One-Pot Synthesis and Molecular Modeling Studies of New Bioactive Spiro-Oxindoles Based on Uracil Derivatives as SARS-CoV-2 Inhibitors Targeting RNA Polymerase and Spike Glycoprotein
Abstract
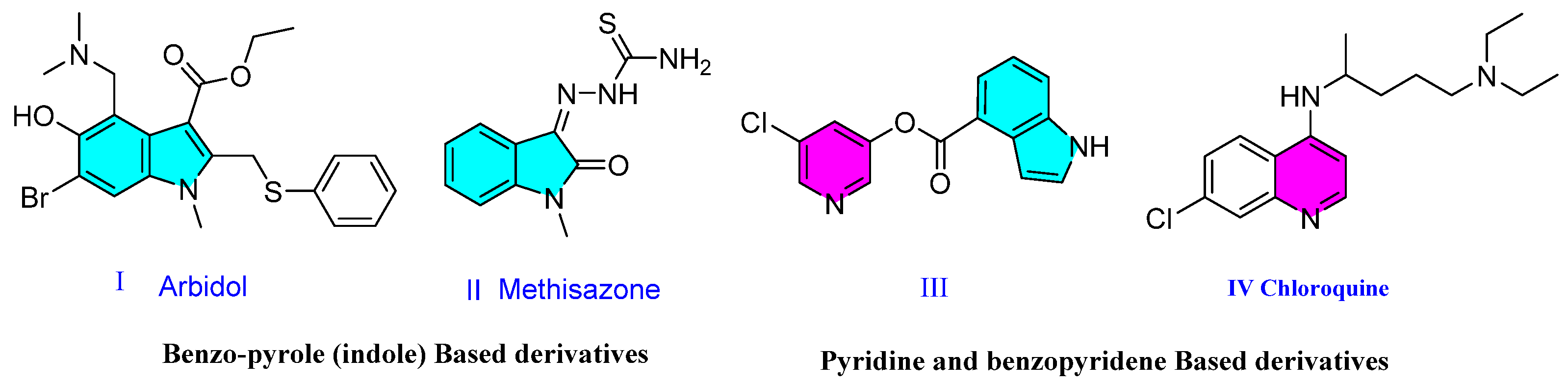
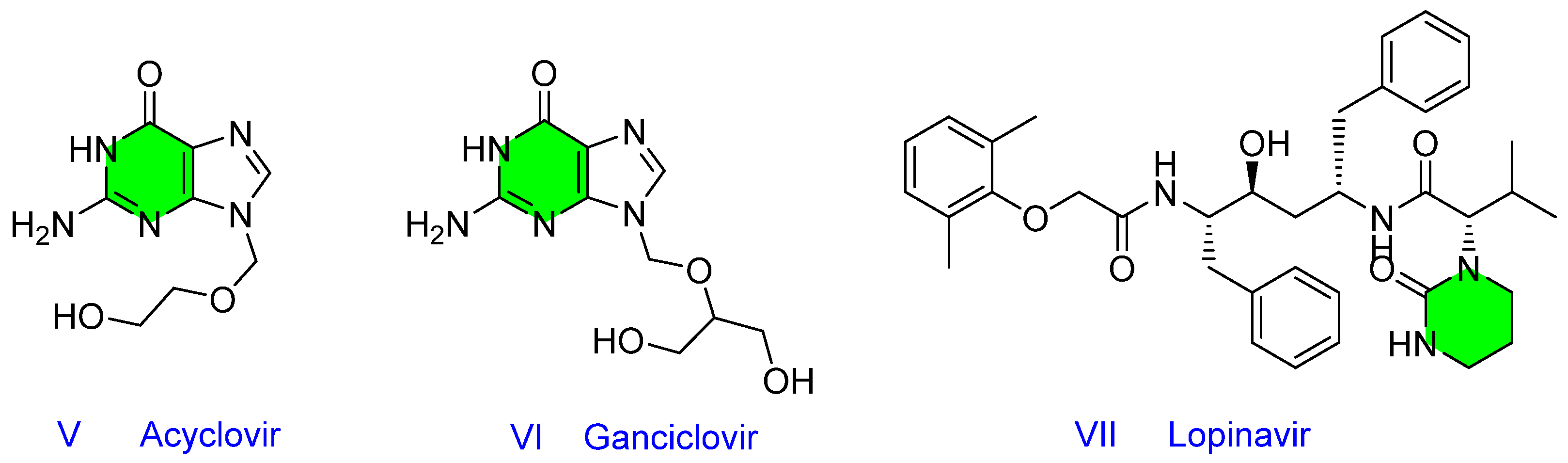
1. Introduction
2. Results and Discussion
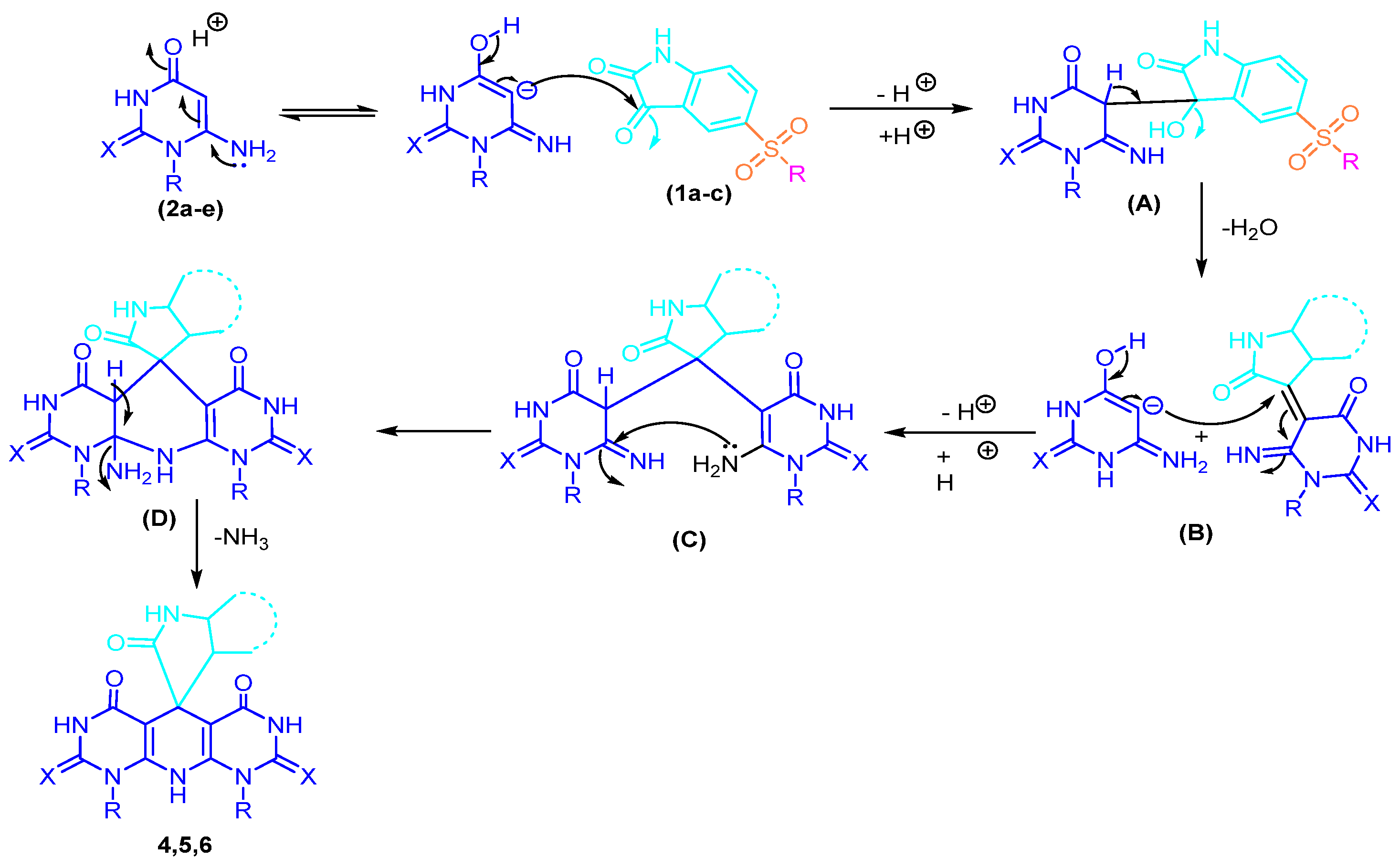
2.1. Chemistry
2.2. Antiviral Activity
2.2.1. The Half-Maximal Inhibitory Concentration (IC50)
2.2.2. Plaque Reduction Assay (% of Inhibition SARS-CoV2)
2.2.3. In Vitro Enzymes Assay
2.3. Computational Study
2.3.1. Computational Study of the Binding Mode
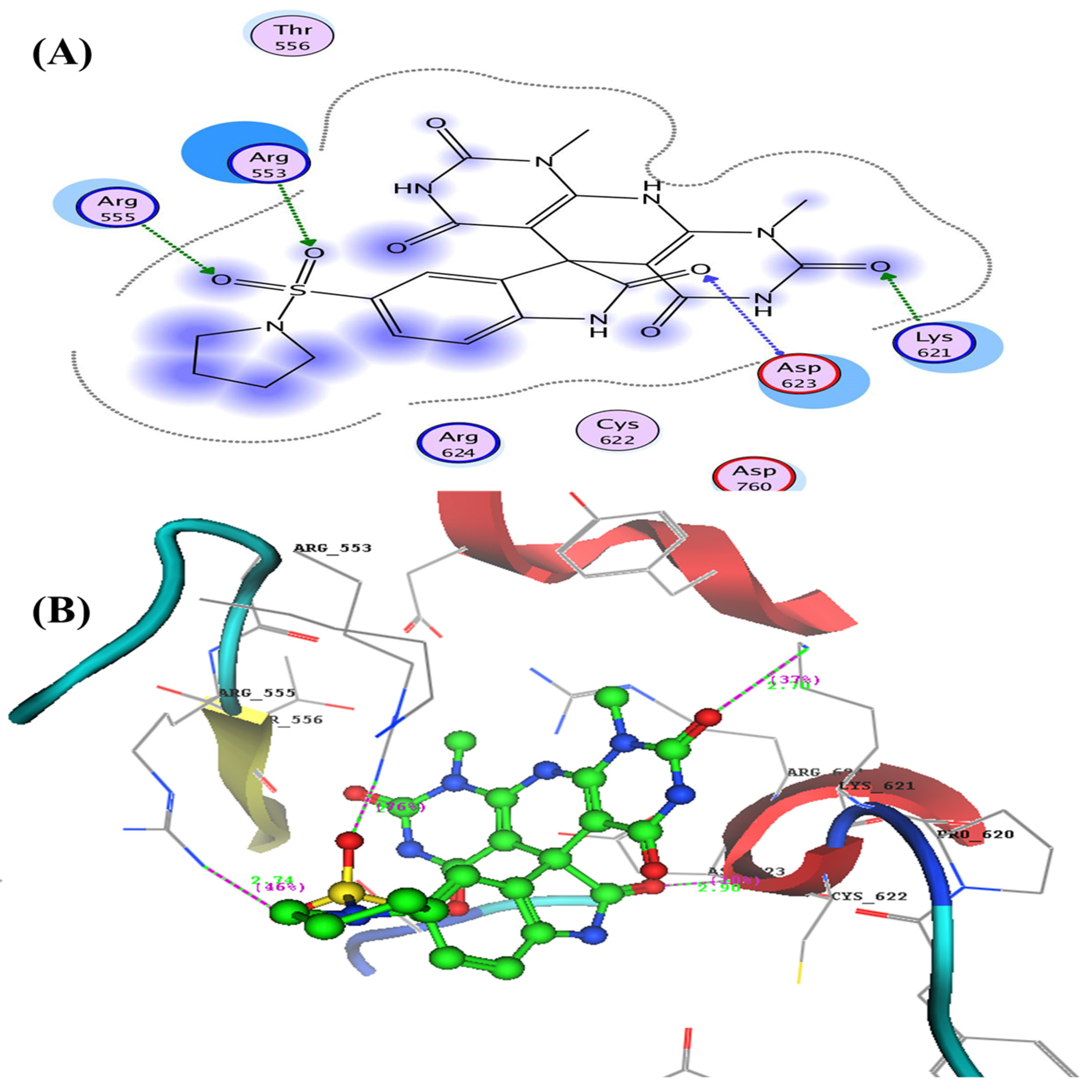
- Docking Study Inside RNA-Dependent RNA Polymerase (RdRp) (PDB: 6m71)
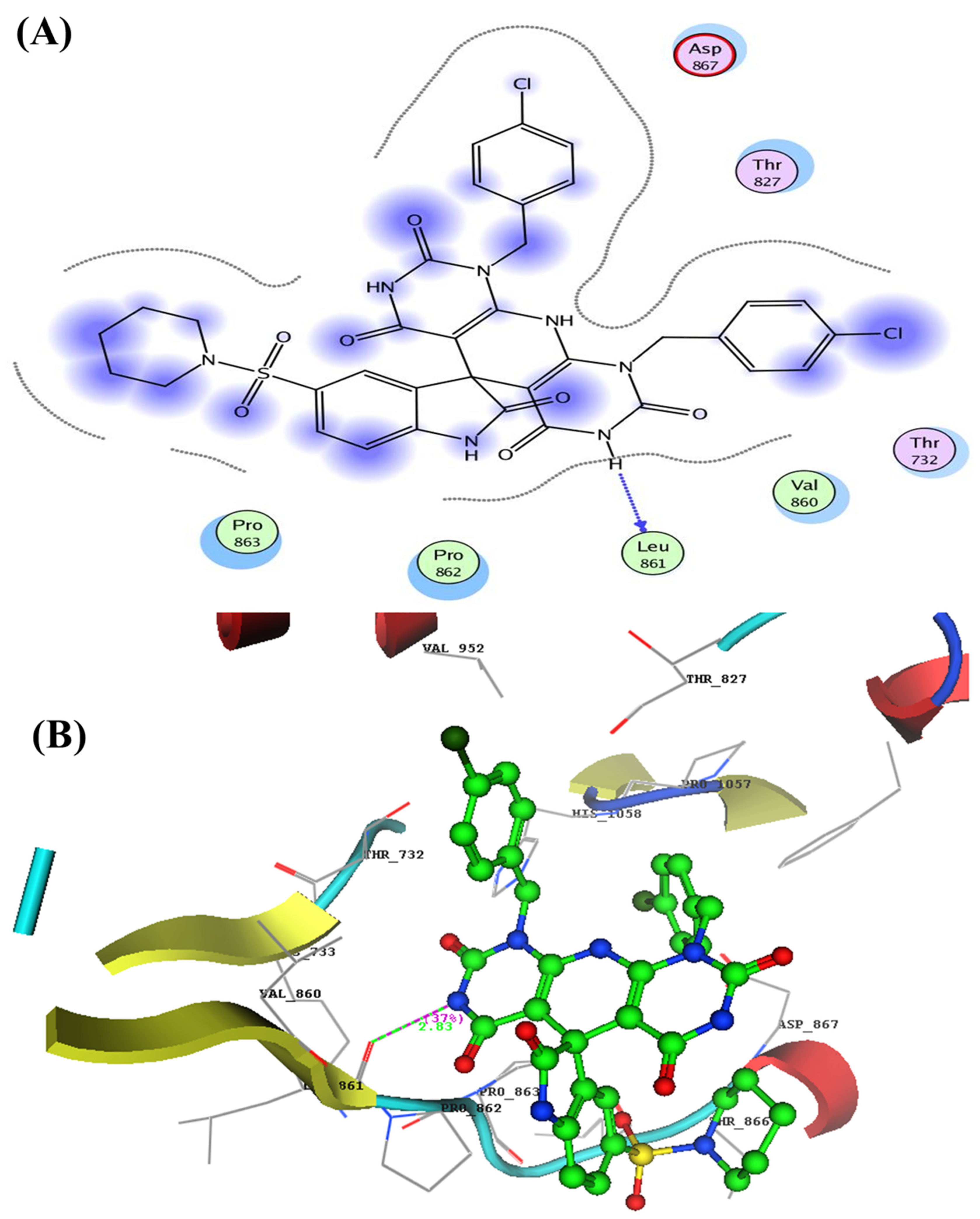
- Docking Study Inside Spike Glycoprotein (SGp) (PDB: 6VXX)
2.3.2. Geometrical Optimization and Molecular Parameters
2.3.3. Molecular Electrostatic Potential (MEP)
3. Materials and Methods
3.1. Chemistry
- Synthesis of 1H-spiro[indoline-3,5′-pyrido[2,3-d:6,5-d′]dipyrimidine derivatives
- 1′,9′-Dimethyl-5-(pyrrolidin-1-ylsulfonyl)-1′H-spiro[indoline-3,5′-pyrido[2,3-d:6,5-d′]dipy-rimidine]-2,2′,4′,6′,8′(3′H,7′H,9′H,10′H)-pentaone (3a)
- 1′,9′-Dibenzyl-5-(pyrrolidin-1-ylsulfonyl)-1′H-spiro[indoline-3,5′-pyrido[2,3-d:6,5-d′]dipy-rimidine]-2,2′,4′,6′,8′(3′H,7′H,9′H,10′H)-pentaone (3b)
- 1′,9′-Bis(4-chlorobenzyl)-5-(pyrrolidin-1-ylsulfonyl)-1′H-spiro[indoline-3,5′-pyrido[2,3-d:-6,5-d′]dipyrimidine]-2,2′,4′,6′,8′(3′H,7′H,9′H,10′H)-pentaone (3c)
- 1′,9′-Dimethyl-5-(pyrrolidin-1-ylsulfonyl)-2′,8′-dithioxo-2′,3′,8′,9′-tetrahydro-1′H-spiro-[indoline-3,5′-pyrido[2,3-d:6,5-d′]dipyrimidine]-2,4′,6′(7′H,10′H)-trione (3d)
- 5-(Piperidin-1-ylsulfonyl)-1′H-spiro[indoline-3,5′-pyrido[2,3-d:6,5-d′]dipyrimidine]-2,2′,4′,-6′,8′(3′H,7′H,9′H,10′H)-pentaone (4a)
- 1′,9′-Dimethyl-5-(piperidin-1-ylsulfonyl)-1′H-spiro[indoline-3,5′-pyrido[2,3-d:6,5-d′]dipyri-midine]-2,2′,4′,6′,8′(3′H,7′H,9′H,10′H)-pentaone (4b)
- 1′,9′-Dibenzyl-5-(piperidin-1-ylsulfonyl)-1′H-spiro[indoline-3,5′-pyrido[2,3-d:6,5-d′]dipyri-midine]-2,2′,4′,6′,8′(3′H,7′H,9′H,10′H)-pentaone (4c)
- 1′,9′-Bis(4-chlorobenzyl)-5-(piperidin-1-ylsulfonyl)-1′H-spiro[indoline-3,5′-pyrido[2,3-d:6,-5-d′]dipyrimidine]-2,2′,4′,6′,8′(3′H,7′H,9′H,10′H)-pentaone (4d)
- 1′,9′-Dimethyl-5-(piperidin-1-ylsulfonyl)-2′,8′-dithioxo-2′,3′,8′,9′-tetrahydro-1′H-spiro-[indoline-3,5′-pyrido[2,3-d:6,5-d′]dipyrimidine]-2,4′,6′(7′H,10′H)-trione (4e)
- 5-(Morpholinosulfonyl)-1′H-spiro[indoline-3,5′-pyrido[2,3-d:6,5-d′]dipyrimidine]-2,2′,4′,-6′,8′(3′H,7′H,9′H,10′H)-pentaone (5a)
- 1′,9′-Dimethyl-5-(morpholinosulfonyl)-1′H-spiro[indoline-3,5′-pyrido[2,3-d:6,5-d′]dipyrimi-dine]-2,2′,4′,6′,8′(3′H,7′H,9′H,10′H)-pentaone (5b)
- 1′,9′-Dibenzyl-5-(morpholinosulfonyl)-1′H-spiro[indoline-3,5′-pyrido[2,3-d:6,5-d′]dipyrimi-dine]-2,2′,4′,6′,8′(3′H,7′H,9′H,10′H)-pentaone (5c)
- 1′,9′-Bis(4-chlorobenzyl)-5-(morpholinosulfonyl)-1′H-spiro[indoline-3,5′-pyrido[2,3-d:6,5-d′]dipyrimidine]-2,2′,4′,6′,8′(3′H,7′H,9′H,10′H)-pentaone (5d)
- 1′,9′-Dimethyl-5-(morpholinosulfonyl)-2′,8′-dithioxo-2′,3′,8′,9′-tetrahydro-1′H-spiro[indo-line-3,5′-pyrido[2,3-d:6,5-d′]dipyrimidine]-2,4′,6′(7′H,10′H)-trione (5e)
3.2. Antiviral Activity
3.2.1. Cytotoxicity Assay
3.2.2. Plaque Reduction Assay
3.3. Molecular Docking Study
3.4. Computational Study
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shao, Y.-M.; Yang, W.-B.; Kuo, T.-H.; Tsai, K.-C.; Lin, C.-H.; Yang, A.-S.; Liang, P.-H.; Wong, C.-H. Design, synthesis, and evaluation of trifluoromethyl ketones as inhibitors of SARS-CoV 3CL protease. Bioorg. Med. Chem. 2008, 16, 4652–4660. [Google Scholar] [CrossRef] [PubMed]
- Ali, G.S.; Ozdemir, B.; Selamoglu, Z. A Review of Severe Acute Respiratory Syndrome Coronavirus 2 and Pathological Disorders in Patients. J. Pharm. Care 2021, 9, 141–147. [Google Scholar] [CrossRef]
- Russell, C.D.; Millar, J.E.; Baillie, J.K. Clinical evidence does not support corticosteroid treatment for 2019-nCoV lung injury. Lancet 2020, 395, 473–475. [Google Scholar] [CrossRef]
- World Health Organization (WHO). Novel Coronavirus; Situation Report 22; World Health Organization: Geneva, Switzerland, 2020. [Google Scholar]
- Andersen, K.G.; Rambaut, A.; Lipkin, W.I.; Holmes, E.C.; Garry, R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020, 26, 450–452. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Guo, H.; Zhou, P.; Shi, Z.-L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021, 19, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Prajapat, M.; Sarma, P.; Shekhar, N.; Avti, P.; Sinha, S.; Kaur, H.; Kumar, S.; Bhattacharyya, A.; Kumar, H.; Bansal, S.; et al. Drug targets for corona virus: A systematic review. Indian J. Pharmacol. 2020, 52, 56–65. [Google Scholar] [CrossRef]
- Kong, R.; Yang, G.; Xue, R.; Liu, M.; Wang, F.; Hu, J.; Guo, X.; Chang, S. COVID-19 Docking Server: A meta server for docking small molecules, peptides and antibodies against potential targets of COVID-19. Bioinformatics 2020, 36, 5109–5111. [Google Scholar] [CrossRef]
- Shah, B.; Modi, P.; Sagar, S.R. In silico studies on therapeutic agents for COVID-19: Drug repurposing approach. Life Sci. 2020, 252, 117652. [Google Scholar] [CrossRef]
- Savarino, A.; Boelaert, J.R.; Cassone, A.; Majori, G.; Cauda, R. Effects of chloroquine on viral infections: An old drug against today’s diseases. Lancet Infect. Dis. 2003, 3, 722–727. [Google Scholar] [CrossRef]
- Devaux, C.A.; Rolain, J.-M.; Colson, P.; Raoult, D. New insights on the antiviral effects of chloroquine against coronavirus: What to expect for COVID-19? Int. J. Antimicrob. Agents 2020, 55, 105938. [Google Scholar] [CrossRef]
- Liu, C.; Zhou, Q.; Li, Y.; Garner, L.V.; Watkins, S.P.; Carter, L.J.; Smoot, J.; Gregg, A.C.; Daniels, A.D.; Jervey, S.; et al. Research and Development on Therapeutic Agents and Vaccines for COVID-19 and Related Human Coronavirus Diseases. ACS Cent. Sci. 2020, 6, 315–331. [Google Scholar] [CrossRef]
- Negi, M.; Chawla, P.A.; Faruk, A.; Chawla, V. Role of heterocyclic compounds in SARS and SARS CoV-2 pandemic. Bioorg. Chem. 2020, 104, 104315. [Google Scholar] [CrossRef] [PubMed]
- Wassel, M.M.; Ammar, Y.A.; Ali, G.A.E.; Belal, A.; Mehany, A.B.; Ragab, A. Development of adamantane scaffold containing 1,3,4-thiadiazole derivatives: Design, synthesis, anti-proliferative activity and molecular docking study targeting EGFR. Bioorg. Chem. 2021, 110, 104794. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Anand, A.; Kumar, V. Recent developments in biological activities of chalcones: A mini review. Eur. J. Med. Chem. 2014, 85, 758–777. [Google Scholar] [CrossRef] [PubMed]
- Nowakowska, Z. A review of anti-infective and anti-inflammatory chalcones. Eur. J. Med. Chem. 2007, 42, 125–137. [Google Scholar] [CrossRef]
- El-Sharief, A.M.S.; Ammar, Y.A.; Mohamed, Y.A.; El-Gaby, M.S.A. A comparative study of the behavior of cyanothioformamide and oxazolidine (thiones or iminothiones) towards some binucleophiles. Heteroat. Chem. 2002, 13, 291–298. [Google Scholar] [CrossRef]
- Ammar, Y.A.; Ismail, M.M.F.; El-Gaby, M.S.A.; Zahran, M.A. Some reactions with quinoxaline-2, 3-dicarboxylic acid anhydride: Novel synthesis of thieno[2,3-d]pyrimidines and pyrrolo[3,4-b]quinoxalines as antimicrobial agents. Indian J. Chem. 2002, 41B, 1486–1491. [Google Scholar] [CrossRef]
- Ibrahim, S.A.; Fayed, E.A.; Rizk, H.F.; Desouky, S.E.; Ragab, A. Hydrazonoyl bromide precursors as DHFR inhibitors for the synthesis of bis-thiazolyl pyrazole derivatives; antimicrobial activities, antibiofilm, and drug combination studies against MRSA. Bioorg. Chem. 2021, 116, 105339. [Google Scholar] [CrossRef]
- Ibrahim, S.A.; Rizk, H.F.; Aboul-Magd, D.S.; Ragab, A. Design, synthesis of new magenta dyestuffs based on thiazole azomethine disperse reactive dyes with antibacterial potential on both dyes and gamma-irradiated dyed fabric. Dyes Pigm. 2021, 193, 109504. [Google Scholar] [CrossRef]
- El-Kalyoubi, S.A.; Fayed, E.A.; Abdel-Razek, A.S. One pot synthesis, antimicrobial and antioxidant activities of fused uracils: Pyrimidodiazepines, lumazines, triazolouracil and xanthines. Chem. Cent. J. 2017, 11, 66. [Google Scholar] [CrossRef]
- El-Kalyoubi, S.; Agili, F. Synthesis, In Silico Prediction and In Vitro Evaluation of Antitumor Activities of Novel Pyrido[2,3-d]pyrimidine, Xanthine and Lumazine Derivatives. Molecules 2020, 25, 5205. [Google Scholar] [CrossRef] [PubMed]
- Fayed, E.A.; Ragab, A.; Eldin, R.R.E.; Bayoumi, A.H.; Ammar, Y.A. In Vivo Screening and Toxicity Studies of Indolinone Incorporated Thiosemicarbazone, Thiazole and Piperidinosulfonyl Moieties as Anticonvulsant Agents. Bioorg. Chem. 2021, 116, 105300. [Google Scholar] [CrossRef] [PubMed]
- Alzahrani, A.Y.; Ammar, Y.A.; Abu-Elghait, M.; Salem, M.A.; Assiri, M.A.; Ali, T.E.; Ragab, A. Development of novel indolin-2-one derivative incorporating thiazole moiety as DHFR and quorum sensing inhibitors: Synthesis, antimicrobial, and antibiofilm activities with molecular modelling study. Bioorg. Chem. 2022, 119, 105571. [Google Scholar] [CrossRef] [PubMed]
- Caly, L.; Druce, J.D.; Catton, M.G.; Jans, D.A.; Wagstaff, K.M. The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antivir. Res. 2020, 178, 104787. [Google Scholar] [CrossRef] [PubMed]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Meng, X.-Y.; Zhang, H.-X.; Mezei, M.; Cui, M. Molecular docking: A powerful approach for structure-based drug discovery. Curr. Comput. Aided Drug Des. 2011, 7, 146–157. [Google Scholar] [CrossRef] [PubMed]
- Rehman, M.F.U.; Akhter, S.; Batool, A.I.; Selamoglu, Z.; Sevindik, M.; Eman, R.; Mustaqeem, M.; Akram, M.S.; Kanwal, F.; Lu, C.; et al. Effectiveness of Natural Antioxidants against SARS-CoV-2? Insights from the In-Silico World. Antibiotics 2021, 10, 1011. [Google Scholar] [CrossRef]
- Mohamed, H.A.; Ammar, Y.A.; Elhagali, G.A.; Eyada, H.A.; Aboul-Magd, D.S.; Ragab, A. In Vitro Antimicrobial Evaluation, Single-Point Resistance Study, and Radiosterilization of Novel Pyrazole Incorporating Thiazol-4-one/Thiophene Derivatives as Dual DNA Gyrase and DHFR Inhibitors against MDR Pathogens. ACS Omega 2022, 7, 4970–4990. [Google Scholar] [CrossRef]
- Wang, G.; Zhu, W. Molecular docking for drug discovery and development: A widely used approach but far from perfect. Future Med. Chem. 2016, 8, 1707–1710. [Google Scholar] [CrossRef]
- Hassan, A.S.; Morsy, N.M.; Awad, H.M.; Ragab, A. Synthesis, molecular docking, and in silico ADME prediction of some fused pyrazolo[1,5-a]pyrimidine and pyrazole derivatives as potential antimicrobial agents. J. Iran. Chem. Soc. 2022, 19, 521–545. [Google Scholar] [CrossRef]
- Ammar, Y.A.; El-Hafez, S.M.A.A.; Hessein, S.A.; Ali, A.M.; Askar, A.A.; Ragab, A. One-pot strategy for thiazole tethered 7-ethoxy quinoline hybrids: Synthesis and potential antimicrobial agents as dihydrofolate reductase (DHFR) inhibitors with molecular docking study. J. Mol. Struct. 2021, 1242, 130748. [Google Scholar] [CrossRef]
- Wang, L.-L.; Battini, N.; Bheemanaboina, R.R.Y.; Ansari, M.F.; Chen, J.-P.; Xie, Y.-P.; Cai, G.-X.; Zhang, S.-L.; Zhou, C.-H. A new exploration towards aminothiazolquinolone oximes as potentially multi-targeting antibacterial agents: Design, synthesis and evaluation acting on microbes, DNA, HSA and topoisomerase IV. Eur. J. Med. Chem. 2019, 179, 166–181. [Google Scholar] [CrossRef] [PubMed]
- Ramadan, A.M.; Elsamra, R.M.I.; Bondock, S. New pyrazole-4-carbothioamide-based metal complexes: Synthesis, spectral characterization, computational, antimicrobial, and antitumor investigations. Appl. Organomet. Chem. 2021, 35, e6102. [Google Scholar] [CrossRef]
- Erol, M.; Celik, I.; Kuyucuklu, G. Synthesis, antimicrobial and in silico studies of new 2.5-disubstituted benzoxazole derivative. Med. Sci. Int. Med. J. 2021, 10, 400. [Google Scholar] [CrossRef]
- Hadda, T.B.; Berredjem, M.; Almalki, F.A.; Rastija, V.; Jamalis, J.; Emran, T.B.; Abu-Izneid, T.; Esharkawy, E.; Rodriguez, L.C.; Alqahtani, A.M. How to face COVID-19: Proposed treatments based on remdesivir and hydroxychloroquine in the presence of zinc sulfate. Docking/DFT/POM structural analysis. J. Biomol. Struct. Dyn. 2021, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Hayden, F.G.; Cote, K.M.; Douglas, R.G. Plaque inhibition assay for drug susceptibility testing of influenza viruses. Antimicrob. Agents Chemother. 1980, 17, 865–870. [Google Scholar] [CrossRef]
- Elfiky, A.A.; Ismail, A. Molecular dynamics and docking reveal the potency of novel GTP derivatives against RNA dependent RNA polymerase of genotype 4a HCV. Life Sci. 2019, 238, 116958. [Google Scholar] [CrossRef]
- Fayed, E.A.; Ammar, Y.A.; Saleh, M.A.; Bayoumi, A.H.; Belal, A.; Mehany, A.B.M.; Ragab, A. Design, synthesis, antiproliferative evaluation, and molecular docking study of new quinoxaline derivatives as apoptotic inducers and EGFR inhibitors. J. Mol. Struct. 2021, 1236, 130317. [Google Scholar] [CrossRef]
- Ragab, A.; Elsisi, D.M.; Ali, O.A.A.; Abusaif, M.S.; Askar, A.A.; Farag, A.A.; Ammar, Y.A. Design, synthesis of new novel quinoxalin-2(1H)-one derivatives incorporating hydrazone, hydrazine, and pyrazole moieties as antimicrobial potential with in-silico ADME and molecular docking simulation. Arab. J. Chem. 2022, 15, 103497. [Google Scholar] [CrossRef]
- SARS-CoV-2 RNA-Dependent RNA Polymerase in Complex with Cofactors. Available online: https://www.rcsb.org/structure/6M71 (accessed on 4 October 2021).
- Structure of the SARS-CoV-2 Spike Glycoprotein (Closed State). Available online: https://www.rcsb.org/structure/6VXX (accessed on 4 October 2021).
- Fukuda, T.; Umeki, T.; Tokushima, K.; Xiang, G.; Yoshida, Y.; Ishibashi, F.; Oku, Y.; Nishiya, N.; Uehara, Y.; Iwao, M. Design, synthesis, and evaluation of A-ring-modified lamellarin N analogues as noncovalent inhibitors of the EGFR T790M/L858R mutant. Bioorg. Med. Chem. 2017, 25, 6563–6580. [Google Scholar] [CrossRef]
- Coulibaly, W.K.; N’dri, J.S.; Koné, M.G.-R.; Dago, C.D.; Ambeu, C.N.; Bazureau, J.-P.; Ziao, N. Studies of the Chemical Reactivity of a Series of Rhodanine Derivatives by Approaches to Quantum Chemistry. Comput. Mol. Biosci. 2019, 9, 49–62. [Google Scholar] [CrossRef][Green Version]
- Alzahrani, A.Y.; Ammar, Y.A.; Salem, M.A.; Abu-Elghait, M.; Ragab, A. Design, synthesis, molecular modeling, and antimicrobial potential of novel 3-[(1H-pyrazol-3-yl)imino]indolin-2-one derivatives as DNA gyrase inhibitors. Arch. Pharm. 2022, 355, e2100266. [Google Scholar] [CrossRef] [PubMed]

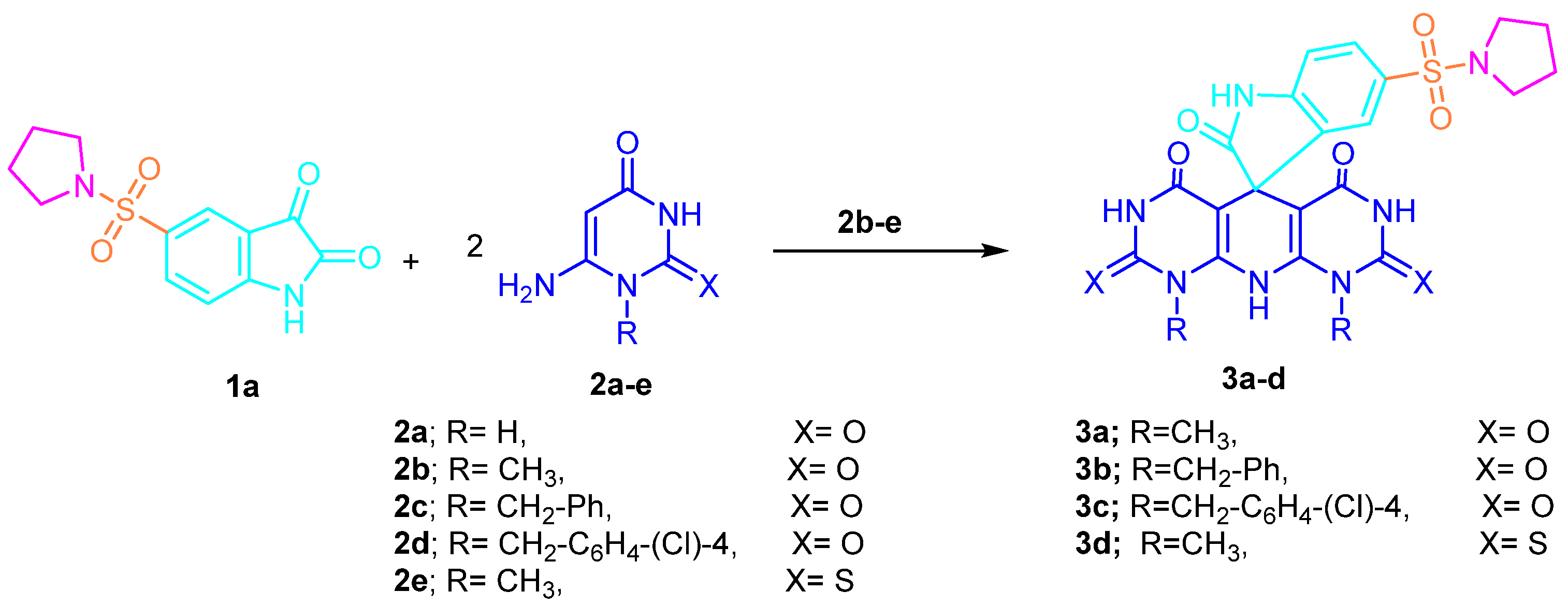

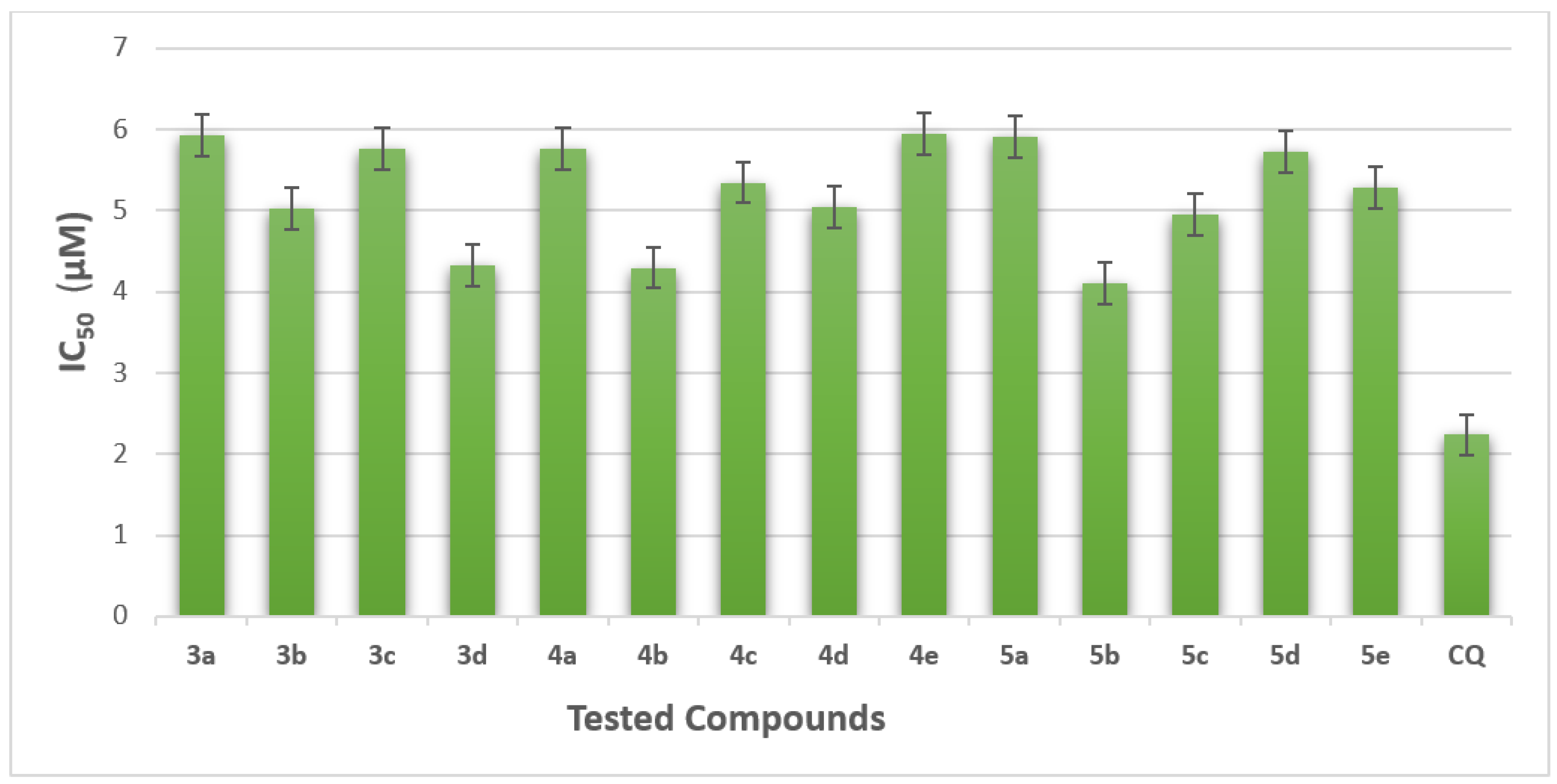




| Cpd. No. | Concentrations (µM) (Mean ± SEM) | IC50 µM | |||||
|---|---|---|---|---|---|---|---|
| 0.312 | 0.625 | 1.25 | 2.5 | 5 | 10 | ||
| 3a | 5.35 ± 0.01 | 12.13 ± 0.46 | 15.21 ± 0.40 | 28.88 ± 0.99 | 50.26 ± 2.40 | 74.95 ± 3.00 | 5.93 ± 0.05 |
| 3b | 4.66 ± 0.04 | 17.11 ± 0.62 | 22.09 ± 0.75 | 39.20 ± 1.23 | 53.88 ± 2.53 | 83.63 ± 3.11 | 5.03 ± 0.01 |
| 3c | 6.02 ± 0.17 | 11.62 ± 0.37 | 15.95 ± 0.41 | 29.24 ± 0.93 | 51.24 ± 2.44 | 76.79 ± 2.91 | 5.76 ± 0.07 |
| 3d | 9.20 ± 0.33 | 21.20 ± 0.90 | 32.77 ± 1.11 | 44.27 ± 2.01 | 56.1 ± 2.54 | 89.56 ± 2.93 | 4.33 ± 0.01 |
| 4a | 6.17 ± 0.11 | 11.87 ± 0.10 | 14.98 ± 0.30 | 29.78 ± 0.90 | 52.88 ± 2.34 | 75.98 ± 2.78 | 5.77 ± 0.01 |
| 4b | 19.18 ± 0.31 | 25.17 ± 1.01 | 37.76 ± 1.20 | 49.12 ± 2.05 | 58.20 ± 2.46 | 77.00 ± 2.69 | 4.30 ± 0.01 |
| 4c | 15.02 ± 0.55 | 23.12 ± 0.99 | 28.12 ± 1.01 | 39.13 ± 1.25 | 41.45 ± 1.67 | 79.12 ± 2.87 | 5.35 ± 0.05 |
| 4d | 2.19 ± 0.15 | 11.19 ± 0.38 | 23.20 ± 0.97 | 30.20 ± 0.95 | 55.67 ± 2.66 | 88.02 ± 2.95 | 5.05 ± 0.02 |
| 4e | 2.30 ± 0.91 | 11.33 ± 0.37 | 14.32 ± 0.32 | 22.45 ± 0.87 | 50.99 ± 2.01 | 77.09 ± 2.43 | 5.95 ± 0.06 |
| 5a | 5.83 ± 0.15 | 13.12 ± 0.45 | 16.88 ± 0.53 | 28.24 ± 0.96 | 50.13 ± 2.01 | 74.87 ± 2.36 | 5.92 ± 0.05 |
| 5b | 16.19 ± 0.50 | 21.14 ± 0.78 | 30.01 ± 1.19 | 44.30 ± 1.80 | 60.30 ± 2.36 | 90.98 ± 3.42 | 4.10 ± 0.02 |
| 5c | 10.65 ± 0.35 | 12.98 ± 0.32 | 28.01 ± 1.00 | 37.12 ± 1.10 | 50.11 ± 1.54 | 85.30 ± 3.12 | 4.96 ± 0.01 |
| 5d | 8.91 ± 0.19 | 14.79 ± 0.41 | 18.08 ± 0.80 | 29.09 ± 0.99 | 56.78 ± 2.02 | 73.14 ± 2.13 | 5.73 ± 0.07 |
| 5e | 5.19 ± 0.04 | 10.98 ± 0.21 | 11.33 ± 0.35 | 22.22 ± 0.79 | 56.23 ± 1.79 | 88.14 ± 3.01 | 5.29 ± 0.01 |
| Chloroquine * | 2.24 | ||||||
| Cpd. No. | Conc (µM) | Viral Count (Untreated) (PFU/mL) | Viral Count (Treated) (PFU/mL) | Inhibition % |
|---|---|---|---|---|
| 3a | 5 | 11 × 105 | 1.76 × 105 | 84 |
| 2.5 | 1.98 × 105 | 82 | ||
| 1.25 | 3.3 × 105 | 70 | ||
| 0.625 | 3.85 × 105 | 65 | ||
| 3b | 5 | 10 × 105 | 2.5 × 105 | 75 |
| 2.5 | 4.3 × 105 | 57 | ||
| 1.25 | 5 × 105 | 50 | ||
| 0.625 | 8.3 × 105 | 17 | ||
| 3c | 5 | 5 × 105 | 5 × 105 | 0 |
| 2.5 | 5 × 105 | 0 | ||
| 1.25 | 5 × 105 | 0 | ||
| 0.625 | 5 × 105 | 0 | ||
| 3d | 5 | 11 × 105 | 4.95 × 105 | 55 |
| 2.5 | 6.6 × 105 | 40 | ||
| 1.25 | 7.7 × 105 | 30 | ||
| 0.625 | 9.9 × 105 | 10 | ||
| 4a | 5 | 10 × 105 | 10 × 105 | 0 |
| 2.5 | 10 × 105 | 0 | ||
| 1.25 | 10 × 105 | 0 | ||
| 0.625 | 10 × 105 | 0 | ||
| 4b | 5 | 9 × 105 | 0.09 × 105 | 99 |
| 2.5 | 0.9 × 105 | 90 | ||
| 1.25 | 1.8 × 105 | 80 | ||
| 0.625 | 2.7 × 105 | 70 | ||
| 4c | 5 | 11 × 105 | 2.86 × 105 | 74 |
| 2.5 | 3.85 × 105 | 65 | ||
| 1.25 | 4.4 × 105 | 60 | ||
| 0.625 | 9.68 × 105 | 12 | ||
| 4d | 5 | 11 × 105 | 2.2 × 105 | 80 |
| 2.5 | 4.4 × 105 | 60 | ||
| 1.25 | 6.6 × 105 | 40 | ||
| 0.625 | 7.26 × 105 | 34 | ||
| 4e | 5 | 9 × 105 | 0.81 × 105 | 91 |
| 2.5 | 1.17 × 105 | 87 | ||
| 1.25 | 1.53 × 105 | 83 | ||
| 0.625 | 3.96 × 105 | 56 | ||
| 5a | 5 | 9 × 105 | 7.2 × 105 | 20 |
| 2.5 | 8.28 × 105 | 8 | ||
| 1.25 | 8.73 × 105 | 3 | ||
| 0.625 | 9 × 105 | 0 | ||
| 5b | 5 | 5 × 105 | 1.65 × 105 | 67 |
| 2.5 | 2.65 × 105 | 47 | ||
| 1.25 | 4.45 × 105 | 11 | ||
| 0.625 | 5 × 105 | 0 | ||
| 5c | 5 | 11 × 105 | 6.6 × 105 | 40 |
| 2.5 | 8.14 × 105 | 26 | ||
| 1.25 | 8.8 × 105 | 20 | ||
| 0.625 | 11 × 105 | 0 | ||
| 5d | 5 | 10 × 105 | 1.8 × 105 | 82 |
| 2.5 | 4.9 × 105 | 51 | ||
| 1.25 | 6 × 105 | 40 | ||
| 0.625 | 6 × 105 | 40 | ||
| 5e | 5 | 5 × 105 | 1.5 × 105 | 70 |
| 2.5 | 3 × 105 | 40 | ||
| 1.25 | 3.35 × 105 | 33 | ||
| 0.625 | 4.5 × 105 | 10 | ||
| Chloroquine | 5 | 6 × 104 | 0 | >99 |
| 2.5 | 0 | >99 | ||
| 1.25 | 0 | >99 | ||
| 0.625 | 0 | >99 |
| Cpd. No. | IC50 (Mean ± SEM) (nM) | |
|---|---|---|
| RdRp | Spike Glycoprotein | |
| 3a | 40.23 ± 0.09 | 40.27 ± 0.17 |
| 4b | 44.90 ± 0.08 | 44.83 ± 0.16 |
| 4d | 41.26 ± 0.25 | 42.27 ± 0.31 |
| 4e | 41.23 ± 0.12 | 42.43 ± 0.26 |
| Chloroquine | 45.00 ± 0.02 | 45.00 ± 0.06 |
| Untreated | 17.73 ± 0.12 | 18.23 ± 0.12 |
| Cpd. No. | S Kcal/mol | Residues | Ligand Atoms | Distance (˚A) | Strength (%) |
|---|---|---|---|---|---|
| (1) RNA-dependent RNA polymerase (RdRp) (PDB: 6m71) | |||||
| 3a | −17.38 | Asp623 | C=O of isatin der. | 2.90 | 10 |
| Lys621 | C=O of pyridino-pyrimidine | 2.70 | 37 | ||
| Arg553 | Oxygen of SO2 | 2.83 | 76 | ||
| Arg555 | Oxygen of SO2 | 2.74 | 46 | ||
| 4b | −17.49 | Lys621 | C=O of isatin der. | 2.88 | 30 |
| Asp623 | NH of pyridino-pyrimidine | 2.56 | 70 | ||
| Arg553 | Oxygen of SO2 | 2.73 | 32 | ||
| 4d | −18.48 | Lys551 | C=O of pyridino-pyrimidine | 2.63 | 44 |
| Lys708 | Phenyl of benzyl der. | - | - | ||
| Arg553 | Phenyl of benzyl der. | - | - | ||
| 4e | −15.38 | Asp760 | NH of pyridino-pyrimidine | 2.38 | 48 |
| Asp623 | NH of pyridino-pyrimidine | 2.81 | 16 | ||
| Arg553 | Oxygen of SO2 | 2.53 | 71 | ||
| CQ | −14.94 | Asp760 | NH of quaternary salt | 2.42 | 70 |
| Arg553 | Phenyl of quinoline | - | - | ||
| RDV | −16.09 | Arg555 | Oxygen of phosphate | 2.93 | 24 |
| Arg553 | Oxygen of furane | 3.11 | 31 | ||
| Arg553 | Nitrogen of cyano group | 3.01 | 52 | ||
| Arg553 | Pyrole of pyrolo[1,2-f]triazine | - | - | ||
| Asp623 | Hydroxy at C4 of furane | 2.36 | 44 | ||
| Thr556 | Nitrogen of cyano group | 3.44 | 12 | ||
| (2) Spike glycoprotein (SGp) (PDB: 6VXX) | |||||
| 3a | −17.67 | His1058 | C=O of pyridino-pyrimidine | 2.64 | 35 |
| Thr732 | NH of pyridino-pyrimidine | 2.73 | 73 | ||
| Thr732 | C=O of pyridino-pyrimidine | 2.66 | 77 | ||
| 4b | −15.22 | His1058 | C=O of isatin | 2.72 | 59 |
| Asp867 | NH of pyridino-pyrimidine | 2.39 | 23 | ||
| 4d | −16.48 | Leu861 | NH of pyridino-pyrimidine | ||
| 4e | −15.92 | His1058 | C=O of isatin | 2.66 | 69 |
| Thr827 | NH of pyridino-pyrimidine | 2.54 | 33 | ||
| CQ | −15.71 | Asp867 | NH of chloroquine C4 | 2.71 | 11 |
| Asp867 | NH of quaternary salt | 2.48 | 72 | ||
| His1058 | Nitrogen of tertiary amine | - | - | ||
| RDV | −15.67 | Phe823 | NH2 of pyrrolo[2,1-f] [1,2,4]triazin | 3.01 | 22 |
| CQ = Chloroquine; RDV = Remdesivir; (-) arene-cation interaction | |||||
| Electronic Parameters | 3a | 4b | 4d | 4e | CQ | RDV |
|---|---|---|---|---|---|---|
| Energy (Hartree) | −2159.03 | −2198.35 | −3579.64 | −2844.24 | −1326.03 | −2321.61 |
| Dipole moment (Debye) | 6.718 | 6.687 | 4.739 | 4.028 | 5.62 | 4.44 |
| E (HOMO) eV | −6.00 | −6.05 | −6.01 | −6.17 | −5.66 | −6.11 |
| E (LUMO) eV | −1.74 | −1.79 | −1.79 | −2.13 | −1.17 | −1.27 |
| ΔE (eV) | 4.26 | 4.26 | 4.23 | 4.035 | 4.49 | 4.83 |
| IP(eV) | 6.00 | 6.05 | 6.01 | 6.17 | 5.66 | 6.11 |
| EA(eV) | 1.74 | 1.79 | 1.79 | 2.13 | 1.17 | 1.27 |
| X(eV) | 3.87 | 3.93 | 3.90 | 4.15 | 3.41 | 3.69 |
| ɳ(eV) | 2.13 | 2.13 | 2.11 | 2.01 | 2.24 | 2.41 |
| S(eV−1) | 0.469 | 0.469 | 0.473 | 0.495 | 0.446 | 0.41 |
| µ(eV) | −3.87 | −3.93 | −3.90 | −4.15 | −3.41 | −3.69 |
| ω(eV) | 3.52 | 3.617 | 3.604 | 4.27 | 2.598 | 2.81 |
| Cpd. No. | E (HOMO) | E (LUMO) |
|---|---|---|
| 3a | 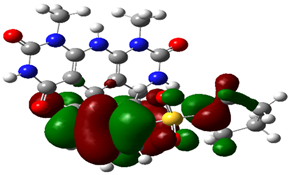 | 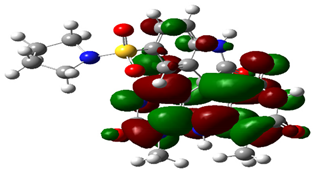 |
| 4b | 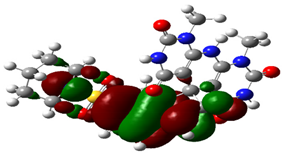 |  |
| 4d | 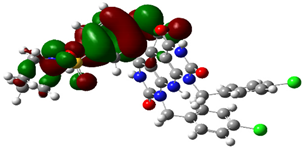 | 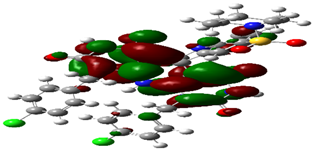 |
| 4e |  | 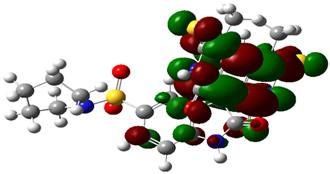 |
| C.Q |  |  |
| RDV | 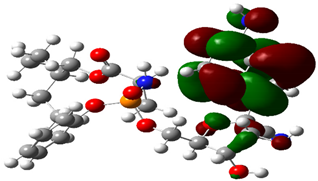 | 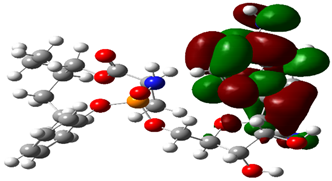 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
El-Kalyoubi, S.A.; Ragab, A.; Abu Ali, O.A.; Ammar, Y.A.; Seadawy, M.G.; Ahmed, A.; Fayed, E.A. One-Pot Synthesis and Molecular Modeling Studies of New Bioactive Spiro-Oxindoles Based on Uracil Derivatives as SARS-CoV-2 Inhibitors Targeting RNA Polymerase and Spike Glycoprotein. Pharmaceuticals 2022, 15, 376. https://doi.org/10.3390/ph15030376
El-Kalyoubi SA, Ragab A, Abu Ali OA, Ammar YA, Seadawy MG, Ahmed A, Fayed EA. One-Pot Synthesis and Molecular Modeling Studies of New Bioactive Spiro-Oxindoles Based on Uracil Derivatives as SARS-CoV-2 Inhibitors Targeting RNA Polymerase and Spike Glycoprotein. Pharmaceuticals. 2022; 15(3):376. https://doi.org/10.3390/ph15030376
Chicago/Turabian StyleEl-Kalyoubi, Samar A., Ahmed Ragab, Ola A. Abu Ali, Yousry A. Ammar, Mohamed G. Seadawy, Aya Ahmed, and Eman A. Fayed. 2022. "One-Pot Synthesis and Molecular Modeling Studies of New Bioactive Spiro-Oxindoles Based on Uracil Derivatives as SARS-CoV-2 Inhibitors Targeting RNA Polymerase and Spike Glycoprotein" Pharmaceuticals 15, no. 3: 376. https://doi.org/10.3390/ph15030376
APA StyleEl-Kalyoubi, S. A., Ragab, A., Abu Ali, O. A., Ammar, Y. A., Seadawy, M. G., Ahmed, A., & Fayed, E. A. (2022). One-Pot Synthesis and Molecular Modeling Studies of New Bioactive Spiro-Oxindoles Based on Uracil Derivatives as SARS-CoV-2 Inhibitors Targeting RNA Polymerase and Spike Glycoprotein. Pharmaceuticals, 15(3), 376. https://doi.org/10.3390/ph15030376







