Gene 2017, 627, 114–122. [Google Scholar] [CrossRef]
- Shang, A.Q.; Wu, J.; Bi, F.; Zhang, Y.J.; Xu, L.R.; Li, L.L.; Chen, F.F.; Wang, W.W.; Zhu, J.J.; Liu, Y.Y. Relationship between HER2 and JAK/STAT-SOCS3 signaling pathway and clinicopathological features and prognosis of ovarian cancer. Cancer Biol. Ther. 2017, 18, 314–322. [Google Scholar] [CrossRef] [Green Version]
- Dai, L.; Li, Z.; Tao, Y.; Liang, W.; Hu, W.; Zhou, S.; Fu, X.; Wang, X. Emerging roles of suppressor of cytokine signaling 3 in human cancers. Biomed. Pharm. 2021, 144, 112262. [Google Scholar] [CrossRef]
- Barclay, J.L.; Anderson, S.T.; Waters, M.J.; Curlewis, J.D. SOCS3 as a tumor suppressor in breast cancer cells, and its regulation by PRL. Int. J. Cancer 2009, 124, 1756–1766. [Google Scholar] [CrossRef]
- Hill, G.R.; Kuns, R.D.; Raffelt, N.C.; Don, A.L.; Olver, S.D.; Markey, K.A.; Wilson, Y.A.; Tocker, J.; Alexander, W.S.; Clouston, A.D.; et al. SOCS3 regulates graft-versus-host disease. Blood 2010, 116, 287–296. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.C.; Lin, C.K.; Tsai, Y.H.; Weng, H.H.; Li, Y.C.; You, L.; Chen, J.K.; Jablons, D.M.; Yang, C.T. Adenovirus-mediated SOCS3 gene transfer inhibits the growth and enhances the radiosensitivity of human non-small cell lung cancer cells. Oncol. Rep. 2010, 24, 1605–1612. [Google Scholar] [CrossRef] [Green Version]
- Speth, J.M.; Penke, L.R.; Bazzill, J.D.; Park, K.S.; de Rubio, R.G.; Schneider, D.J.; Ouchi, H.; Moon, J.J.; Keshamouni, V.G.; Zemans, R.L.; et al. Alveolar macrophage secretion of vesicular SOCS3 represents a platform for lung cancer therapeutics. JCI Insight 2019, 4, e131340. [Google Scholar] [CrossRef] [Green Version]
- Tang, H.; Long, Q.; Zhuang, K.; Yan, Y.; Han, K.; Guo, H.; Lu, X. miR-665 promotes the progression of gastric adenocarcinoma via elevating FAK activation through targeting SOCS3 and is negatively regulated by lncRNA MEG3. J. Cell Physiol. 2020, 235, 4709–4719. [Google Scholar] [CrossRef] [PubMed]
- Ying, M.Z.; Li, D.W.; Yang, L.J.; Wang, M.; Wang, N.; Chen, Y.; He, M.X.; Wang, Y.J. Loss of SOCS3 expression is associated with an increased risk of recurrent disease in breast carcinoma. J. Cancer Res. Clin. 2010, 136, 1617–1626. [Google Scholar] [CrossRef] [PubMed]
- Dominguez, E.; Mauborgne, A.; Mallet, J.; Desclaux, M.; Pohl, M. SOCS3-mediated blockade of JAK/STAT3 signaling pathway reveals its major contribution to spinal cord neuroinflammation and mechanical allodynia after peripheral nerve injury. J. Neurosci. 2010, 30, 5754–5766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tao, H.; Shi, P.; Zhao, X.D.; Xuan, H.Y.; Gong, W.H.; Ding, X.S. DNMT1 deregulation of SOCS3 axis drives cardiac fibroblast activation in diabetic cardiac fibrosis. J. Cell Physiol. 2021, 236, 3481–3494. [Google Scholar] [CrossRef]
- Wang, T.; Zhou, P.; Xie, X.; Tomita, Y.; Cho, S.; Tsirukis, D.; Lam, E.; Luo, H.R.; Sun, Y. Myeloid lineage contributes to pathological choroidal neovascularization formation via SOCS3. EBioMedicine 2021, 73, 103632. [Google Scholar] [CrossRef]
- La Manna, S.; De Benedictis, I.; Marasco, D. Proteomimetics of Natural Regulators of JAK-STAT Pathway: Novel Therapeutic Perspectives. Front. Mol. Biosci. 2021, 8, 792546. [Google Scholar] [CrossRef]
- La Manna, S.; Lopez-Sanz, L.; Bernal, S.; Fortuna, S.; Mercurio, F.A.; Leone, M.; Gomez-Guerrero, C.; Marasco, D. Cyclic mimetics of kinase-inhibitory region of Suppressors of Cytokine Signaling 1: Progress toward novel anti-inflammatory therapeutics. Eur. J. Med. Chem. 2021, 221, 113547. [Google Scholar] [CrossRef]
- La Manna, S.; Lopez-Sanz, L.; Bernal, S.; Jimenez-Castilla, L.; Prieto, I.; Morelli, G.; Gomez-Guerrero, C.; Marasco, D. Antioxidant Effects of PS5, a Peptidomimetic of Suppressor of Cytokine Signaling 1, in Experimental Atherosclerosis. Antioxidants 2020, 9, 754. [Google Scholar] [CrossRef]
- La Manna, S.; Lopez-Sanz, L.; Leone, M.; Brandi, P.; Scognamiglio, P.L.; Morelli, G.; Novellino, E.; Gomez-Guerrero, C.; Marasco, D. Structure-activity studies of peptidomimetics based on kinase-inhibitory region of suppressors of cytokine signaling 1. Biopolymers 2017, 110, e23082. [Google Scholar] [CrossRef]
- Doti, N.; Scognamiglio, P.L.; Madonna, S.; Scarponi, C.; Ruvo, M.; Perretta, G.; Albanesi, C.; Marasco, D. New mimetic peptides of the kinase-inhibitory region (KIR) of SOCS1 through focused peptide libraries. Biochem. J. 2012, 443, 231–240. [Google Scholar] [CrossRef]
- Ahmed, C.M.; Massengill, M.T.; Brown, E.E.; Ildefonso, C.J.; Johnson, H.M.; Lewin, A.S. A cell penetrating peptide from SOCS-1 prevents ocular damage in experimental autoimmune uveitis. Exp. Eye Res. 2018, 177, 12–22. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, C.M.; Patel, A.P.; Ildefonso, C.J.; Johnson, H.M.; Lewin, A.S. Corneal Application of R9-SOCS1-KIR Peptide Alleviates Endotoxin-Induced Uveitis. Transl. Vis. Sci. Technol. 2021, 10, 25. [Google Scholar] [CrossRef] [PubMed]
- Recio, C.; Oguiza, A.; Lazaro, I.; Mallavia, B.; Egido, J.; Gomez-Guerrero, C. Suppressor of cytokine signaling 1-derived peptide inhibits Janus kinase/signal transducers and activators of transcription pathway and improves inflammation and atherosclerosis in diabetic mice. Arter. Thromb. Vasc. Biol. 2014, 34, 1953–1960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernal, S.; Lopez-Sanz, L.; Jimenez-Castilla, L.; Prieto, I.; Melgar, A.; La Manna, S.; Martin-Ventura, J.L.; Blanco-Colio, L.M.; Egido, J.; Gomez-Guerrero, C. Protective effect of suppressor of cytokine signalling 1-based therapy in experimental abdominal aortic aneurysm. Br. J. Pharm. 2020, 178, 564–581. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Sanz, L.; Bernal, S.; Recio, C.; Lazaro, I.; Oguiza, A.; Melgar, A.; Jimenez-Castilla, L.; Egido, J.; Gomez-Guerrero, C. SOCS1-targeted therapy ameliorates renal and vascular oxidative stress in diabetes via STAT1 and PI3K inhibition. Lab. Investig. 2018, 98, 1276–1290. [Google Scholar] [CrossRef]
- Opazo-Rios, L.; Sanchez Matus, Y.; Rodrigues-Diez, R.R.; Carpio, D.; Droguett, A.; Egido, J.; Gomez-Guerrero, C.; Mezzano, S. Anti-inflammatory, antioxidant and renoprotective effects of SOCS1 mimetic peptide in the BTBR ob/ob mouse model of type 2 diabetes. BMJ Open Diabetes Res. Care 2020, 8, e001242. [Google Scholar] [CrossRef]
- Recio, C.; Lazaro, I.; Oguiza, A.; Lopez-Sanz, L.; Bernal, S.; Blanco, J.; Egido, J.; Gomez-Guerrero, C. Suppressor of Cytokine Signaling-1 Peptidomimetic Limits Progression of Diabetic Nephropathy. J. Am. Soc. Nephrol. 2017, 28, 575–585. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Wu, Y.; Li, K.; Currie, I.; Keating, N.; Dehkhoda, F.; Grohmann, C.; Babon, J.J.; Nicholson, S.E.; Sleebs, B.E. Optimization of Phosphotyrosine Peptides that Target the SH2 Domain of SOCS1 and Block Substrate Ubiquitination. ACS Chem. Biol. 2022, 17, 449–462. [Google Scholar] [CrossRef]
- Madonna, S.; Scarponi, C.; Morelli, M.; Sestito, R.; Scognamiglio, P.L.; Marasco, D.; Albanesi, C. SOCS3 inhibits the pathological effects of IL-22 in non-melanoma skin tumor-derived keratinocytes. Oncotarget 2017, 8, 24652–24667. [Google Scholar] [CrossRef] [Green Version]
- La Manna, S.; Lee, E.; Ouzounova, M.; Di Natale, C.; Novellino, E.; Merlino, A.; Korkaya, H.; Marasco, D. Mimetics of suppressor of cytokine signaling 3: Novel potential therapeutics in triple breast cancer. Int. J. Cancer 2018, 143, 2177–2186. [Google Scholar] [CrossRef]
- La Manna, S.; Lopez-Sanz, L.; Mercurio, F.A.; Fortuna, S.; Leone, M.; Gomez-Guerrero, C.; Marasco, D. Chimeric Peptidomimetics of SOCS 3 Able to Interact with JAK2 as Anti-inflammatory Compounds. ACS Med. Chem. Lett. 2020, 11, 615–623. [Google Scholar] [CrossRef] [PubMed]
- La Manna, S.; Scognamiglio, P.L.; Di Natale, C.; Leone, M.; Mercurio, F.A.; Malfitano, A.M.; Cianfarani, F.; Madonna, S.; Caravella, S.; Albanesi, C.; et al. Characterization of linear mimetic peptides of Interleukin-22 from dissection of protein interfaces. Biochimie 2017, 138, 106–115. [Google Scholar] [CrossRef] [PubMed]
- OECD. Guidelines for the Testing of Chemicals, Test No. 107: Partition Coefficient (n-Octanol/Water): Shake Flask Method. Available online: https://www.oecd.org/chemicalsafety/testing/21047299.pdf (accessed on 2 March 2022).
- Griesinger, C.; Otting, G.; Wuthrich, K.; Ernst, R.R. Clean TOCSY for proton spin system identification in macromolecules. J. Am. Chem. Soc. 1988, 110, 7870–7872. [Google Scholar] [CrossRef]
- Kumar, A.; Ernst, R.R.; Wuthrich, K. A two-dimensional nuclear Overhauser enhancement (2D NOE) experiment for the elucidation of complete proton-proton cross-relaxation networks in biological macromolecules. Biochem. Biophys. Res. Commun. 1980, 95, 1–6. [Google Scholar] [CrossRef]
- Bax, A.; Davis, D.G. Practical aspects of two-dimensional transverse NOE spectroscopy. J. of Magn. Reson. 1969 1985, 63, 207–213. [Google Scholar] [CrossRef]
- Piantini, U.; Sorensen, O.W.; Ernst, R.R. Multiple quantum filters for elucidating NMR coupling networks. J. Am. Chem. Soc. 1982, 104, 6800–6801. [Google Scholar] [CrossRef]
- Hwang, T.L.; Shaka, A.J. Water suppression that works. Excitation sculpting using arbitrary waveforms and pulsed field gradients. J. Magn. Reson. Ser. A 1995, 112, 275–279. [Google Scholar] [CrossRef]
- Wuthrich, K. NMR of Proteins and Nucleic Acids; Wiley: New YorK, NY, USA, 1986. [Google Scholar]
- Bartels, C.; Xia, T.; Billeter, M.; Güntert, P.; Wüthrich, K. The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J. Biomol. NMR 1995, 6, 1–10. [Google Scholar] [CrossRef]
- Kjaergaard, M.; Brander, S.; Poulsen, F.M. Random coil chemical shift for intrinsically disordered proteins: Effects of temperature and pH. J. Biomol. NMR 2011, 49, 139–149. [Google Scholar] [CrossRef]
- Herrmann, T.; Guntert, P.; Wuthrich, K. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 2002, 319, 209–227. [Google Scholar] [CrossRef] [Green Version]
- Yilmaz, E.M.; Guntert, P. NMR structure calculation for all small molecule ligands and non-standard residues from the PDB Chemical Component Dictionary. J. Biomol. NMR 2015, 63, 21–37. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; Rullmannn, J.A.; MacArthur, M.W.; Kaptein, R.; Thornton, J.M. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 1996, 8, 477–486. [Google Scholar] [CrossRef] [PubMed]
- Koradi, R.; Billeter, M.; Wuthrich, K. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 1996, 14, 51–55. [Google Scholar] [CrossRef]
- Hellinen, L.; Bahrpeyma, S.; Rimpela, A.K.; Hagstrom, M.; Reinisalo, M.; Urtti, A. Microscale Thermophoresis as a Screening Tool to Predict Melanin Binding of Drugs. Pharmaceutics 2020, 12, 554. [Google Scholar] [CrossRef]
- Scognamiglio, P.L.; Di Natale, C.; Perretta, G.; Marasco, D. From peptides to small molecules: An intriguing but intricated way to new drugs. Curr. Med. Chem. 2013, 20, 3803–3817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- La Manna, S.; Di Natale, C.; Florio, D.; Marasco, D. Peptides as Therapeutic Agents for Inflammatory-Related Diseases. Int. J. Mol. Sci. 2018, 19, 2714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Russo, A.; Aiello, C.; Grieco, P.; Marasco, D. Targeting "Undruggable" Proteins: Design of Synthetic Cyclopeptides. Curr. Med. Chem. 2016, 23, 748–762. [Google Scholar] [CrossRef] [PubMed]
- Moiola, M.; Memeo, M.G.; Quadrelli, P. Stapled Peptides—A Useful Improvement for Peptide-Based Drugs. Molecules 2019, 24, 3654. [Google Scholar] [CrossRef] [Green Version]
- Fodje, M.N.; Al-Karadaghi, S. Occurrence, conformational features and amino acid propensities for the pi-helix. Protein Eng. 2002, 15, 353–358. [Google Scholar] [CrossRef]
- Cooley, R.B.; Arp, D.J.; Karplus, P.A. Evolutionary origin of a secondary structure: Pi-helices as cryptic but widespread insertional variations of alpha-helices that enhance protein functionality. J. Mol. Biol. 2010, 404, 232–246. [Google Scholar] [CrossRef] [Green Version]
- Andersson, D.; Carlsson, U.; Freskgard, P.O. Contribution of tryptophan residues to the CD spectrum of the extracellular domain of human tissue factor: Application in folding studies and prediction of secondary structure. Eur. J. Biochem. 2001, 268, 1118–1128. [Google Scholar] [CrossRef]
- Mercurio, F.A.; Pirone, L.; Di Natale, C.; Marasco, D.; Pedone, E.M.; Leone, M. Sam domain-based stapled peptides: Structural analysis and interaction studies with the Sam domains from the EphA2 receptor and the lipid phosphatase Ship2. Bioorg. Chem. 2018, 80, 602–610. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Sykes, B.D.; Richards, F.M. Relationship between nuclear magnetic resonance chemical shift and protein secondary structure. J. Mol. Biol. 1991, 222, 311–333. [Google Scholar] [CrossRef]
- Wishart, D.S.; Sykes, B.D.; Richards, F.M. The chemical shift index: A fast and simple method for the assignment of protein secondary structure through NMR spectroscopy. Biochemistry 1992, 31, 1647–1651. [Google Scholar] [CrossRef] [PubMed]
- Asmari, M.; Ratih, R.; Alhazmi, H.A.; El Deeb, S. Thermophoresis for characterizing biomolecular interaction. Methods 2018, 146, 107–119. [Google Scholar] [CrossRef] [PubMed]
- Jerabek-Willemsen, M.; Wienken, C.J.; Braun, D.; Baaske, P.; Duhr, S. Molecular interaction studies using microscale thermophoresis. Assay Drug Dev. Technol. 2011, 9, 342–353. [Google Scholar] [CrossRef] [Green Version]
- Verstovsek, S.; Kantarjian, H.; Mesa, R.A.; Pardanani, A.D.; Cortes-Franco, J.; Thomas, D.A.; Estrov, Z.; Fridman, J.S.; Bradley, E.C.; Erickson-Viitanen, S. Safety and efficacy of INCB018424, a JAK1 and JAK2 inhibitor, in myelofibrosis. N. Engl. J. Med. 2010, 363, 1117–1127. [Google Scholar] [CrossRef] [Green Version]
- Park, J.S.; Hong, M.H.; Chun, Y.J.; Kim, H.R.; Cho, B.C. A phase Ib study of the combination of afatinib and ruxolitinib in EGFR mutant NSCLC with progression on EGFR-TKIs. Lung Cancer 2019, 134, 46–51. [Google Scholar] [CrossRef]
- Walensky, L.D.; Bird, G.H. Hydrocarbon-stapled peptides: Principles, practice, and progress. J. Med. Chem. 2014, 57, 6275–6288. [Google Scholar] [CrossRef] [Green Version]





| Name | Sequence | KD (µM) | LogP |
|---|---|---|---|
| KIRCONGchim [51] |  | (1.1 ± 0.3) × 10 | −1.32 |
| KIRCONG amide |  | (2.0 ± 0.9) × 102 | −0.92 |
| KIRCONG disulfide |  | (6 ± 3) × 102 | −1.46 |
| KIRCONG i/i+5 |  | No binding | −0.27 |
| KIRCONG i/i+7 |  | No binding | 0.29 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
La Manna, S.; Leone, M.; Mercurio, F.A.; Florio, D.; Marasco, D. Structure-Activity Relationship Investigations of Novel Constrained Chimeric Peptidomimetics of SOCS3 Protein Targeting JAK2. Pharmaceuticals 2022, 15, 458. https://doi.org/10.3390/ph15040458
La Manna S, Leone M, Mercurio FA, Florio D, Marasco D. Structure-Activity Relationship Investigations of Novel Constrained Chimeric Peptidomimetics of SOCS3 Protein Targeting JAK2. Pharmaceuticals. 2022; 15(4):458. https://doi.org/10.3390/ph15040458
Chicago/Turabian StyleLa Manna, Sara, Marilisa Leone, Flavia Anna Mercurio, Daniele Florio, and Daniela Marasco. 2022. "Structure-Activity Relationship Investigations of Novel Constrained Chimeric Peptidomimetics of SOCS3 Protein Targeting JAK2" Pharmaceuticals 15, no. 4: 458. https://doi.org/10.3390/ph15040458
APA StyleLa Manna, S., Leone, M., Mercurio, F. A., Florio, D., & Marasco, D. (2022). Structure-Activity Relationship Investigations of Novel Constrained Chimeric Peptidomimetics of SOCS3 Protein Targeting JAK2. Pharmaceuticals, 15(4), 458. https://doi.org/10.3390/ph15040458